Abstract
Coconut rhinoceros beetle (Oryctes rhinoceros; CRB) was discovered in 2015 in a small outbreak in Honiara, Guadalcanal, Solomon Islands. This was the first record of CRB from Solomon Islands and a response plan was prepared. An awareness programme was launched and where CRB sightings were confirmed, delimitation surveys were carried out. Following the launch of the awareness programme, later in 2015, CRB was also reported from the Shortland Islands in the Western Province. Other islands were surveyed from 2015 to 2020. If CRB presence was confirmed, beetles were collected and analysed for haplotype and presence of the classical biological control agent, Oryctes rhinoceros nudivirus (OrNV). A distribution map and timeline of invasion were developed. The initial populations belonged to two distinct haplotypes: CRB-G (clade IA) in Honiara and CRB-S (clade II) in Shortland Islands. Despite control measures, by 2020 CRB-G had spread to islands in eight provinces and CRB-S had spread to islands in seven provinces. CRB-S and CRB-G co-occur in Guadalcanal and Malaita provinces. In 2019, OrNV was detected from field collected CRB from Guadalcanal and has since spread to Malaita Island. In both cases the virus was detected where CRB-G and CRB-S co-occur. Other outbreak areas in Solomon Islands remain OrNV-free. The two haplotypes appear to have spread following patterns of air and sea movement between the point of origin (CRB-G from Honiara and CRB-S from the Shortland Islands) to other islands/provinces.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Coconut rhinoceros beetle (Oryctes rhinoceros; CRB) is one of the most damaging pests to coconut and oil palms in Asia and the Pacific Islands (Bedford 1980). The adult beetle bores into the palm crown, damaging the developing fronds and creating distinctive V-shaped notches (Fig. 1), which affect tree development and yield. Severe attack will kill the palm. Larvae feed on rotting organic matter (green waste, manure, dead palm trunks, etc.) but do not cause damage to living palms. The insect is native to South and Southeast Asia and was inadvertently introduced to the Samoan islands in 1909 (Catley 1969; Paudel et al. 2022) and spread to other nearby island nations in the south-west Pacific resulting in significant economic impact on palm production (Jackson 2009; Young 1986).
The early pest outbreaks were brought under control by releasing biological control agents, particularly Oryctes rhinoceros nudivirus (OrNV) and phytosanitary measures (Bedford 2013; Paudel et al. 2021a). Such was the success of the virus, that damage was reduced to manageable levels and no significant new invasions were reported for a period of more than 30 years (Huger 2005). Recently a new wave of invasions has occurred with beetle outbreaks reported in Guam, Hawaii, Solomon Islands, and the mainland of PNG (Ero et al. 2016; Marshall et al. 2017; Moore 2011). The initial response to the CRB outbreak detected in Guam during 2007 was to introduce previously successful OrNV isolates as biocontrol agents, but these were unsuccessful as the virus could not be established in the population (Marshall et al. 2017; Moore 2018). Using the mitochondrial DNA marker, cytochrome c oxidase subunit I (COI), Marshall et al. (2017) recognized that the outbreak belonged to a distinct population which was designated as CRB-G.
By examining CRB samples from the Asia/Pacific region, Marshall et al. (2017) differentiated the CRB population into four clades (Clade I, II, III and IV) based on geographical origin and invasion pathway. Three of these clades (clade II, III, and IV), while genetically distinct, were characterized biologically by susceptibility to the original strain of OrNV used to control the earlier wave of CRB invasion in the Pacific. These three clades included native populations, and populations from regions affected by the previous CRB invasions, and were collectively named CRB-S. The fourth clade (clade I), which included populations from the newly invaded regions (Guam, Papua New Guinea, Hawaii, Solomon Islands) and from parts of the native range (Philippines, Indonesia), was named CRB-G. Thus, two distinct levels of population structure within CRB were identified: the first grouping (CRB-G and CRB-S) is associated with susceptibility to the original strain of OrNV introduced to the Pacific, although there is no evidence for a causal relationship between the mitochondrial haplotype marker and virus susceptibility. The second, more complex, clade grouping reflects geographic populations in the native and invasive range of CRB and presumably relates to historic patterns of dispersal by CRB. Haplotype analysis using the COI gene has been proven to be an effective tool to identify genetically distinct populations and to analyze invasion pathways of several pest species (Abdallah et al. 2013; Doorenweerd et al. 2020; Gariepy et al. 2021; Machado et al. 2020; Rollins et al. 2011; Schlum et al. 2021; Sun et al. 2020) and was used to suggest CRB invasion of Hawaii was a relatively recent invasion likely to be human mediated (Reil et al. 2016). Improved understanding of different invasion pathways of a pest assists towards developing a biosecurity plan and subsequent implementation of effective management programs (Brookes et al. 2020). This study uses haplotype and virus analysis to follow invasion pathways and spread of CRB in the Solomon Islands.
In January 2015, the first official record of a CRB incursion in Solomon Islands was reported from the capital, Honiara, Guadalcanal (SIBC 2015), and was identified as CRB-G clade IA (Marshall et al. 2017). After awareness was raised of the presence of CRB in Solomon Islands, CRB was also reported from Shortland Islands (Western Province) but identified as belonging to CRB-S (clade II) (Marshall et al. 2017; SIBC 2018c; Tsatsia et al. 2018). The presence of CRB in the Solomon Islands poses a major threat as the country is highly dependent on coconut and oil palm for economic development and food security in the territory which spreads over 28,400 km2 and comprises over 900 islands, with limited air and sea connections between islands (Bennett 2018). The separation of the islands provides a barrier to spread, but once the pest has spread to new islands, communication and implementation of controls are difficult. Unprecedented damage from CRB has already been documented from Honiara and its surrounds, therefore, failure to control the pest is anticipated to have a significant impact on the country's SBD$140 million coconut industry (~ 12% of GDP) and food security of more than 40,000 rural households (Tsatsia et al. 2018; Vaqalo et al. 2017).
In response to reports of the CRB incursion, Biosecurity Solomon Islands (BSI), a division of the Ministry of Agriculture and Livestock (MAL), established a CRB Task Force and with the assistance of local stakeholders, the Land Resources Division of the Pacific Community (SPC), and AgResearch, New Zealand, started a campaign to contain and control the pest. An awareness campaign was launched and extension officers in the provinces were asked to look for the beetle and send information to Honiara (SIBC 2016b; 2017a). It was important to differentiate CRB attack from that of the native Scapanes australis, which causes similar damage on young palms (Stapley 1973). Based on reports from the extension officers, delimitation surveys as well as exploratory visits were conducted by MAL officers to determine the infestation boundaries within each province and to collect samples for analysis of CRB haplotype and the presence of virus.
In this report we summarize data collected from exploratory and delimitation field surveys for CRB in Solomon Islands to determine distribution of invasive CRB populations and the presence of the biological control agent, OrNV. A more detailed analysis of results from Guadalcanal province for a longer timeframe (2015 to 2020) is also provided. We postulate a timeline for CRB spread within Solomon Islands and produce distribution maps. The results are analysed, and recommendations provided for stemming the spread and limiting damage by the pest based on haplotype and virus occurrence.
