Abstract
In an effort to halt the global decline of large carnivores, reintroductions have become increasingly popular to establish satellite populations and reduce the risk of stochastic events. These artificial range expansions are typically formed by a small number of founders, which can lead to changes in population genetic structure. For instance, serial founder events can lead to neutral and even deleterious alleles reaching higher than expected frequencies along the front end of an expansion, referred to as gene surfing. One of the world’s most extensive range expansion programmes has been for endangered African wild dogs (Lycaon pictus). In this study, we examine the effect of continent-wide translocations on spatial genetic diversity, by determining what effect genetic surfing has on population structure in wild dogs, and measuring how long it will take for population structure to homogenize in the face of ongoing dispersal. We used a set of microsatellite loci to look at surfing alleles in five populations across southern Africa, and simulated the movement of these alleles forward in time under the current demographic scenario. We found that it would take about 150 generations for the expanding population to be 50% introgressed with genes from the free-roaming population. With the current rate of translocations, genetic differentiation in southern Africa will disappear, overturning the effects of genetic drift or surfing alleles. Understanding genetic patterns in expanding populations is of great interest to conservation, and we demonstrate that reintroduction programmes can help restore genetic diversity, and consequently adaptive potential, in recovering wildlife populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Demographic reductions in wildlife populations are occurring at alarming rates around the globe, primarily driven by habitat loss, agricultural expansion, and persecution by humans (Johnson et al. 2017). This has been particularly evident for apex predators, which are one of the most challenging groups to conserve due to their large area requirements, low densities and reproduction rates, and common conflicts with humans over resources (Ripple et al. 2014). In an effort to halt the global declines of large carnivores, reintroductions have become increasingly popular to establish satellite populations and reduce the risk of stochastic events, such as disease outbreaks, which may also speed up the process of natural dispersal to unoccupied habitat fragments (Wolf and Ripple 2018). This falls within the original rewilding framework, which focussed on establishing core areas and connecting corridors, and releasing keystone carnivores (Soulé and Noss 1998). When reserves have become isolated, augmentations can help to compensate for the lack of natural dispersal, increase genetic heterogeneity, and avoid inbreeding in small remnant populations, thereby increasing their reproductive and survival success (Moritz 1999). Large carnivore reintroductions and augmentations have become frequent in southern Africa (Miller et al. 2013; Davies-Mostert et al. 2015; Boast et al. 2018), where there are less spatial constraints compared to other continents (Hayward and Somers 2009). Furthermore, population recovery and range expansions have occurred within fenced reserves for many large carnivores in recent decades, partly as a result of changing attitudes and protective legislation (Chapron et al. 2014), such as lions (Panthera leo) in Gorongosa-Marromeu, Mozambique (Bouley et al. 2018), and brown hyenas (Parahyena brunnea) in Namibia (Edwards et al. 2019).
Because expansion fronts are typically populated by a small number of founders, reintroductions and natural range expansions may lead to changes in population genetic structure and diversity, different from genetic changes caused by demographic growth alone (Hagen et al. 2015). Due to a bottleneck effect and genetic drift, there will initially be an increase in genetic structuring and loss of diversity, which will be reversed over time due to balancing gene flow and genetic homogenization (Ray et al. 2003). Another consequence of range expansions is that some neutral and even deleterious alleles reach higher than expected frequencies along the front end of an expansion, because of repeated founder events, which is referred to as genetic surfing (Excoffier and Ray 2008). This term is based on the idea that mutations ‘surf on the wave’ of a range expansion, causing rapid genetic change over time (Edmonds et al. 2004; Klopfstein et al. 2006). As such, genetic surfing can be considered a spatial analogue of genetic drift, nonbeneficial to the viability of populations. It is unrelated to natural selection on beneficial or deleterious alleles, such as genetic hitchhiking, in which alleles under selection influence allele frequencies of nearby loci due to chromosomal linkage (Friedlander and Steinrücken 2022). Nonetheless, population structure as a result of allele surfing and genetic drift may be mistaken as local adaptation or spatial variation of clades that resemble expansion from refugia, which may change genetic composition in a similar way (Ibrahim et al. 1996; Currat et al. 2006). Besides some alleles reaching near-fixation, a major difference is that there are no or only few private alleles present in expanding populations, as illustrated by theoretical simulations (Hallatschek and Nelson 2008) and empirical cases of expanding wolves Canis lupus (Szewczyk et al. 2019), coyotes C. latrans (Heppenheimer et al. 2018), and brown bears Ursus arctos (Hagen et al. 2015).
The effect of genetic surfing is initially deleterious, before genetic diversity may recover in the face of ongoing gene flow (Ibrahim et al. 1996). Whether population structure is the result of neutral processes such as genetic drift and surfing in small and expanding populations or a result of local adaptation and historic refugia is important for conservation programmes that rely on reintroductions and translocations (Frankham 2009). For instance, if genetic variability in source populations has been compromised by low allelic richness caused by either genetic drift or surfing, it would make admixture with recipient populations undesirable in case of augmentation if genetic diversity cannot be restored during future interventions (Woodworth et al. 2002; Malone et al. 2018). Another concern that is often raised is that historic or adaptive genetic differentiation may exist between the source and recipient population, if they were isolated or exposed to different environmental and selective pressures (Weeks et al. 2015). Admixing genetically diverged populations, referred to as evolutionary significant units (ESUs) (Crandall et al. 2000), may disrupt spatial patterns of genetic variation and homogenize unique evolutionary trajectories (Bertola et al. 2021). This leads to the overall loss of adaptive potential, and in extreme cases outbreeding depression, due to which it is recommended to find populations with similar evolutionary histories for reintroduction programmes (Becker et al. 2022). However, in small and isolated populations the risk of inbreeding depression far outweighs the risk posed by outbreeding (Edmands 2007; Ralls et al. 2020). Furthermore, when genetic differentiation is caused by recent habitat fragmentation or population bottlenecks, it is advised to counteract this by enhancing gene flow, either by translocations or restoring habitat connectivity (Frankham 2015; McLennan et al. 2020).
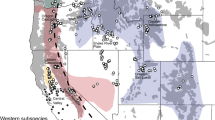
One of the world’s most extensive reintroduction and translocation efforts has been for the endangered African wild dog (Lycaon pictus), also known as painted dogs, which suffered population losses throughout their range and went extinct in 15 out of 39 countries where they historically occurred (Woodroffe and Sillero-Zubiri 2020). In an attempt to revive this species in South Africa, where they had gone extinct outside Kruger National Park (NP), wild dogs were first reintroduced into Hluhluwe-iMfolozi Park in 1980 (Mills et al. 1998) (Fig. 1A). The network of private reserves subsequently grew with an additional twelve reserves, and translocations between these have been ongoing to mimic natural dispersal and maintain genetic diversity (Tensen et al. 2019). Hereby, the African wild dog fits the rewilding concept (Wolf and Ripple 2018), which focuses on reintroducing carnivores into their former range to restore top-down regulation in ecosystems. Due to the success of the programme, wild dogs became available for introductions outside South Africa, which became known as the range expansion programme. As part of this programme, wild dogs have recently been reintroduced in Mozambique and Malawi and translocated to Zambia, which were thought to have historically formed one metapopulation (Tensen et al. 2022) and are part of the Southern African Conservation Strategy (IUCN/SSC 2015). Wild dogs were only sporadically seen in Mozambique, and became functionally extinct in Malawi since land reform programmes resulted in the loss of many game farms and conservancies (IUCN/SSC 2015). There is much debate on whether animals should be translocated over such large distances, particularly when facing the possibility of admixing genetically differentiated populations and reducing intrapopulation diversity (Mona et al. 2014). In wild dogs, it has already been illustrated that the managed metapopulation in South Africa has its own genetic signature, as well as Kruger NP and the Lowveld of Zimbabwe (Tensen et al. 2022). Although they are expected to have resulted from anthropogenic habitat fragmentation (Marsden et al. 2012), it is not yet clear why the managed metapopulation genetically differs, considering it was sourced from Kruger NP and Botswana, and no inbreeding was detected (Tensen et al. 2019).