Materials and methods
Beetle collection
Beetles were field collected either during exploratory visits, usually to confirm first sighting in a location, or by delimitation survey using pheromone traps or collection from breeding sites. Hygiene was maintained and tools were cleaned between sites. In all cases, live beetles were counted from a site, sexed, and individuals each placed in a clean, labelled container. Site location was recorded, by GPS on-site where possible, or estimated from maps after return to the office. Pheromone traps were cleared frequently. From the surveys, at least 2–3 beetles were taken from each trap sampled and dissected. Gut condition was described, and gut tissue segments placed in preservative for dispatch.
Delimitation surveys
Delimitation surveys were conducted on twenty-eight islands across the nine provinces of Solomon Islands to demarcate the spread and severity of the CRB incursion (Table 1 and Online Resource 1). Survey sites were selected based on local reports of putative CRB damage. Led by BSI, the surveys were organized in coordination with local extension officers and AgResearch, New Zealand. These surveys gathered snapshots of CRB activity from each area visited (with GPS and locality information) and collected beetle samples for subsequent haplotype identification and virus testing (detailed protocol in Online Resource 1). Briefly, at each survey location the team established pheromone traps to monitor adult CRB activity, then conducted visual assessment of palm damage and inspection of potential breeding sites (i.e., primarily dead coconut/other palm trunks and accumulations of organic wastes including chicken manure) to check for CRB larvae. The survey team also met with local extension staff, local communities, and farmers to raise awareness of CRB and to advise on sanitation efforts to reduce damage and control the spread of CRB.
Pheromone traps
For the delimitation surveys, at least three bucket traps baited with aggregation pheromone, oryctalure (ethyl 4-methyloctanoate; ChemTica Internacional, Costa Rica) were established at each reported invasion site (ground zero) and spaced at least 100 m apart. Traps were hung in the tree canopy at approximately 1.5–1.8 m above the ground and were in operation for approximately 2–10 days. Captured CRB were either placed into individual containers (with GPS coordinates, collection dates, etc.) for transport back to the MAL laboratory in Honiara or dissected locally and tissue preserved for transport (see Online Resource 1 for details). In Honiara, a network of 29 bucket traps was established to monitor CRB activity in the area, as part of local control efforts, and checked every 7–14 days (Paudel et al. 2021b). Up to two beetles per trap were collected for dissection in September 2019 and February and June 2020, (Table 1) and tissue preserved for analysis.
CRB detection and breeding sites
From the ground zero point, the survey team travelled along the roads or tracks to search for damaged palms and breeding sites (see Online Resource 1 for detailed methods). Initially, records (photos, GPS, etc.) were taken of what was seen every 1 km (up to 3 km from ground zero). If no damage was detected along the first route, the team searched down another road/track, until all possible routes from ground zero had been searched.
When breeding sites were identified, the date and site details were recorded (with GPS coordinates) and up to 10 CRB adults per site were collected in labelled individual containers. Either whole CRB were transported back to the MAL laboratory in Honiara, or they were dissected locally and then the preserved tissue was sent for further analysis.
Beetle dissection to collect gut tissue
Training in the dissection and preservation of CRB specimens was provided to the BSI staff by Marshall, Jackson, and Mansfield during in-country workshops prior to the surveys (see Online Resource 1 for detailed methods). After dissection, tissue samples were sent to AgResearch, New Zealand to determine CRB haplotype (G or S) and geographic clade (according to Marshall et al. 2017), and presence or absence of OrNV using DNA analysis and histology (as described in Sect. "Determination of haplotype (G or S) and presence/absence of OrNV").
Determination of haplotype (G or S) and presence/absence of OrNV
Haplotype (G or S) and presence/absence of OrNV were determined from CRB tissue using methods described by Marshall et al. (2017). The methods are described briefly below.
Genomic DNA extraction from CRB gut tissue
DNA was extracted from the gut tissue supplied (preserved and shipped in monopropylene glycol) using the Tissue Genomic DNA Mini Kit (Geneaid) column system following manufacturer instructions. DNA elution was carried out using 100 µl of elution buffer and aliquots of eluted DNA samples were subsequently used for further analyses.
PCR–RFLP method for detecting the CRB-G haplotype
The following primer pair was designed and used to amplify a 524 bp fragment of the O. rhinoceros COI gene: C1-J-1718Oryctes (5 ‘- GGAGGTTTCGGAAATTGACTTGTTCC -3‘) and C1-N-2191Oryctes (5’- CCAGGTAGAATTAAAATRTATACCTC -3‘) (Marshall et al. 2017). A unique Tru1I restriction site polymorphism within this amplified region allows the CRB-G haplotype to be identified. Note that Tru1I is an isoschizomer of the MseI restriction enzyme reported in Marshall et al. 2017, and therefore recognizes the same DNA site. Each 20 μL PCR reaction contained: 10 μL VitaTaq® 2X HS Mastermix Gold (Procomcure), 0.4 μL C1-J-1718Oryctes (10 μM), 0.4 μl C1-N-2191Oryctes (10 μM), 1.5 μL 100-fold diluted O. rhinoceros DNA template, and 7.7 μL sterile distilled water. PCR amplifications were performed in an Eppendorf Mastercycler Gradient with a cycling profile of 30 cycles of 94 °C denaturation (30 s), 50 °C annealing (45 s), 72 °C extension (1 min) with an initial denaturation of 3 min at 94 °C and a final extension of 5 min at 72 °C. A 5 μL aliquot of each PCR reaction was checked on a 1% agarose gel. For RFLP analysis, successfully amplified COI PCR products (5 μL) were each combined with 0.25 μL Tru1I (10 U/μL; Thermo Fisher Scientific), 1.5 μL 10 × Buffer R, and 8.25 μL sterile distilled water, and incubated at 65 °C for 1 h. The digested samples (15 μL) still contained enough loading dye from the VitaTaq® 2X HS Mastermix Gold to be directly loaded on a 2% agarose gel.
Gel electrophoresis and UV visualization of DNA
Aliquots of PCR reactions were checked by agarose gel electrophoresis (1%, 0.5X TBE) alongside a 100 bp DNA Ladder (GeneRuler™, Thermo Scientific). All PCR runs included a template-free reaction as a negative control. RedSafe (iNtRON Biotechnology) was included in all agarose gels and allowed DNA fluorescence to be visualized over UV light. Photographs were recorded using an UVIdoc HD2 gel doc (UVItech).
COI sequencing for determining CRB geographic clade
Barcode primers, LCO1490 (GGTCAACAAATCATAAAGATATTGG) and HCO2198 (TAAACTTCAGGGTGACCAAAAAATCA) (Folmer et al. 1994), were used to amplify the 5’ end of the COI gene for a subset of DNA samples. Each 40 μL PCR reaction contained: 20 μL VitaTaq® 2X HS Mastermix Gold (Procomcure), 0.8 μL LCO1490 (10 μM), 0.8 μl HCO2198 (10 μM), 3.0 μL 100-fold diluted O. rhinoceros DNA template, and 15.4 μL sterile distilled water. PCR amplifications were performed in an Eppendorf Mastercycler Gradient with a cycling profile of 40 cycles of 94 °C denaturation (30 s), 45 °C annealing (30 s), 72 °C extension (45 s) with an initial denaturation of 3 min at 94 °C and a final extension of 5 min at 72 °C. A 5 μL aliquot of each PCR reaction was checked on a 1% gel. Sanger sequencing (BigDye®v 3.1, Applied Biosystems) of the amplified COI product was conducted at Macrogen (Korea). Sequences were tidied and analysed using Geneious version Prime (http://www.geneious.com) (Kearse et al. 2012). Multiple alignments of the CRB sequences were generated using the program MUSCLE (Edgar 2004) in Geneious version Prime. Nucleotide-based analyses were calculated using distance-based (neighbor joining) methods and allowed visualization of the CRB geographic clades based on the reference sequences reported in Marshall et al. (2017).