Wild dog (Lycaon pictus) populations across Africa. (A) Four genetic clusters identified in southern Africa based on Tensen et al. (2022): Kafue Zambezi transfrontier conservation areas (KAZA-TFCA); the Lowveld (including the free-roaming population in the Waterberg); Kruger National Park; and a managed metapopulation (as part of the range expansion programme). (B) Three main phylogenetic clusters in Africa, based on Girman et al. (2001) and Marsden et al. (2012). West Central Africa has never been genetically examined, but likely forms a unique lineage (Woodroffe et al. 1999). (C) All known records of dispersing and translocated wild dogs in Africa, based on literature and Wild Dog Advisory Group (WAG) minutes (Table S1)
Recovering and reintroduced populations of large carnivores offer a unique opportunity for investigating the genetic effects of range expansions on population structure (Hagen et al. 2015; McLennan et al. 2020). Currently, there is limited knowledge on the population genetic consequences of translocations and demographic recovery, especially across broad geographical scales. In this study, we will use wild dogs as a model species to investigate the genetic consequences of the range expansion programme, in which individuals from the management metapopulation in South Africa are used to repopulate other areas in Africa. We combine long-term demographic and genetic data and use population simulations to examine the effect of genetic surfing, by asking the following questions: (i) what effect does genetic surfing have on population structure in African wild dogs; and (ii) how long will it take for population structure to homogenize with ongoing dispersal and translocations? According to theory, we predict that the range expansion from South Africa will initially cause a steep cline in population structure, due to shifting allele frequencies, which will eventually become introgressed by genes of local remnant populations in southern Africa and lead to an increase genetic variation in subsequent generations (Ibrahim et al. 1996; Hallatschek and Nelson 2008; Ralls et al. 2020). As the loss of genetic diversity through genetic drift and surfing can negatively impact population viability, it is important to include these neutral processes in the genetic management and monitoring of species. Furthermore, given the increasing threat of habitat fragmentation and application of reintroductions, the African wild dog as a natural experiment could provide important lessons for other range expansion programmes.
Materials and methods
Study species
The African wild dog is listed as Endangered by the International Union for Conservation of Nature, with approximately 7,700 individuals (adults and yearlings) left in the wild, which are scattered across the African continent (Kuiper et al. 2018; Woodroffe and Sillero-Zubiri 2020). Habitat destruction and fragmentation have been the main drivers of their population decline (Davies and du Toit 2004), and they have also been intentionally eliminated in the past due to the negative attitude towards this predator (Fanshawe et al. 1997; Woodroffe et al. 1997; Lindsey et al. 2005). The current distribution has become highly fragmented as savannah grasslands and woodlands are progressively being threatened by livestock overgrazing and urbanisation (Riggio et al. 2013). One population stronghold, consisting of approximately 17% of its historical range, remains in southern Africa, where roughly 4,400 wild dogs are distributed over reserves extending from southern Angola across Botswana to western Zimbabwe (IUCN/SSC 2015). In East Africa, roughly 3.300 wild dogs are spread across Ethiopia, Sudan, Kenya and Tanzania, where they have strongholds in the Selous and Serengeti-Masai Mara ecosystems (IUCN/SCC 2008). Wild dogs have been exterminated in West Central Africa except for a small population in Senegal, due to which they are now listed as Critically Endangered in this region (Woodroffe and Sillero-Zubiri 2020).
Demographic history
Due to several reintroductions, South Africa now holds a viable population of circa 350 wild dogs in Kruger NP, private reserves, and unprotected areas (WAG-SA 2022). Because maintaining genetic diversity forms an essential part of the management efforts, translocations between private reserves are carried out to mimic natural dispersal (Spiering et al. 2011). Genetic monitoring has illustrated that reserves remain connected as part of a managed metapopulation (Tensen et al. 2019). Wild dogs were first introduced into Hluhluwe-iMfolozi in 1980, and the population has since grown to 120 individuals spread among 12 reserves (WAG-SA 2022). As part of the range expansion programme, wild dogs from South Africa have been used to repopulate areas in southern Africa where they have gone extinct or no longer occur at viable population densities (Fig. 1C). Thus far, eleven translocations were sourced from the managed metapopulation in South Africa, four from Zimbabwe, and one from the augmented population in Gorongosa, Mozambique. New populations have been established in Karingani Game Reserve in Mozambique, and Liwonde National Park and Majete Wildlife Reserve in Malawi. There has also been a recovery of free-roaming wild dogs in the northern part of South Africa, which dispersed from southeast Botswana and Zimbabwe (Davies-Mostert et al. 2012; Tensen et al. 2022). Long-distance dispersal still occurs freely across southern Africa in Namibia, Botswana and Zimbabwe (Fig. 1C), partly due to the establishment of large transfrontier conservation areas (Davies-Mostert et al. 2015; Cozzi et al. 2020; Hofmann et al. 2021). In this study, we deploy information from natural dispersal events of wild dogs on the African continent, as well as augmentations and reintroductions that have taken place outside the managed metapopulation in South Africa, where animals are translocated frequently (i.e. 22 annually, Tensen et al. 2019). Data were obtained from the scientific literature (Table S1), as well as the Wild Dog Advisory Group of South Africa (WAG-SA), who have been documenting translocations of wild dogs in southern Africa since the start of the range expansion programme.
Genetic analysis
African wild dogs from various areas in southern Africa have been genotyped for twenty short-tandem repeats (for extraction methods, PCR protocols and fragment analysis see Tensen et al. 2022). We have used a subset of the previously collected data (of 125 individuals from five populations) to test for the possibility of genetic surfing. The five populations are: (i) the central population that extends from Angola to western Zimbabwe across the Kafue Zambezi transfrontier conservation area (KAZA-TFCA), (ii) the Lowveld population in southern Zimbabwe, and (iii) the free-roaming Waterberg population in northern South Africa, (iv) Kruger NP in South Africa, and (v) the managed metapopulation in private reserves outside Kruger NP (Fig. 1; Table S2). In this study, we consider the presence of three genetic clades on the African continent (Fig. 1B), based on previous genetic studies (e.g. Girman et al. 2001; Marsden et al. 2012; Tensen et al. 2022), due to which southern and eastern Africa are not combined in our analyses. To test the possibility that genetic surfing has occurred, we calculated allelic richness and number of private alleles with HP-rare v1.1 (Kalinowski 2005). We visualized the distribution of alleles by code-colouring the ten most variable loci, and tested whether allele frequencies significantly deviated from expected ratios with the Cochran-Mantel-Haenszel (CMH) test in Excel, using XLSTAT (Addinsoft, Paris, France) applied to a 2 × 66 genotype contingency table, for each pairwise population comparison separately. The CMH test is an extension of Chi-squared tests for multiple biological replicates, and can identify differences in allele frequencies that are consistent across populations (Wiberg et al. 2017). The genetic spatial pattern characterizing genetic surfing is that shifted alleles are heading towards their maximum frequency as the expansion proceeds.
Ecological modelling
Virtual landscape
To predict the movement of surfing alleles during range expansions, we used the programme SPLATCHE3 (Currat et al. 2019), which simulates the demography of populations and molecular diversity under a wide range of evolutionary scenarios. We included 33 populations in the simulation, to represent the geographic distribution of wild dogs in southern Africa, based on IUCN/SCC (2015) and WAG-SA minutes (for all input parameters, see Table S3). The continental surface of southern Africa was represented by a grid of more than 700 demes, each representing an area of 7,000 km2. The initial metapopulation was assumed to be at carrying capacity, and we applied a two-dimensional stepping-stone grid (30 × 35 demes) from which range expansions took place, starting at generation 5 (Fig S1). All range expansions occurred from the managed metapopulation (met) into the free-roaming population (wild) in other parts of southern Africa. To predict the level of admixture between genetically differentiated wild dog populations, we performed a series of simulations under the two-layer admixture model, allowing the simulation of dispersal events between any deme (occupied or empty). One layer represents the free-roaming populations (Nwild) while the other layer represents the metapopulation (Nmet). The density of all demes was logistically regulated, with carrying capacity K set to 0.1-1 per 100 km2 across four vegetation types, and an intrinsic growth rate r set to 0.10. We performed 1000 independent stochastic simulations to get average introgression proportion and variance.
Introgression rate
We simulated neutral genetic diversity at 20 independent short tandem repeats (STRs), with a mutation rate of 0.001 (Cullingham et al. 2020). Gene flow between demes of free-roaming populations was controlled by the parameter mwild (migration rate) and mmet between demes of the managed metapopulation (translocation rate), while gene flow between the managed metapopulation (Nmet) and free-roaming populations (Nwild) was controlled by the parameter γi,j (assimilation rate from population i to j). The migrant rate m was set to 0.05 (met) and to 0.01 (wild), from each deme to adjacent demes at each generation, and γi,j varies depending on the scenario (see below). Note that γi,j is the proportion of contacts between individuals of both populations (met and wild) which results in gene flow from i to j. It is thus a measure of interbreeding success rate which could represent assimilation of individuals i in j, but also disassortative mating or hybrid relative fitness of admixed individuals. In SPLATCHE3, the level of introgression from population i (met) to j (wild) is considered to be represented by the proportion of genes from autosomal markers that are present in population i (met) at time t but sampled in population j (wild) at time t + 1. Within the density-dependent admixture model for two populations, each of the Nmet individuals has a probability to reproduce successfully with Nwild individuals from the same deme, following the assimilation model equation:
We set the value of αmet,wild and αwild,met to 0, which means that there is no competition between Nmet and Nwild individuals. For more details on the algorithms see SPLATCHE3 user manual (Currat et al. 2019). We ran genetic simulations for 500 generations (i.e. 2000 years; Girman et al. 2001), to measure how many generations it would take to achieve full genetic homogenization between Nmet and Nwild after range expansion (Quilodrán et al. 2020). We consider the population to be homogenized when 50% of the genome in the expanding metapopulation is introgressed with genes from the free-roaming populations. Introgression rates for both populations were visualized using the R package ggplot2 (Wickham et al. 2016), averaged over 1,000 stochastic simulations and 95% confidence intervals (CI).
Scenario comparisons
To test how different assimilation rates (from assortative mating to random mating) influence the genomic introgression rate, we ran simulations under different scenarios. The first is the baseline scenario (1), with strong assortative mating and asymmetric assimilation (1% of Nwild in Nmet, versus 5% assimilation of Nmet in Nwild). Under this scenario, we expect that individuals with similar genotypes are more likely to mate, and that metapopulation genes are more likely to spread into southern Africa than the other way around. Following, we have the scenarios: (2) strong assortative mating with less asymmetric assimilation (2.5% vs. 5%); (3) strong assortative mating with symmetric assimilation (5% vs. 5%); (4) less assortative mating with asymmetric assimilation (10% vs. 50%); (5) less assortative mating with symmetric assimilation (50% vs. 50%); and (6) random mating between both populations (i.e. full panmixia, 100% assimilation). Although full random mating is not expected in wild dogs, they do display strong inbreeding avoidance (Leigh et al. 2012), increasing the chance that Nmet in Nwild will admix in the future.
Population structure
To measure the effect of ongoing translocations on Nei’s F-statistics (Nei and Tajima, 1981), we used the simulation programme EASYPOP 2.01 (Balloux 2001), which is a software for population genetics that includes species-specific life-history traits. In this simulation tally, we adjusted the male-to-female ratio to 2:1 (McNutt 1996). We applied the polygynous mating system, with a 0.5 proportion of matings between subordinate males (Spiering et al. 2010). For the migration model, we chose a two-dimensional spatial model, which integrates isolation by distance. We tested for three different dispersal rates: 0.01, 0.05, 0.1. Mean dispersal distance was set to 19 for females and 41 for males, as wild dogs form single-sex dispersal coalitions (McNutt 1996). We followed a mixed mutation model with 75% of mutations following a stepwise mutation model (SMM) and 25% following a KAM model, and a mutation rate of 0,001 (Cullingham et al. 2020). We simulated 20 independent loci with a maximum possible number of alleles of K = 20, and a minimum variability of the initial population. Each parameter set was replicated 10 times, and for each replicate, we sampled 10 individuals in 5 subpopulations. In total, we performed 420 simulations. All scenarios and input parameters used for EASYPOP are listed in Table S4. Using the simulation output files, Nei’s F-statistics (Nei and Tajima 1981) were retrieved with the programme FSTAT 2.9.3.2 (Goudet 1995). Changes in genetic differentiation (FST) under all dispersal scenarios were visualized with the R package ggplot2 (Wickham et al. 2016), after they were calibrated to observed heterozygosity levels retrieved from our microsatellite dataset, by adjusting the first year of the simulation to the level of heterozygosity measured in Arlequin 3.5.2 (Excoffier and Lischer 2010). The EASYPOP output files were also used for downstream analyses performed in STRUCTURE 2.3.4 (Pritchard et al. 2000). Structure analyses were run for K = 1–5, correlated allele frequencies, a sampling of 250.000, burn-in of 100.000, and five iterations. Clumpak 1.1 (Kopelman et al. 2015) was used to summarize and visualize the structure runs at generation 30.