PCR detection of OrNV infected CRB beetles
DNA was extracted from the gut tissue supplied in monopropylene glycol as described above. The PCR protocol for detection of OrNV was based on a previous study (Richards et al. 1999) and has been modified subsequently by using diluted DNA template (down to 1 in 5000) to better distinguish active infection from mere presence due to cross-contamination between infected and uninfected beetles in the pheromone traps. The primer pairs used to amplify a 945 base pair (bp) fragment of the OrNV genome were OrNV15a (5‘-ATTACGTCGTAGAGGCAATC-3‘) and OrNV15b (5’-ATGATCGATTCGTCTATGG-3 ‘) (Richards et al. 1999). Each 20 μL PCR reaction contained: 10 μL VitaTaq® 2X HS Mastermix Gold (Procomcure), 0.4 μL OrNV15a (10 μM), 0.4 μL OrNV15b (10 μM), 1.5 μL of the 100-fold or 5,000-fold diluted DNA and 7.7 μL sterile distilled water. PCR amplifications were performed in an Eppendorf Mastercycler Gradient with a cycling profile of 30 cycles of 94 °C denaturation (30 s), 50 °C annealing (45 s), 72 °C extension (1 min) with an initial denaturation of 3 min at 94 °C and a final extension of 5 min at 72 °C. A 10 μL aliquot of each PCR reaction was checked on a 1% agarose gel. Detection of OrNV PCR product in the 1 in 5,000 dilution was considered here as indicative of OrNV infection.
Histology
Gut tissue samples were preserved in FAA fixative (5% formaldehyde, 2.5% acetic acid, 50% ethanol as an aqueous solution) and sent to Gribbles Veterinary, an IANZ (International Accreditation New Zealand) accredited pathology lab, for paraffin embedding, serial sectioning, and hematoxylin and eosin (H & E) staining (Kiernan 1990). Slides of gut tissue were examined under bright-field and differential interference contrast (DIC) optics with observations of OrNV infection status recorded based on pathology (Huger 2005). A subset of beetles was subjected to histological examination due to limited availability of the FAA fixative (Online Resource 2).
Data analysis
Records of CRB-G and CRB-S haplotypes, clade affiliation and OrNV infection from the beetle samples were summarized using descriptive statistics (frequencies and percentages). Maps of CRB and virus distribution were produced using ArcGIS software (ArcGISPro 2021).
Results
Overview of CRB distribution within Solomon Islands
Based on the field surveys and sample analyses, CRB is confirmed on twenty-eight islands, from the Shortland Islands in the northwestern region bordering Papua New Guinea’s Autonomous Region of Bougainville to Temotu, the easternmost province of Solomon Islands (Online Resource 2 and 3). Using RFLP, two haplotypes were detected, CRB-G and CRB-S. In total, 396 beetles were analysed from multiple islands from > 100 independent sample collections. Sequencing of the COI gene from all 396 samples showed that all CRB-G (257) were identical to the Solomon Islands sample described in clade IA of Marshall et al. (2017). The majority of CRB-S (131/139) were identical to the PNG samples categorized in clade II from Marshall et al. (2017); the exception being a new single nucleotide polymorphism (SNP) variant found in seven Choiseul samples and one from Shortland Islands (Western Province) (Online Resource 2). Representative COI sequences have been deposited into GenBank as the following accessions: OL546395-OL546457.
Since first reported in 2015, CRB has spread through the Solomon Islands. CRB-G is now present in eight out of nine provinces, whereas CRB-S is present in seven provinces, with both haplotypes present in six provinces (Table 2). Closer examination, however, shows that haplotypes are mostly present in distinct populations (Fig. 2 and 3). All CRB samples from Makira-Ulawa and Rennell and Bellona Provinces were determined as CRB-G whereas all the specimens from Choiseul Province were CRB-S.
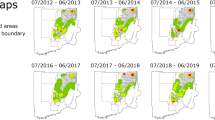
Timeline of Oryctes rhinoceros (CRB-G haplotype) detection across Solomon Islands with confirmed year of detection (in different colors) and presence or absence of Oryctes nudivirus (OrNV) (+ : present, o: absent) for each island. Each point represents the site of sample collection plotted using GPS coordinates. The inset at the top right shows the location of submaps A-D within a complete Solomon Islands map
Timeline of Oryctes rhinoceros (CRB-S haplotype) detection across Solomon Islands with confirmed year of detection (in different colors) and presence or absence of Oryctes nudivirus (OrNV) (+ : present, o: absent) for each island. Each point represents the site of sample collection plotted using GPS coordinates. The inset at the top right shows the location of submaps A-D within a complete Solomon Islands map
In Western Province, CRB-S was found on the islands of Shortland Islands and Ghizo while CRB-G was found on New Georgia. In Temotu Province, CRB-S was found from one location on Santa Cruz Island; CRB-G was found from two locations on Vanikoro Island. In Malaita province, CRB-G and CRB-S were present on the main island of Malaita but in different outbreak areas; whereas only CRB-S were found on Maramasike Island and CRB-G on Ontong Java Atoll (Fig. 2 and 3, Online Resource 3). The geographic distributions of the two haplotypes overlap in Guadalcanal and Central Provinces, although there are more site records for CRB-G than CRB-S (Fig. 2–5). CRB-G were confirmed for 85% and 67% of the samples from Guadalcanal and Central Province, respectively (Table 2).
Timeline of CRB-G invasion in Solomon Islands
CRB-G was first detected in Honiara (Guadalcanal Province), early in 2015 (Fig. 2, Table 3, Online Resource 2). At that time, it was centered in Honiara but already spreading in plantations east of the town towards the oil palm plantations 20 km away (personal communication, Bob Macfarlane). By June 2017, CRB damage was evident along 50 km of the northern Guadalcanal coastline, extending in both directions from the original site of infestation in Honiara (SIBC 2016a; Vaqalo et al. 2017). The more recent data from this province is presented in Sect. "CRB spread on Guadalcanal".
In 2017, MAL extension officers reported that CRB was present on northern Malaita Island (Malaita Province) (SIBC 2017b), although no samples were available to confirm haplotype at that time (Table 3, Online Resource 3).
In 2018, CRB invasion was detected in three further provinces (Fig. 2, Table 3). Two adult CRB-G were captured from a single trap on Ulawa Island (Makira-Ulawa Province), out of 20 traps established during the delimitation survey. Palm damage characteristic of CRB was limited to an area of approximately one hectare on this island, suggesting the infestation was comparatively recent (Online Resource 3). Two adult CRB-G were also collected from Rennell Island (Rennell and Bellona Province). Adult CRB (n = 72) were captured in pheromone traps from three sites on Nggela Island (Central Province), although no samples were collected to confirm the haplotype during this survey. CRB damage was also recorded from three small islands of the Russell Islands group in this province (Online Resource 3).