Results
Surfing alleles
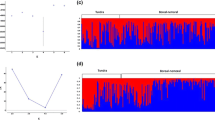
To assess spatial genetic diversity, we compared allele frequencies of 20 hypervariable STRs across five population clusters (Table S5). In the Kafue Zambezi area (KAZ) that stretches from Angola to eastern Zimbabwe, we found the highest number of alleles (140) and private alleles (17). Only four private alleles were found in all other populations, of which the managed metapopulation (MET) owned three, and the free-roaming population in South Africa (FRM) none. The total number of alleles was similar among these four populations, ranging from 74 to 89. When code-colouring various alleles at the 10 most variable loci found in the five populations, some waving patterns could be detected (Fig. 2A). What is mainly evident by this visualization is that alleles do not differentiate between the expanding populations (LOW, FRM, KNP, and MET), but merely display shifted allele frequencies, compared to the central wild dog population in the Kafue Zambezi area, which has a visibly higher allelic richness. When comparing ‘expected’ allele frequencies (from KAZ) to the ‘observed’ expanding populations, we found that an average of 13.5 (20.5%) alleles were significantly lower, and 11.25 (17.1%) were significantly higher (Fig. 2B).
Introgression following range expansion
The movement of the range expansion and level of introgression were simulated forward in time, from the managed metapopulation Nmet (second virtual layer, Fig S2), into the free-roaming populations Nwild (first virtual layer). Overall, we measured a carrying capacity of 20,100 wild dogs in southern Africa, if the area would solely consist of natural vegetation. The Nmet expanded outward from South Africa, Malawi, and Mozambique (m = 0.05), and the Nwild expanded outwards from Angola, Zambia, Namibia, central Botswana and Zimbabwe (m = 0.01), connecting populations in Zambia and Malawi (Fig S2). Under the baseline scenario, spatially explicit simulations showed that in the face of ongoing dispersal / translocations, it would take approximately 150 generations for Nmet to have been (50%) introgressed with alleles from Nwild population (Fig. 2C). Despite a larger assimilation rate from Nmet to Nwild than the reverse, the Nmet was expected to be introgressed faster with Nwild genes than the other way around, due to ongoing translocations from Nmet into southern Africa but less dispersal in return, until it slowly reaches a plateau. When two populations were treated as one, the relative proportion of Nmet genes tended to slowly increase with time.
The effect of range expansions on population genetic structure. (A) Microsatellite alleles in five African wild dog (Lycaon pictus) populations in southern Africa: the central population across the Kafue Zambezi transfrontier conservation area (KAZ); Zimbabwean Lowveld (LOW), free-roaming population in the Waterberg (FRM), Kruger NP (KNP), and the managed metapopulation (MET) in South Africa. All alleles are colour-coded to visualize their frequencies. White circles are missing alleles. (B) We tested for significant shifts in allele frequencies with a Cochran-Mantel-Haenszel test, and found that 9 to 13 alleles occurred in higher proportions in the expanding ‘observed’ populations (LOW, FRM, KNP, and MET), compared the original ‘expected’ population (KAZ), which could be an indication of surfing alleles. (C) The proportion of introgressed genes in a simulated African wild dog population, in the face of ongoing dispersal and translocations. Introgression values are retrieved by SPLATCHE3, and averaged over 1,000 stochastic simulations and 95% CI. The dark blue curve (1) represents managed metapopulation Nmet genes from South Africa into the free-roaming populations Nwild in southern Africa. The dark green curve (4) represents the proportion of Nwild genes into Nmet. The light blue curve (2) represents the average introgression in both populations, and the light green (3) curve the total proportion of Nmet genes in both populations considered as one. After 150 generations, Nmet has been 50% introgressed with alleles from Nwild population. D. The influence of dispersal on genetic differentiation (FST) of wild dogs in southern Africa. Results are based on forward-time genetic simulations, using EASYPOP. The simulation outputs were used to run STRUCTURE analysis, which decreased from K = 4 to K = 2 with increasing dispersal rate
Comparison between various assimilation rates
The assimilation rate was low under the baseline scenario, assuming that it is more likely that individuals from the same population mate. Hence, it implies that there is strong assortative mating between the expanding managed metapopulation (Nmet) and the free-roaming populations (Nwild). To allow for comparison, we also tested other assimilation scenarios between Nmet and Nwild, including a random mating scenario, to see how this would influence the introgression rate (Fig S2). In all scenarios, introgression started to occur after approximately 50 generations in southern Africa, between Nmet and Nwild. The average introgression rate varied between ~ 60% (scenario 1) to < 5% (scenario 6) after 250 generations. Under all scenarios, introgression rate was predicted to be higher in Nmet compared to Nwild, due to population dynamics. To elaborate, a smaller number of Nmet individuals enter areas where there are resident wild populations at demographic equilibrium, resulting in asymmetric introgression from the local (Nwild) to the arriving population (Nmet), see Currat et al. (2008) for a detailed explanation of the underlying mechanisms. This introgression asymmetry is reduced when there is strong assortative mating, with much less Nwild individuals entering Nmet, which allows Nmet time to grow locally without much initial introgression. We found that with random mating, there was much faster genomic admixing, which means that the contribution of Nmet genes in the overall population was greatly reduced, from 45 to 5% after 500 generations (Fig S2).
Population structure
Changes in genetic differentiation (FST) were simulated under three dispersal / translocation scenarios over 30 generations (Fig. 2D). Scenario 1 (0.01 dispersal rate) caused an increase of FST over time, whereas scenario 3 (0.1 dispersal rate) followed the expected reduction in genetic differentiation. In the most realistic scenario (0.1 dispersal rate; Tensen et al. 2019), the FST dropped below 0.05 within 10 and 15 generations, after which no population structure was observed. This means that with the current rate of translocations (10% from the managed metapopulation), genetic differentiation based on neutral loci in southern Africa will disappear, overturning the effects of genetic drift or surfing alleles associated with range expansions. To the contrary, the different scenarios had little effect on observed heterozygosity (HO), which increased under all dispersal scenarios (Table S6). In scenario 1, HO was 0.566 after 30 generations (1% increase), in scenario 2 HO was 0.589 (3% increase), and in scenario 3 HO was 0.596 after 30 generations (4% increase) as a result of population admixture.
Discussion
In this study, we used the African wild dog as a model species to test genetic theory on allelic surfing after reintroductions. To do so, we presented an overview of all translocated and dispersing wild dogs across southern Africa, and combined these with a genetic dataset to make predictions about the effects of range expansions on intraspecific genetic variation. We also simulated the movement of surfing alleles forward in time under the current demographic scenario. Our results show that management interventions by means of translocation or facilitation of natural dispersal can help restore genetic diversity in fragmented and expanding populations. The range expansion of wild dogs is currently one of the world’s largest rewilding efforts for an endangered large carnivore, which makes the evaluation of the conservation value of this programme, including the preservation of genetic integrity, desirable.