Presence of CRB-G in Central Province was confirmed in 2019 from several sites in the Russell Islands (Fig. 2, Table 3). The discovery of CRB-G in 2019 (n = 10 collected in June, n = 4 collected in November) near the airport at Munda, New Georgia Island (Western Province), marked a significant expansion of its geographic range in Solomon Islands (Fig. 2, Table 3). The delimitation surveys from Rennell Island (Rennell and Bellona Province), conducted in mid-2019, found CRB damage at 21 out of 35 sites visited (Online Resource 3), with adult CRB-G collected from one site (n = 6). The delimitation survey of Malaita and Maramasike Islands (Malaita Province) conducted from late October to early November 2019, confirmed that CRB-G (n = 26) was established at multiple sites in the north and west of Malaita Island (Fig. 2, Table 3). During the survey, officers noted that CRB was often present near forestry operations on Malaita (Online Resource 3). Finally, the first evidence of CRB presence was recorded in November 2019 from Vanikoro Island, in remote Temotu Province, although samples were not collected for confirmation. CRB was not detected on nearby Utupua Island during this delimitation survey.
Early in 2020, a delimitation survey confirmed that CRB-G had spread to West Guadalcanal at two (Tiaro and Tangarare) out of eight sites surveyed (Fig. 2, Table 3), with probable CRB damage noted at a third location, Duidui (Online Resource 3). Several follow-up visits were made during 2020 to provinces where CRB-G was known to be present. The first such visit in January to Ulawa Island (Makira-Ulawa Province), captured three adult CRB-G and found CRB damage over a larger area compared with the 2018 survey (Online Resource 2). More CRB-G (n = 3) were collected in July 2020 from New Georgia (Western Province). CRB-G (n = 8) had expanded its range to five locations on Bellona Island (Rennell and Bellona Province) by early August 2020, (Fig. 2, Table 3). On Nggela Island (Central Province), CRB-G (n = 24) was present at 11 locations altogether by mid-October 2020, including two sites originally surveyed in 2018 (Fig. 2).
The first record of CRB-G (n = 1) from the southeast of Santa Isabel Island (Isabel Province), was confirmed in August 2020. In mid-October, presence of CRB-G was confirmed at two locations on Vanikoro Island (Temotu Province) (Fig. 2, Table 3). Finally in November 2020, a delimitation survey confirmed the presence of CRB-G (n = 4) on the remote Ontong Java Atoll (also known as Outer Malaita Islands; Malaita Province). CRB damage was first observed in October that year (Fig. 2, Table 3, Online Resource 2). CRB damage was found in the southeast region of the Atoll, around Luaniua, whereas no evidence of CRB damage was found to the northwest. The survey officer noted that coconut planting materials were imported to this region from Guadalcanal earlier in 2020 and were the presumed source of the CRB infestation (Online Resource 3).
Timeline of CRB-S invasion in Solomon Islands
CRB was first reported from the Shortland Islands (Western Province) in 2015. Samples collected from Fauro and Alu islands (n = 6) in 2016 were identified as CRB-S; one adult CRB-S was collected from Ghizo in the New Georgia group (Fig. 3, Table 4, Online Resource 2). During 2016, live CRB-S beetles were imported from the Shortland Islands to the MAL laboratory in Honiara (Guadalcanal), for research purposes as part of the CRB response (Marshall et al. 2016).
As noted above in Sect. "Timeline of CRB-G invasion in Solomon Islands", the presence of CRB was reported in 2017 on northern Malaita Island (Malaita Province) (SIBC 2017b), but the haplotype was not known at that time.
The delimitation survey of Nggela Island (Central Province) also reported CRB presence and associated damage in 2018, but the haplotype was not determined (see Sect. "Timeline of CRB-G invasion in Solomon Islands" above). During 2018, follow-up visits to the Shortland Islands (Western Province), collected CRB-S (n = 43) from 10 sites altogether, including both islands visited previously during 2016.
More CRB-S were collected from Ghizo (Western Province; n = 5 in June and n = 2 in November). The spread of CRB-S was confirmed during 2019. In July, the delimitation survey recorded CRB damage and collected one adult CRB-S from one out of 26 sites on Santa Isabel Island (Isabel Province; Fig. 3, Table 4, Online Resource 2). CRB-S (n = 9) were collected from several sites in the Russell Islands (Central Province; Fig. 3, Table 4) during October 2019. The delimitation survey conducted from late October to early November 2019 collected CRB-S (n = 21) from sites on Malaita and Maramasike Islands (Malaita Province; Fig. 3, Table 4). In Guadalcanal Province, CRB-S was confirmed from the Honiara trap network and the oil palm plantations at GPPOL in September 2019 (see Sect. "CRB spread on Guadalcanal").
Early in 2020, the delimitation survey of West Guadalcanal collected one CRB-S from Tangarare out of eight sites surveyed (Fig. 3, Table 4). In September 2020, the first CRB-S (n = 7) were collected from the north-western tip of Choiseul Island (Choiseul Province), during the delimitation survey (Fig. 3, Table 4). The survey officer noted that CRB damage was more severe near logging sites in the survey area (Online Resource 3). The first record of CRB-S (n = 4) in Temotu province was from Santa Cruz Island in October 2020 (Fig. 3, Table 4). Also in October, CRB-S (n = 4) was found at four sites on Nggela Island (Central Province), including three originally surveyed in 2018 where damage was observed, and beetles collected without haplotype determination (Fig. 3, Online Resource 3).
CRB spread on Guadalcanal
From the center of Honiara in Guadalcanal Province, CRB spread first to the massive oil palm plantations of GPPOL, then as far as Aola in the east; to Lambi, Tiaro and Tangarare in the west and up to Duidui in the south (Figs. 4 and 5, see Online Resource 3 for detailed survey results). The initial delimitation survey carried out by MAL in Jan 2015 showed that heavy damage was limited to an area of 3 × 2 km in the suburbs of Panatina and Vura with light damage surrounding this area and reports of fresh damage as far as Tenaru (10 km to the east) and with suspected CRB damage as far as Visale in the west (Online Resource 4). In 2016, minor CRB damage at the Ngalimbiu oil palm plantation on the Guadalcanal plains were observed, but by 2017 damage was severe with high numbers of beetles reported throughout the GPPOL plantations (SIBC 2016a; 2017c; SPC 2017). At the same time a survey showed that CRB was widespread and highly damaging throughout Honiara and had spread along the northwest coast (Vaqalo et al. 2017). By 2018, damage was very heavy in the central zone from Honiara to the GPPOL plantations and evidence of CRB was obvious along the northwest coast of Guadalcanal from Tetere, GPPOL, to Verenasso, more than 80 km (Online Resource 5). Intense damage has continued with many dead palms in the core outbreak area, despite clean up and control attempts. Beetle populations now extend at least from Aola in the east, to Lamuas on the west coast of Guadalcanal with some reports of damage from the southern (Weather) coast (Figs. 4 and 5).
Timeline of Oryctes rhinoceros (CRB-G haplotype) detection from field surveys across Guadalcanal Province. The central map covers the whole province with a boxed summary of results from Honiara and surrounds expanded in the lower map. The maps show confirmed year of detection (in different colors) and presence or absence of Oryctes nudivirus (OrNV) (+ : present, o: absent) for each site. Each point represents the site of sample collection plotted using GPS coordinates whereas the size of the dot or cross is proportionate to the number of samples collected or the number of OrNV positive CRB from that site
Timeline of Oryctes rhinoceros (CRB-S haplotype) detection from field surveys across Guadalcanal Province. The central map covers the whole province with a boxed summary of results from Honiara and surrounds expanded in the lower map. The maps show confirmed year of detection (in different colors) and presence or absence of Oryctes nudivirus (OrNV) (+ : present, o: absent) for each site. Each point represents the site of sample collection plotted using GPS coordinates whereas the size of the dot or cross is proportionate to the number of samples collected or the number of OrNV positive CRB from that site
Throughout the study (2015–2020), 182 CRB samples were collected from exploratory visits and delimitation surveys. CRB-G was dominant throughout the collection period (2015 to 2020) with 85% of the specimens (155 insects) determined as CRB-G vs. 15% (27 samples) as CRB-S (Table 2). Note that only CRB-G samples were collected from Guadalcanal field surveys from 2015 to 2018 (Online Resource 2 but see haplotype detections from bioassays reported below). When compared across different regions within the Guadalcanal province, CRB-G remains the dominant haplotype (> 80% of samples) from the Honiara trap network, the GPPOL plantations and the West Guadalcanal delimitation survey (Online Resource 6). The most recent collection from the Honiara trap network was taken from late June to early July 2020. Again, CRB-G was dominant with 90% of the samples whereas 10% were CRB-S. When compared geographically, CRB-G is more widespread across Guadalcanal Province than CRB-S (Figs. 4 and 5). CRB-G has been collected from 35 sites altogether whereas CRB-S was collected from only 16 sites.
In response to the CRB invasion, bioassays were initiated in Honiara for laboratory screening of OrNV isolates against CRB (SIBC 2018a). In the first year (2016) locally collected CRB-G and CRB-S imported from the Shortland Islands were used for bioassays, but from 2017 onwards, only locally collected CRB from Guadalcanal were used for bioassays (Online Resource 7). As part of these bioassays, all beetles were tested to determine haplotype. All beetles used for bioassays in 2017 were CRB-G. The first CRB-S haplotype originating from a local site collection and used in these bioassays was detected as a single insect in April 2018 with a further nine CRB-S collected between August and October 2018. From January to July 2019, CRB-S was found at low levels (11%) among 186 CRB used for bioassays from three collections in the surrounds of Honiara (Online Resource 7). CRB-S were not collected from the Honiara pheromone trap network until September 2019.
Overview and timeline of OrNV prevalence in field collected CRB samples
Although OrNV was imported for use in laboratory bioassays from 2016 onwards (SIBC 2018a), OrNV was not detected from field collected CRB samples until 2019. In June 2019, OrNV was detected in CRB-G samples (6 out of 10; Online Resource 2) collected from the GPPOL plantations (Guadalcanal province). In September 2019, CRB-G and CRB-S samples from the Honiara trap network (Guadalcanal province) tested positive for OrNV (Fig. 4 and 5, Online Resource 2). Later in 2019, OrNV was also detected in CRB-S beetles collected from GPPOL (Online Resource 2). During the same year, OrNV was detected in two CRB-G beetles collected from Malaita Island (Malaita Province) (Fig. 2, Online Resource 2). OrNV was not detected in CRB samples (n = 6 CRB-G and 1 CRB-S) collected from West Guadalcanal (Guadalcanal province) in January 2020 (Figs. 4 and 5, Online Resource 2). In Guadalcanal province, 32.3.% (50 from 155) and 48.1.0% (13 from 25) of field collected CRB-G and CRB-S were OrNV positive, respectively. Of the 77 gut samples that were analysed for virus symptoms using histology, 71 of these samples produced OrNV PCR results that matched the histology results (Online Resource 2). Five samples were negative by OrNV PCR but showed virus symptoms by histology; these samples were positive for virus when the 1:100 dilution of extracted DNA was used. One sample was weakly positive for OrNV using PCR but no virus symptoms were observed for the histology slide. OrNV has not been detected from field collected CRB samples from Central, Makira-Ullawa, Rennell and Bellona, Western, Isabel or Temotu Provinces (Fig. 2, Online Resource 2).
Discussion
We identified two invasions of CRB into Solomon Islands, one starting in Honiara, Guadalcanal, and the other in the Shortland Islands, Western Province. The invasive CRB populations belong to two haplotypes, CRB-G and CRB-S. COI sequencing showed that these invasive populations align with clade IA (CRB-G) and clade II (CRB-S), using reference samples described by Marshall et al. 2017. Clade IA (CRB-G) is the haplotype that has invaded Guam, Hawaii and PNG mainland. CRB-G was first confirmed from Honiara in 2015 and has since spread widely in Guadalcanal, Malaita and Central provinces. CRB-S was first confirmed from Shortland Islands (Western Province) and is dominant in the Northwestern region of the country (e.g., Shortland Islands, Ghizo, Choiseul). The two haplotypes appear to have spread following patterns of air and sea movement between the point of origin (CRB-G from Honiara and CRB-S from the Shortland Islands) and other islands/provinces. Haplotype analysis conforms with local observations of the pattern of spread within Solomon Islands.
CRB-G
In 2015, the distribution of CRB-G haplotype was limited to a relatively small area in Honiara. But it has now invaded across much of the productive coast of Guadalcanal and rapidly established on other surrounding islands (Malaita, Ngella) resulting in high levels of damage. This mirrors CRB-G invasion events elsewhere in the Pacific Island countries and territories (e.g., Guam, Hawaii, Papua New Guinea) (Ero et al. 2016; Marshall et al. 2017; Reil et al. 2018). Sequencing of the COI gene revealed that CRB-G specimens sampled from across Solomon Islands were identical and corresponded to the invasive haplotype (clade IA) as described previously (Marshall et al. 2017). While first reported in 2015, CRB-G is believed to have entered Solomon Islands two or more generations earlier, possibly during the ‘11th Festival of Pacific Arts’ in 2012, which was held in Honiara, Solomon Islands (personal communication, Bob Macfarlane). The early stages of the outbreak could have been missed as damage similar to CRB is also caused by a native scarab beetle, Scapanes australis, although this usually attacks younger palms (Bedford 1980; Stapley 1973).
We suggest that CRB-G spread gradually from the initial invasion site along the coconut lined northwest coast of Guadalcanal by natural spread, and perhaps assisted by human transport of goods and machinery by road or along the coast by small boats. The beetle was first reported from Panatina, 5 km from central Honiara, in 2015 (Tsatsia et al. 2018; Vaqalo et al. 2017). By June 2017, CRB extended along a 100 km front along the northern Guadalcanal coastline (Vaqalo et al. 2017). Considering the extensive CRB damage observed on the Okea block of GPPOL during Jan 2018, it is likely that the pest had arrived more than one year previously, perhaps sometime in 2016 (SPC 2017). CRB-G was reported from west Guadalcanal (Tiaro and Tangarare) in February 2020 and is likely to have been assisted by small boat travel to these remote villages.
Inadvertent transport on small boats probably led to the outbreaks on Malaita, Russell Islands and Ngella (SIBC 2018b). There are regular crossings from Honiara and boats often carry materials, such as chicken manure and plant materials, which are attractive to the beetles. Distances are greater to outer provinces such as Temotu, and the remote Ontong Java Atoll, so it is likely that larger ships or even aircraft were unwittingly involved in transportation. CRB adults are large, long-lived beetles, attracted to light, fruits and decaying organic matter (Bedford 1980). Rapid spread on a contiguous landmass is not surprising as CRB movement of 1–3 km per month has been reported (Gorick 1980; Jacob 1996; Young 1974). Flight across short sections of ocean is possible but for widely separated islands human mediated transport is more likely.