A stepping-stone mode of dispersal over multiple generations allows wild dogs to cover large distances, connecting multiple countries (Davies-Mostert et al. 2012; Tensen et al. 2022). For example, dispersal events have been recorded between Savé Valley Conservancy in Zimbabwe and Northern Tuli in Botswana (~ 350 km), the Waterberg region in South Africa and the Central Kalahari in Botswana (~ 400 km), and the Okavango Delta in Botswana and Khaudum NP in Namibia (~ 250 km). Nonetheless, radio telemetry and genetic studies remain scarce for wild dogs, due to which our understanding of large-scale connectivity on the African continent still suffers from a lack of information (Abrahms et al. 2017). The translocation history of wild dogs, on the other hand, is well-documented. We recorded sixteen translocation events within southern Africa, of which ten were reintroductions, totalling 144 individuals. The longest translocation distances were from South Africa to Zambia (~ 1200 km) and Malawi (~ 1000 km). All translocations were considered successful, based on post-release survival, social integration, and reproduction (Gusset et al. 2006). The only reintroductions that failed were to Etosha NP in Namibia, which occurred between 1978 and 1989, due to low prey density and high lion density at the release site (Scheepers and Venzke 1995). Reintroduction techniques and guidelines have been optimized rigorously since then (Hayward and Somers 2009; Gusset et al. 2010; Bouley et al. 2021). Another incident took place in Malawi, where a pack of 18 wild dogs died from unintended poisoning in Liwonde NP, which was translocated from South Africa and Mozambique (African Parks 2022). This highlights the need to create satellite populations of wild dogs to reduce the impact of stochastic events (Davies-Mostert et al. 2009).
Marsden et al. (2012) found that populations that had recently experienced recolonization, such as the Lowveld and Laikipia, consistently showed lower genetic diversity, which may lead to population structure (Excoffier et al. 2009). In this study, we also found that allelic richness was lower in recently expanding populations compared to the central cluster in KAZA-TFCA. Furthermore, some alleles reached high frequencies, and only four private alleles were present outside KAZA-TFCA, which is indicative of genetic surfing (Edmonds et al. 2004). Genetic surfing as a result of a range expansion can cause rapid genetic change over time and space (Klopfstein et al. 2006), which has been observed in many recovering large carnivores. In coyotes, populations colonized eastern North America along two expansion fronts, which resulted in genetic differences among groups of conspecifics at a fine temporal scale (Heppenheimer et al. 2018). After an initial bottleneck, the population quickly recovered its genetic diversity due to increased population size and connectivity. The grey wolf recently recolonized areas in Europe, and a strong west-east population structure has occurred due to founder-flush and genetic surfing (Szewczyk et al. 2019). In Finland, brown bears have been expanding their range, which led to strong initial genetic changes, which gradually disappeared after ongoing population growth and admixture (Hagen et al. 2015). Likewise, we believe that surfing alleles after range expansion have led to the population genetic structure observed in wild dogs in southern Africa. Shifting allele frequencies can arise even within continuous populations and in the absence of dispersal barriers between demes (Paulose and Hallatschek 2020), and spatially clustered allele distributions can persist for hundreds of generations (Ibrahim et al. 1996). Because the effect of genetic surfing can be deleterious, similar to genetic drift, it is important to understand how to overcome this unintended effect of range expansion programmes.
Currently, there is limited knowledge on the genetic consequences of translocations in large carnivores, especially across broad geographical scales (Hagen et al. 2015). Using population genetic simulations, we illustrated that with current levels of dispersal and translocations, the FST observed in wild dogs across southern Africa will drop below 0.05 within 15 generations. We also measured that it would take 200 generations to become genomically homogenised, i.e. when about 40% of the genome of each population would be introgressed by genes from other populations. Thus, the negative effects of genetic surfing and drift will gradually disappear as recipient or local populations become admixed with donor or expanding populations. It is known that few migrants are capable of reversing the stochastic process of genetic drift on allele frequencies (Vilà et al. 2003). For wild dogs, introgression was always predicted to be higher in the expanding metapopulation compared to the free-roaming population due to population dynamics, as a smaller number of reintroduced (Nmet) individuals enter areas where there are already established (Nwild) populations at demographic equilibrium. This introgression asymmetry is reduced only when there is strong assortative mating, which allows the expanding population to grow locally without initial extensive introgression. With full panmixia, the contribution of reintroduced genes in the overall population would have reduced to less than 5% after 500 generations. These general trends could help predict genetic recovery in expanding populations with ongoing conservation interventions, based on the assumption that the demographic parameters are constant over time.
Generally, it is advised that individuals should not be translocated between evolutionary significant units (ESUs), to conserve ecological and evolutionary processes (Bertola et al. 2021). ESUs have evolved independently as a result of local adaptation (Moritz et al. 1999), which is relevant for translocation programmes where the risk arises that unique lineages become admixed, reducing interspecific diversity of the overall population (Edwards 2007; Weeks et al. 2015). In wild dogs, two main phylogenetic clades have previously been distinguished, in eastern Africa (E genotypes) and southern Africa (S genotypes), based on both genetic and morphological differences (Girman et al. 1993, 2001). Limited genetic data indicate that wild dogs from West Central Africa may be distinct as well. However, since there are no viable wild dog populations in this region, future reintroductions may have to harvest animals from non-native lineages (Woodroffe et al. 1999). The phylogenetic clades were suggested to have originated from an extinction event in eastern Africa and subsequent recolonization from western or central Africa (Marsden et al. 2012), with secondary migration between eastern and southern Africa that may have taken place during Pleistocene climatic changes (Girman et al. 2001). There are large admixture zones between the two clades, which indicates that long-distance dispersal (> 400 km) has occurred until recently (Girman et al. 2001). Consistent with many other savannah species, admixture occurs from Ethiopia to Sudan, and Zambia to Malawi and Mozambique (Lorenzen et al. 2012; Bertola et al. 2021).
It has been suggested that, historically, wild dogs within the southern and eastern units were panmictic, with no pronounced population structure (Girman et al. 2001). Genetic clustering has occurred since, as a result of reduced effective population sizes and habitat fragmentation (Marsden et al. 2012; Tensen et al. 2022). Although Moritz (1999) and Szewzcyk et al. (2019) argue that population structure based on diverged allele frequencies needs to be maintained regardless of phylogenetic distinctiveness of the alleles, we dispute this notion. Genetic drift and surfing reduce genetic diversity, and the risks associated with low genetic diversity are well established (Frankham 2015). Because population differentiation resulting from recent habitat fragmentation or other human-mediated processes through genetic drift is considered harmful, unlike ESUs, it is advised to restore gene flow when possible (Frankham et al. 2017; Liddell et al. 2021), particularly for endangered species (Edmands 2007; McLennan et al. 2020). Maintaining large metapopulations is widely considered the most beneficial and natural scenario for wild animals, which will inevitably lead to genetic homogenisation at small spatial scales (Frankham et al. 2017).
The stark relationship between genetic differentiation and genetic diversity highlights the value of translocation programmes in increasing population versatility (Coleman et al. 2013). This is particularly relevant for augmentation programmes, where the goal is to increase population fitness by reintroducing new alleles, often referred to as genetic rescue (Coates et al. 2018; Ralls et al. 2020). Our results suggest that demographic histories and genetic surfing shape some of the current population structure of wild dogs in southern Africa. We propose that the rapid genetic changes following local extirpation and recolonization require an ongoing effort to maintain geographical connectivity and demographic recovery of African wild dogs. Genomic introgression during the course of range expansions will cause a gradual disappearance of population structure within southern Africa, and increase genetic diversity without risking the loss of evolutionary distinct units.
Data Availability
The datasets generated during and/or analysed during the current study are made available in the Supplementary Material.
References
Abrahms B, Sawyer SC, Jordan NR, McNutt JW, Wilson AM, Brashares JS (2017) Does wildlife resource selection accurately inform corridor conservation? J Appl Ecol 54:412–422. https://doi.org/10.1111/1365-2664.12714
African P (2022) African wild dogs: poisoned in Liwonde National Parj, Malawi. African Wild Dogs poisoned in Liwonde National Park, Malawi | African Parks Accessed 28 November 2022
Balloux F (2001) EASYPOP (version 1.7): a computer program for population genetics simulations. J Hered 92:301–302. https://doi.org/10.1093/jhered/92.3.301
Becker MS, Almeida J, Begg C, Bertola L, Breitenmoser C, Breitenmoser U, Coals P, Funston P, Gaylard A, Groom R, Henschel PH (2022) Guidelines for evaluating the conservation value of African lion (Panthera leo) translocations. Front Environ Sci 3:963961. https://doi.org/10.3389/fcosc.2022.963961
Bertola LD, Miller SM, Williams VL et al (2021) Genetic guidelines for translocations: maintaining intraspecific diversity in the lion (Panthera leo). Evol Appl 15:22–39. https://doi.org/10.1111/eva.13318
Boast LK, Chelysheva EV, van der Merwe V, Schmidt-Küntzel A, Walker EH, Cilliers D, Gusset M, Marker L (2018) Cheetah translocation and reintroduction programs: past, present, and future. In: Nyhus PJ, Marker L, Boast LK, Schmidt-Küntzel A (eds) Cheetahs: biology and conservation. Elsevier, Germany, pp 275–289
Bouley P, Poulos M, Branco R, Carter NH (2018) Post-war recovery of the African lion in response to large-scale ecosystem restoration. Biol Conserv 227:233–242. https://doi.org/10.1016/j.biocon.2018.08.024
Bouley P, Paulo A, Angela M, Du Plessis C, Marneweck DG (2021) The successful reintroduction of African wild dogs (Lycaon pictus) to Gorongosa National Park, Mozambique. PLoS ONE 16:e0249860. https://doi.org/10.1371/journal.pone.0249860
Chapron G, Kaczensky P, Linnell JD, von Arx M, Huber D, Andrén H, López-Bao JV, Adamec M, Álvares F, Anders O, Balčiauskas L (2014) Recovery of large carnivores in Europe’s modern human-dominated landscapes. Science 346:1517–1519. https://doi.org/10.1126/science.1257553
Coates DJ, Byrne M, Moritz C (2018) Genetic diversity and conservation units: dealing with the species-population continuum in the age of genomics. Front Ecol Evol 6:165. https://doi.org/10.3389/fevo.2018.00165
Coleman RA, Weeks AR, Hoffmann AA (2013) Balancing genetic uniqueness and genetic variation in determining conservation and translocation strategies: a comprehensive case study of threatened dwarf galaxias, Galaxiella pusilla (Mack)(Pisces: Galaxiidae). Mol Ecol 22:1820–1835. https://doi.org/10.1111/mec.12227
Cozzi G, Behr DM, Webster HS, Claase M, Bryce CM, Modise B, McNutt JW, Ozgul A (2020) African wild dog dispersal and implications for management. J Wildl Man 84:614–621. https://doi.org/10.1002/jwmg.21841
Crandall KA, Bininda-Emonds OR, Mace GM, Wayne RK (2000) Considering evolutionary processes in conservation biology. Trends Ecol Evol 15:290–295. https://doi.org/10.1016/S0169-5347(00)01876-0
Cullingham CI, Miller JM, Peery RM, Dupuis JR, Malenfant RM, Gorrell JC, Janes JK (2020) Confidently identifying the correct K value using the ∆K method: when does K = 2? Mol Ecol 29:862–869. https://doi.org/10.1111/mec.15374
Currat M, Excoffier L, Maddison W, Otto SP, Ray N, Whitlock MC, Yeaman S (2006) Comment on ongoing adaptive evolution of ASPM, a brain size determinant in Homo sapiens and Microcephalin, a gene regulating brain size, continues to evolve adaptively in humans. Science 313:172–172. https://doi.org/10.1126/science.1113722
Currat M, Arenas M, Quilodran CS, Excoffier L, Ray N (2019) SPLATCHE3: simulation of serial genetic data under spatially explicit evolutionary scenarios including long-distance dispersal. Bioinformatics 35:4480–4483. https://doi.org/10.1093/bioinformatics/btz311
Davies HT, Du Toit JT (2004) Anthropogenic factors affecting wild dog Lycaon pictus reintroductions: a case study in Zimbabwe. Oryx 38(1):32–39. https://doi.org/10.1017/S0030605304000067
Davies-Mostert HT, Mills MGL, Macdonald DW (2009) A critical assessment of South Africa’s managed metapopulation recovery strategy for African wild dogs. In: Hayward MW, Somers MJ (eds) Reintroduction of top-order predators. Blackwell, London, pp 10–42
Davies-Mostert HT, Kamler JF, Mills MG, Jackson CR, Rasmussen GS, Groom RJ, Macdonald DW (2012) Long‐distance transboundary dispersal of African wild dogs among protected areas in southern Africa. Afr J Ecol 50:500
Davies-Mostert HT, Mills MG, Macdonald DW (2015) The demography and dynamics of an expanding, managed African wild dog metapopulation. Afr J Wildl Res 45:258–273
Edmands S (2007) Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation and management. Mol Ecol 16:463–475. https://doi.org/10.1111/j.1365-294X.2006.03148.x
Edmonds CA, Lillie AS, Cavalli-Sforza LL (2004) Mutations arising in the wave front of an expanding population. Proc Nat Acad Sci 101:975–979. https://doi.org/10.1073/pnas.0308064100
Edwards S, Noack J, Heyns L, Rodenwoldt D (2019) Evidence of a high-density brown hyena population within an enclosed reserve: the role of fenced systems in conservation. Mammal Res 64(4):519–527. https://doi.org/10.1007/s13364-019-00432-7
Excoffier L, Lischer HE (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res 10(3):564–567. https://doi.org/10.1111/j.1755-0998.2010.02847.x
Excoffier L, Ray N (2008) Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol Evol 23:347–351. https://doi.org/10.1016/j.tree.2008.04.004
Excoffier L, Foll M, Petit RJ (2009) Genetic consequences of range expansions. Annu Rev Ecol Evol Syst 40:481. https://doi.org/10.1146/annurev.ecolsys.39.110707.173414
Fanshawe JH, Ginsberg JR, Sillero-Zubiri C, Woodroffe R (1997) The status and distribution of remaining wild dog populations. In: Woodroffe R, Ginsberg JR (eds) The African Wild Dog: Status Survey and Conservation Action Plan. IUCN, Gland
Frankham R (2009) Genetic considerations in reintroduction programmes for top-order terrestrial predators. In: Hayward MW, Somers M (eds) Reintroduction of Top-Order predators. Blackwell Publishing, Oxford, UK, pp 371–387