CRB-S
The invasion front from CRB-S haplotype appears to have started in the Shortland Islands, most likely spreading from Papua New Guinea (PNG). CRB has been present in the outer island provinces of PNG since 1940s (Bedford 1976) and in the Autonomous Region of Bougainville since 1980s (Moxon & Hela 1989). Subsequently, on Bougainville Islands we found CRB-S (clade II) and a new variant of CRB-S (clade II) with a single SNP difference (GenBank accessions: OL546395-OL546457). Invasion of CRB-S into Solomon Islands is not entirely surprising as the Shortland Islands are adjacent to the Bougainville Islands.
COI sequences for 139 CRB-S samples from across Solomon Islands corresponded to the clade II haplotype with the majority (131/139) being identical to the PNG reference sequences found in New Britain, New Ireland, and New Guinea Island (Marshall et al. 2017). The remaining eight CRB matched to the new CRB-S clade II variant. It is likely that the initial invasive CRB population in Western Province was present before the start of this study.
On Guadalcanal, one CRB-S beetle was found among field collected beetles that were used for bioassays in 2018 with subsequent detections of CRB-S from the Honiara pheromone traps later in 2019. This agrees with Etebari et al. 2021 who reported CRB-S from Guadalcanal in 2019. CRB-S were likely imported through commercial shipping services to Honiara and then subsequently spread to Malaita and Isabel provinces, although accidental escape cannot be ruled out because live CRB-S beetles were imported from Western Province for research purposes in 2016. The establishment of CRB-S in Honiara has provided the opportunity for interbreeding between haplotypes and development of mixed populations.
Haplotypes and interbreeding
In Solomon Islands, we have only found two haplotypes from 396 samples (257 clade IA and 139 clade II) except for the single minor variant within clade II for 8 specimens. This indicates that there have only been two invasive episodes which is consistent with local knowledge of origins and the pattern of spread. Surprisingly, Etebari et al. (2021) reported two CRB clade III samples collected from Guadalcanal (Genbank MN809505 SOL GC hap 4) with sequences identical to the Marshall et al. (2017) reference sequences found in Samoa, Fiji, India and Diego Garcia. Despite extensive sampling on Guadalcanal, no individuals with haplotypes corresponding to CRB clade III were found in this study. Neither did we find specimens of the 3 additional novel haplotypes of CRB from Solomon Islands proposed by Etebari et al. (2021). The majority of Solomon Islands samples from the Etebari et al. (2021) study matched the reference haplotypes CRB-G clade IA (59/61) and CRB-S clade II (14/20) from Marshall et al. (2017).
The arrival of CRB-S on Guadalcanal and subsequent spread in the central Solomon Islands (Guadalcanal, Ngella, Russell Islands, Malaita, and Isabel) has led to mixed populations (CRB-G/CRB-S). We cannot determine if these haplotypes arrived simultaneously or independently on these islands, but mixed populations raise the possibility of interbreeding. However, as we have shown, CRB exists mostly in monophyletic groups in discrete populations, either CRB-G or CRB-S, through Solomon Islands, with no opportunity for interbreeding. In mixed populations, offspring may carry nuclear traits from either parent. Currently the mixed populations recorded are dominated by the CRB-G haplotype marker. Despite its maternal mode of inheritance, mitochondrial markers (e.g., COI) are effective in detecting invasions (Rubinoff & Holland 2005). The mitochondrial based CRB haplotype marker was developed by Marshall et al. (2017) because it showed strong correlation to resistance to the historic OrNV strain used for biological control of CRB. The nuclear genes responsible for virus resistance are not yet known.
OrNV
All field collected CRB specimens from Solomon Islands were OrNV-free until the virus was first isolated from CRB-G in June 2019 and CRB-S in September 2019 in the vicinity of Honiara (Guadalcanal). In the subsequent surveys, CRB-G samples from Malaita in 2019 were also positive for the virus, indicating that the virus had spread between islands. Notably, all CRB-S and CRB-G samples from seven provinces including Central, Choiseul, Isabel, Makira-Ulawa, Rennell-Bellona, Temotu and Western provinces were OrNV free.
OrNV was imported for research purposes into Guadalcanal from 2016 onwards (SIBC 2018a). The virus circulating in the Honiara population was sequenced by Etebari et al. (2020) but was not compared with virus strains tested against CRB-G during bioassay screening (Marshall et al. 2016) so escape of OrNV cannot be ruled out. While virus has been detected in CRB-G specimens, this has occurred only where there are mixed haplotype populations. Furthermore, the extent of virus infection in the mixed populations is lower than would be expected in pure susceptible populations. Both factors suggest interbreeding of CRB-G and CRB-S on Solomon Islands and require further study. In this study, OrNV positive CRB-G were collected from Guadalcanal and Malaita. Previously all CRB-G collected from outbreak populations in Guam, PNG and the initial samples from Honiara, have been OrNV free. It is possible that the OrNV positive CRB-G from this study were offspring of maternal CRB-G and paternal CRB-S as they were only found in mixed populations. No virus was found in solely (monophyletic) CRB-G (clade IA) populations. High virus infection rates (more than 80%) reported by Etebari et al. (2021) may be due to virus transmission between beetles during the trapping process (Moslim et al. 2011) and are inconsistent with this study, which is based on more than 100 independent beetle collections.
Pest management implications
Two CRB haplotypes continue to spread within Solomon Islands from two centres of origin: CRB-G (clade IA) from Honiara and CRB-S (clade II) from Shortland Islands. Both invasive CRB populations are largely virus-free and highly damaging to palms. The extent and the intensity of CRB incursions, however, vary based on provinces and between locations within each province. By 2020, outbreak areas ranged from the extensive, highly damaging populations on Guadalcanal, Malaita and Nggella to the small outbreak areas on Isabel, Choiseul and Makira-Ulawa with low trap catch numbers and damage limited to a few palms. It is likely that the latter sites represent new outbreak areas with the potential for population growth. CRB is thought to have established in Honiara at least 2–3 years before January 2015, allowing time for large populations to develop. Response plans, therefore, should be tailored to reflect the outbreak levels as well as the presence of CRB-G or CRB-S. To manage small, isolated outbreaks, it is important to locate and destroy CRB breeding sites through surveillance and sanitation activities (including destroying organic waste and dead palm trunks). Management of CRB-G will be difficult without introduction of a self-replicating biological control agent, therefore, local government and international partners should prioritize research on testing native and exotic entomopathogens against CRB-G (Paudel et al. 2021a). Until a viable biological control agent is found, it is important to contain the beetle and stop further spread through surveillance, sanitation, and enforcing strict local quarantines. Efforts should be intensified to prevent transport of the beetle to currently uninfested islands with an awareness campaign to warn locals and stakeholders in potential points of entry and to ensure quick detection of new CRB incursions. Successful control of CRB-S in the past has been achieved using OrNV (Huger 2005; Jackson 2009). Therefore, in areas where only CRB-S is present (e.g., northwestern region), OrNV strains used historically against CRB-S should be tested and released to reduce CRB spread and damage. For CRB-G and mixed (CRB-S/CRB-G) populations a range of strains of OrNV should be tested. The presence of an OrNV strain circulating in the mixed Honiara and Malaita populations suggests some susceptibility to the virus, but to date transmission and spread have been slow. The challenge remains to find a more effective strain of OrNV, especially against the invasive CRB populations in Solomon Islands, and incorporate it into an integrated pest management (IPM) programme for control of the pest.
References
Abdallah Z, Mezghani-Khemakhem M, Bouktila D, Makni H, Makni M (2013) Genetic variation and invasion pattern of the Arabian rhinoceros beetle, Oryctes agamemnon arabicus (Burmeister)(Coleoptera: Scarabaeidae), in Tunisia, deduced from mitochondrial DNA sequences. Afr Entomol 21:362–367. https://doi.org/10.4001/003.021.0201
ArcGISPro (2021), Version 2.7.4, Accessed March 06, 2021 https://www.esri.com/en-us/arcgis/products/arcgis-pro/overview
Bedford GO (1976) Observations of the biology and ecology of Orycetes rhinoceros and Scapanes australis (Coleoptera: Scarabaeidae: Dynastinae): pests of coconut palms in Melanesia. J Aust Entomol Soc 15:241–251
Bedford GO (1980) Biology, ecology, and control of palm rhinoceros beetles. Annu Rev Entomol 25:309–339
Bedford GO (2013) Biology and management of palm dynastid beetles: recent advances. Annu Rev Entomol 58:353–372
Bennett JA (2018) Pacific coconut: comestible, comfort and commodity. J Pac Hist 53:353–374. https://doi.org/10.1080/00223344.2018.1523705
Brookes DR, Hereward JP, Wilson LJ, Walter GH (2020) Multiple invasions of a generalist herbivore—Secondary contact between two divergent lineages of Nezara viridula Linnaeus in Australia. Evol Appl 13:2113–2129. https://doi.org/10.1111/eva.12971
Catley A (1969) The coconut rhinoceros beetle Oryctes rhinoceros (L)[Coleoptera: Scarabaeidae: Dynastinae]. PANS Pest Articles & News Summaries 15: 18-30
Doorenweerd C, San Jose M, Barr N, Leblanc L, Rubinoff D (2020) Highly variable COI haplotype diversity between three species of invasive pest fruit fly reflects remarkably incongruent demographic histories. Sci Rep 10:1–10. https://doi.org/10.1038/s41598-020-63973-x
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Ero MM, Sar S, Kawi A, Tenakanai D, Gende P, Bonneau LJG (2016) Detection of the Guam biotype (CRB-G) Oryctes rhinoceros Linneaus (Coleoptera: Scarabaeidae) in Port Moresby, Papua New Guinea. Planter 92:883–891
Etebari K, Parry R, Beltran MJB, Furlong MJ (2020) Transcription profile and genomic variations of Oryctes rhinoceros nudivirus in coconut rhinoceros beetles. J Virol 94(22):e01097–20. https://doi.org/10.1128/JVI.01097-20
Etebari, K., Hereward, J., Sailo, A., Ahoafi, E. M., Tautua, R., Tsatsia, H., ... & Furlong, M. J. (2021). Examination of population genetics of the Coconut Rhinoceros Beetle (Oryctes rhinoceros) and the incidence of its biocontrol agent (Oryctes rhinoceros nudivirus) in the South Pacific Islands. Current Research in Insect Science, 1, 100015. https://doi.org/10.1016/j.cris.2021.100015
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotech 3:294–299
Gariepy TD, Musolin DL, Konjević A, Karpun NN, Zakharchenko VY, Zhuravleva EN, Tavella L, Bruin A, Haye T (2021) Diversity and distribution of cytochrome oxidase I (COI) haplotypes of the brown marmorated stink bug, Halyomorpha halys Stål (Hemiptera, Pentatomidae), along the eastern front of its invasive range in Eurasia. Neobiota. https://doi.org/10.3897/neobiota.68.68915
Gorick BD (1980) Release and establishment of the baculovirus disease of Oryctes rhinoceros (L.) (Coleoptera: Scarabaeidae) in Papua New Guinea. Bull Entomol Res 70:445–453
Huger AM (2005) The Oryctes virus: its detection, identification, and implementation in biological control of the coconut palm rhinoceros beetle, Oryctes rhinoceros (Coleoptera: Scarabaeidae). J Invertebr Pathol 89:78–84. https://doi.org/10.1016/j.jip.2005.02.010
Jackson TA (2009) The use of Oryctes virus for control of rhinoceros beetle in the Pacific Islands. In: Hajek AE, Glare TR, Oallaghan M (eds) Use of microbes for control and eradication of invasive arthropods. Springer, Netherlands, pp 133–140
Jacob TK (1996) Introduction and establishment of baculovirus for the control of rhinoceros beetle Oryctes rhinoceros (Coleoptera: Scarabaeidae) in the Andaman Islands (India). Bull Entomol Res 86:257–262
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Kiernan J (1990) Methods for connective tissue. Histological and histochemical methods, pp: 119–124
Machado DdN, Costa EC, Guedes JV, Barbosa LR, Martínez G, Mayorga SI, Ramos SO, Branco M, Garcia A, Vanegas-Rico JM (2020) One maternal lineage leads the expansion of Thaumastocoris peregrinus (Hemiptera: Thaumastocoridae) in the new and old worlds. Sci Rep 10:1–12. https://doi.org/10.1038/s41598-020-60236-7
Marshall SDG, Moore A, Vaqalo M, Noble A, Jackson TA (2017) A new haplotype of the coconut rhinoceros beetle, Oryctes rhinoceros, has escaped biological control by Oryctes rhinoceros nudivirus and is invading Pacific Islands. J Invertebr Pathol 149:127–134. https://doi.org/10.1016/j.jip.2017.07.006
Marshall SDG, Moore A, Vaqalo M, Noble A & Jackson TA (2016) Use and production of Oryctes nuduvirus to assist with control of the coconut rhinoceros beetle, Oryctes rhinoceros, in Solomon Islands. Client Report - 5219
Moore A (2011) Containing the rhinoceros beetle outbreak on Guam: phytopathology (ed. American Phytopathological Society, St Paul, MN, USA, pp. Retrieved from: https://aubreymoore.github.io/CRB-Guam-Past-Present-Future/
Moore A (2018) Failed attempts to establish IPM for Asian cycad scale and coconut rhinoceros beetle on Guam: annual meeting of the entomological society of America (ed., Vancouver, Canada, p. Retrieved from: https://zenodo.org/record/2545065#.YUEq2545069rgzZik
Moslim R, Kamarudin N, Ghani IA, Wahid MB, Jackson TA, Tey CC, Ahdly AM (2011) Molecular approaches in the assessment of Oryctes rhinoceros virus for the control of rhinoceros beetle in oil palm plantations. J Palm Oil Res 23:1096–1109
Moxon JE & Hela FE (1989) A new report in North Solomons province of Oryctes rhinoceros: Department of agriculture and livestock, agrictural research division, 1989 annual research report. (ed. Department of agriculture and livestock, agrictural research division, Konedobu, Papua New Guinea
Paudel S, Mansfield S, Villamizar LF, Jackson TA, Marshall SDG (2021a) Can biological control overcome the threat from newly invasive coconut rhinoceros beetle populations (Coleoptera: Scarabaeidae)? a review. Ann Entomol Soc Am 114:247–256
Paudel S, Marshall SDG, Tsatsia F, Fanai C, Kolubalona M, Mansfield S, Jackson TA (2021b) Monitoring an invasive coconut rhinoceros beetle population using pheromone traps in Honiara, Solomon Islands. N Z Plant Prot 74:37–41
Paudel S, Marshall SD, Richards NK, Hazelman G, Tanielu P, Jackson TA (2022) Coconut rhinoceros beetle in Samoa: review of a century-old invasion and prospects for control in a changing future. InSects. https://doi.org/10.3390/insects13050487