Frankham R (2015) Genetic rescue of small inbred populations: meta-analysis reveals large and consistent benefits of gene flow. Mol Ecol 24:2610–2618. https://doi.org/10.1111/mec.13139
Frankham R, Ballou JD, Ralls K, Eldridge M, Dudash MR, Fenster CB, Lacy RC, Sunnucks P (2017) Genetic management of fragmented animal and plant populations. Oxford University Press, London
Friedlander E, Steinrücken M (2022) A numerical framework for genetic hitchhiking in populations of variable size. Genetics 220(3):iyac012. https://doi.org/10.1093/genetics/iyac012
Girman DJ, Kat PW, Mills MGL, Ginsberg JR, Borner M, Wilson V, Fanshawe JH, Fitzgibbon C, Lau LM, Wayne RK (1993) Molecular genetic and morphological analyses of the African wild dog (Lycaon pictus). J Hered 84:450–459. https://doi.org/10.1093/oxfordjournals.jhered.a111371
Girman DJ, Vilà C, Geffen E, Creel S, Mills MGL, McNutt JW, Ginsberg JKPW, Kat PW, Mamiya KH, Wayne RK (2001) Patterns of population subdivision, gene flow and genetic variability in the African wild dog (Lycaon pictus). Mol Ecol 10:1703–1723. https://doi.org/10.1046/j.0962-1083.2001.01302.x
Goudet J (1995) FSTAT (version 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Gusset M, Slotow R, Somers MJ (2006) Divided we fail: the importance of social integration for the re-introduction of endangered African wild dogs (Lycaon pictus). J Zool 270(3):502–511. https://doi.org/10.1111/j.1469-7998.2006.00168.x
Gusset M, Stewart GB, Bowler DE, Pullin AS (2010) Wild dog reintroductions in South Africa: a systematic review and cross-validation of an endangered species recovery programme. J Nat Conserv 18:230–234. https://doi.org/10.1016/j.jnc.2009.11.001
Hagen SB, Kopatz A, Aspi J, Kojola I, Eiken HG (2015) Evidence of rapid change in genetic structure and diversity during range expansion in a recovering large terrestrial Carnivore. Proc R Soc B Biol Sci 282:p20150092. https://doi.org/10.1098/rspb.2015.0092
Hallatschek O, Nelson DR (2008) Gene surfing in expanding populations. Theor Pop Biol 73:158–170. https://doi.org/10.1016/j.tpb.2007.08.008
Hayward MW, Somers M (2009) Reintroduction of top-order predators. John Wiley & Sons
Heppenheimer E, Cosio DS, Brzeski KE, Caudill D, Van Why K, Chamberlain MJ, Hinton JW, vonHoldt B (2018) Demographic history influences spatial patterns of genetic diversity in recently expanded coyote (Canis latrans) populations. Heredity 120(3):183–195. https://doi.org/10.1038/s41437-017-0014-5
Hofmann DD, Behr DM, McNutt JW, Ozgul A, Cozzi G (2021) Bound within boundaries: do protected areas cover movement corridors of their most mobile, protected species? J Appl Ecol 58:1133–1144. https://doi.org/10.1111/1365-2664.13868
Ibrahim KM, Nichols RA, Hewitt GM (1996) Spatial patterns of genetic variation generated by different forms of dispersal during range expansion. Heredity 77:282–291. https://doi.org/10.1038/hdy.1996.142
IUCN/SSC (2008) Regional conservation strategy for the cheetah and wild dog in eastern Africa. IUCN, Gland
IUCN/SSC (2015) Southern African conservation strategy for Cheetah and Wild Dogs. IUCN, Gland
Johnson CN, Balmford A, Brook BW, Buettel JC, Galetti M, Guangchun L, Wilmshurst JM (2017) Biodiversity losses and conservation responses in the Anthropocene. Science 356:270–275. https://doi.org/10.1126/science.aam9317
Kalinowski ST (2005) Hp-rare 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5(1):187–189. https://doi.org/10.1111/j.1471-8286.2004.00845.x
Klopfstein S, Currat M, Excoffier L (2006) The fate of mutations surfing on the wave of a range expansion. Mol Biol Evol 23:482–490. https://doi.org/10.1093/molbev/msj057
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Res 15(5):1179–1191. https://doi.org/10.1111/1755-0998.12387
Kuiper T, Dickman AJ, Hinks AE, Sillero-Zubiri C, Macdonald EA, Macdonald DW (2018) Combining biological and socio‐political criteria to set spatial conservation priorities for the endangered African wild dog. Anim Conserv 21:376–386. https://doi.org/10.1111/acv.12405
Leigh KA, Zenger KR, Tammen I, Raadsma HW (2012) Loss of genetic diversity in an outbreeding species: small population effects in the African wild dog (Lycaon pictus). Conserv Gen 13:767–777. https://doi.org/10.1007/s10592-012-0325-2
Liddell E, Sunnucks P, Cook CN (2021) To mix or not to mix gene pools for threatened species management? Few studies use genetic data to examine the risks of both actions, but failing to do so leads disproportionately to recommendations for separate management. Biol Conserv 256:p109072. https://doi.org/10.1016/j.biocon.2021.109072
Lindsey PA, Du Toit JT, Mills MGL (2005) Attitudes of ranchers towards African wild dogs Lycaon pictus: conservation implications on private land. Biol Conserv 125:113–121. https://doi.org/10.1016/j.biocon.2005.03.015
Lorenzen ED, Heller R, Siegismund HR (2012) Comparative phylogeography of African savannah ungulates. Mol Ecol 21(15):365–. https://doi.org/10.1111/j.1365-294X.2012.05650.x
Malone EW, Perkin JS, Leckie BM, Kulp MA, Hurt CR, Walker DM (2018) Which species, how many, and from where: integrating habitat suitability, population genomics, and abundance estimates into species reintroduction planning. Glob Chang Biol 24:3729–3748. https://doi.org/10.1111/gcb.14126
Marsden CD, Woodroffe R, Mills MG, McNutt JW, Creel S, Groom R, Emmanuel M, Cleaveland S, Kat P, Rasmussen GS, Ginsberg J (2012) Spatial and temporal patterns of Neutral and adaptive genetic variation in the endangered African wild dog (Lycaon pictus). Mol Ecol 21:1379–1393. https://doi.org/10.1111/j.1365-294X.2012.05477.x
McLennan EA, Grueber CE, Wise P, Belov K, Hogg CJ (2020) Mixing genetically differentiated populations successfully boosts diversity of an endangered Carnivore. Anim Conserv 23(6):700–712. https://doi.org/10.1111/acv.12589
McNutt JW (1996) Sex-biased dispersal in African wild dogs, Lycaon pictus. Anim Behav 52(6):1067–1077. https://doi.org/10.1006/anbe.1996.0254
Miller SM, Bissett C, Parker DM et al (2013) Management of reintroduced lions in small, fenced reserves in South Africa: an assessment and guidelines. S Afr J Wildl Res 43(2):138–154
Mills MGL, Ellis S, Woodroffe R, Maddock A, Stander P, Rasmussen G, Pole A, Fletcher P, Bruford M, Wildt D, Macdonald D, Seal U (1998) Population and habitat viability assessment for the African wild dog (Lycaon pictus) in southern Africa. Final workshop report. IUCN/SSC Conservation Breeding Specialist Group, Apple Valley
Mona S, Ray N, Arenas M, Excoffier L (2014) Genetic consequences of habitat fragmentation during a range expansion. Heredity 112:291–299. https://doi.org/10.1038/hdy.2013.105
Moritz C (1999) Conservation units and translocations: strategies for conserving evolutionary processes. Hereditas 130:217–228. https://doi.org/10.1111/j.1601-5223.1999.00217.x
Paulose J, Hallatschek O (2020) The impact of long-range dispersal on gene surfing. Proc Nat Acad Sci 117:7584–7593. https://doi.org/10.1073/pnas.1919485117
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959. https://doi.org/10.1093/genetics/155.2.945
Ralls K, Sunnucks P, Lacy RC, Frankham R (2020) Genetic rescue: a critique of the evidence supports maximizing genetic diversity rather than minimizing the introduction of putatively harmful genetic variation. Biol Conserv 251:108784. https://doi.org/10.1016/j.biocon.2020.108784
Ray N, Currat M, Excoffier L (2003) Intra-deme molecular diversity in spatially expanding populations. Mol Biol Evol 20(1):76–86. https://doi.org/10.1093/molbev/msg009
Riggio J, Jacobson A, Dollar L, Bauer H, Becker M, Dickman A, Funston P, Groom R, Henschel P, de Iongh H, Lichtenfeld L (2013) The size of savannah Africa: a lion’s (Panthera leo) view. Biod Cons 22(1):17–35. https://doi.org/10.1007/s10531-012-0381-4
Ripple WJ, Estes JA, Beschta RL et al (2014) Status and ecological effects of the world’s largest carnivores. Science 343:p1241484. https://doi.org/10.1126/science.124148
Scheepers JL, Venzke KAE (1995) Attempts to reintroduce African wild dogs Lycaon pictus into Etosha National Park, Namibia. S Afr J Wildl Res 25(4):138–140
Soulé ME, Noss R (1998) Rewilding and biodiversity: complementary goals for continental conservation. Wild Earth 8(3):18–28
Spiering PA, Somers MJ, Maldonado JE, Wildt DE, Gunther MS (2010) Reproductive sharing and proximate factors mediating cooperative breeding in the African wild dog (Lycaon pictus). Behav Ecol Sociobiol 64:583–592. https://doi.org/10.1007/s00265-009-0875-6
Spiering PA, Szykman Gunther M, Somers MJ, Wildt DE, Walters M, Wilson AS, Maldonado JE (2011) Inbreeding, heterozygosity and fitness in a reintroduced population of endangered African wild dogs (Lycaon pictus). Conserv Genet 12(2):401–412. https://doi.org/10.1007/s10592-010-0147-z
Szewczyk M, Nowak S, Niedźwiecka N, Hulva P, Špinkytė-Bačkaitienė R, DemjanoviŠová K, Bolfíková Bč, Antal V, Fenchuk V, Figura M, Tomczak P (2019) Dynamic range expansion leads to establishment of a new, genetically distinct wolf population in Central Europe. Sci Rep 9(1):19003. https://doi.org/10.1038/s41598-019-55273-w
Tensen L, van Vuuren BJ, Du Plessis C, Marneweck DG (2019) African wild dogs: genetic viability of translocated populations across South Africa. Biol Conserv 234:131–139. https://doi.org/10.1016/j.biocon.2019.03.033
Tensen L, van Jansen B, Groom R et al (2022) Spatial genetic patterns in African wild dogs reveal signs of effective dispersal across southern Africa. Front Ecol Evol 10:1216. https://doi.org/10.3389/fevo.2022.992389
Vilà C, Sundqvist AK, Flagstad Ø, Seddon J, Björnerfeldt SB, Kojola I, Casulli A, Sand H, Wabakken P, Ellegren H (2003) Rescue of a severely bottlenecked wolf (Canis lupus) population by a single immigrant. Proc Royal Soc B 270(1510):91–97. https://doi.org/10.1098/rspb.2002.2184
WAG-SA (2022) Wild Dog Specialist Group minutes November 2022
Weeks AR, Moro D, Thavornkanlapachai R, Taylor HR, White NE, Weiser EL, Heinze D (2015) Conserving and enhancing genetic diversity in translocation programs. In: Armstrong DP, Hayward MW, Moro D, Seddon PJ (eds) Advances in reintroduction biology of Australian and New Zealand Fauna. CSIRO Publishing, Australia, pp 127–140
Wiberg RA, Gaggiotti OE, Morrissey MB, Ritchie MG (2017) Identifying consistent allele frequency differences in studies of stratified populations. Methods Ecol Evol 8(12):1899–1909. https://doi.org/10.1111/2041-210X.12810
Wickham H, Chang W, Wickham MH (2016) Package ‘ggplot2’. Create Elegant data Visualisations Using the Grammar of Graphics Version 2(1):1–189
Wolf C, Ripple WJ (2018) Rewilding the world’s large carnivores. R Soc Open Sci 5(3):p172235. https://doi.org/10.1098/rsos.172235
Woodroffe R, Ginsberg JR (1999) Conserving the African wild dog Lycaon pictus. II. Is there a role for reintroduction? Oryx 33(2):143–151. https://doi.org/10.1046/j.1365-3008.1999.00053.x
Woodroffe R, Sillero-Zubiri C (2020) Lycaon pictus (amended version of 2012 assessment). The IUCN Red List of Threatened Species 2020: e. T12436A166502262
Woodroffe R, Ginsberg JR, Macdonald DW (1997) The African Wild Dog: Status Survey and Conservation Action Plan. IUCN, Gland, Switzerland
Woodworth LM, Montgomery ME, Briscoe DA, Frankham R (2002) Rapid genetic deterioration in captive populations: causes and conservation implications. Conserv Gen 3:277–288. https://doi.org/10.1023/A:1019954801089
Funding
This work has been supported by research funding from DFG (TE 1502/1–1) granted to L. Tensen.
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
Study design, data collection, and manuscript preparation were performed by L.T. Data analysis was performed by M.C. and L.T. Study conception was performed by H. D.-M. and C.P. Supervision was performed by K.F. All authors have commented on previous versions of the manuscript, and have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
This study has been approved in 2022 by the Endangered Wildlife Trust Ethics Committee (EWTEC2022_016). Samples were collected and transported in 2016 under permit 13/1/1/30/2/0-2015/01/003889 and analyzed at the University of Johannesburg, which was approved by the Faculty Ethics Committee under permit Tensen_201337943.
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Communicated by David Hawksworth.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tensen, L., Currat, M., Davies-Mostert, H. et al. Genetic surfing during the range expansion of an endangered large carnivore. Biodivers Conserv 33, 361–378 (2024). https://doi.org/10.1007/s10531-023-02755-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10531-023-02755-z