Reil JB, San Jose M, Rubinoff D (2016) Low variation in nuclear and mitochondrial DNA inhibits resolution of invasion pathways across the Pacific for the coconut rhinoceros beetle (Scarabeidae: Oryctes rhinoceros). Proc Hawaiian Entomol Soc 48:57–69
Reil JB, Doorenweerd C, San Jose M, Sim SB, Geib SM, Rubinoff D (2018) Transpacific coalescent pathways of coconut rhinoceros beetle biotypes: resistance to biological control catalyses resurgence of an old pest. Mol Ecol 27:4459–4474. https://doi.org/10.1111/mec.14879
Richards NK, Glare TR, Aloali’i I, Jackson TA (1999) Primers for the detection of Oryctes virus from Scarabaeidae (Coleoptera). Mol Ecol 8:1551–1561. https://doi.org/10.1046/j.1365-294X.1999.07072.x
Rollins LA, Woolnough AP, Sinclair R, Mooney NJ, Sherwin WB (2011) Mitochondrial DNA offers unique insights into invasion history of the common starling. Mol Ecol 20:2307–2317. https://doi.org/10.1111/j.1365-294X.2011.05101.x
Rubinoff D, Holland BS (2005) Between two extremes: mitochondrial DNA is neither the panacea nor the nemesis of phylogenetic and taxonomic inference. Syst Biol 54:952–961. https://doi.org/10.1080/10635150500234674
Schlum KA, Lamour K, De Bortoli CP, Banerjee R, Meagher R, Pereira E, Murua MG, Sword GA, Tessnow AE, Dillon DV (2021) Whole genome comparisons reveal panmixia among fall armyworm (Spodoptera frugiperda) from diverse locations. BMC Genom 22:1–12. https://doi.org/10.1186/s12864-021-07492-7
SIBC (2015) World’s most dangerous coconut beetle now in Solomons: Solomon Islands Broadcasting Corporation (SIBC), Voice of the Nation (ed., Retrieved from https://www.sibconline.com.sb/worlds-most-dangerous-coconut-beetle-now-in-solomons/
SIBC (2016a) Invasive coconut beetle reached North-East Guadalcanal: Solomon Islands Broadcasting Corporation (SIBC), Voice of the Nation (ed., Retrieved from https://www.sibconline.com.sb/invasive-coconut-beetle-reached-north-east-guadalcanal/
SIBC (2016b) Ministry of agriculture embarked on rhinoceros beetle: Solomon Islands Broadcasting Corporation (SIBC), Voice of the Nation (ed., Retrieved from https://www.sibconline.com.sb/ministry-of-agriculture-embarked-on-rhinoceros-beetle/
SIBC (2017a) Biosecurity teams mobilise against coconut rhinoceros beetle: Solomon Islands Broadcasting Corporation (SIBC), Voice of the Nation (ed., Retrieved from https://www.sibconline.com.sb/biosecurity-teams-mobilise-against-coconut-rhinoceros-beetle/
SIBC (2017b) Farmers call for action as catastrophic Rhinoceros beetle hits North Malaita: Solomon Islands Broadcasting Corporation (SIBC), Voice of the Nation (ed., Retrieved from https://www.sibconline.com.sb/farmers-call-for-action-as-catastrophic-rhinoceros-beetle-hits-north-malaita/
SIBC (2017c) GPPOL hit with profit loss as rhinoceros beetle invades: Solomon Islands Broadcasting Corporation (SIBC), Voice of the Nation (ed., Retrieved from https://www.sibconline.com.sb/gppol-hit-with-profit-loss-as-rhinoceros-beetle-invades/
SIBC (2018a) Biosecurity preps virus for beetle eradication: Solomon Islands Broadcasting Corporation (SIBC), Voice of the Nation (ed., Retrieved from https://www.sibconline.com.sb/biosecurity-preps-virus-for-beetle-eradication/
SIBC (2018b) Coconut rhinoceros beetle threatens Russell Islands plantation: Solomon Islands Broadcasting Corporation (SIBC), Voice of the Nation (ed., Retrieved from https://www.sibconline.com.sb/coconut-rhinoceros-beetle-threatens-russell-islands-plantation/
SIBC (2018c) Coconut rhinoceros beetle threatens shortland Islands plantations.: Solomon Islands Broadcasting Corporation (SIBC), Voice of the Nation (ed., https://www.sibconline.com.sb/coconut-rhinoceros-beetle-threatens-shortland-islands-plantations/
SPC (2017) Outcomes report for the subregional workshop on coconut Rhinoceros beetle (CRB-G) in the Pacific: South Pacific Community, SPC Narere, Campus, Suva, Fiji
Stapley JH (1973) Insect pests of coconuts in the Pacific region. Outlook Agric 7:211–217
Sun X, Tao J, Roques A, Luo Y (2020) Invasion history of Sirex noctilio based on COI sequence: the first six years in China. InSects. https://doi.org/10.3390/insects11020111
Tsatsia F, Wratten H, Gharuka M, Fanai C, Wate D, Tsatsia H & Macfarlane B (2018) The status of coconut Rhinoceros Beetle, Oryctes rhinoceros (L) Scarabaeidae: dynastinae, in Solomon Islands, Retrieved from: https://devpolicy.org/pdf/blog/Status-of-the-coconut-rhinoceros-beetle.pdf, 01 Feb 2021
Vaqalo M, Timote V, Baiculacula S, Suda G & Kwainarara F (2017) The coconut rhinoceros beetle in solomon islands: a rapid damage assessment of coconut palms on Guadalcanal: Pacific Community, Suva, Fiji, pp. 1–13
Young EC (1974) The epizootiology of two pathogens of the coconut palm rhinoceros beetle. J Invertebr Pathol 24:82–92
Young EC (1986) The rhinoceros beetle project: history and review of the research programme. Agr Ecosyst Environ 15:149–166
Acknowledgements
This work was supported by the New Zealand Ministry of Foreign Affairs and Trade (MFAT project PF9-548). Thanks to Caitlin Hyde for assisting with the haplotype and virus testing, and to Peter Pletnyakov for GIS mapping (AgResearch). We are grateful to Bob Macfarlane for thoughtful comments on this paper. In-country support from MAL and GPPOL (Max Kolubalona, Samuel Hone, Iro Ramoi, George Harunari, Trevor Loloito, Aloysio Kwaierebeu, Jeremy Puisasa, Obed Maneara, Reuben Alipio, Esmond Roba, Patrick Riasi, Cecil Sigiani, Walter Rofo, Timothy Samani, Patrick Maesuba, Arron Taupongi, Florence Kawai, Narelle Wilson, Alfred Pokana, Craig Gibsone, Simon Chris, Paul Gende, Jaydita and Hilary Sau) is greatly appreciated.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. This work was supported by the New Zealand Ministry of Foreign Affairs and Trade (MFAT project PF9-548, ACT-0101031).
Author information
Authors and Affiliations
Contributions
SM and TJ initiated this study. FT, CF and GS were primarily responsible for field surveys and sample collection with support from SM, SM, and TJ. Material preparation and data analysis were performed by NR, SM, SP and SM. SM, SP, SM, NR, and TJ wrote the draft manuscript. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials. The raw data are available from the corresponding author, upon reasonable request.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Marshall, S.D.G., Paudel, S., Mansfield, S. et al. Coconut rhinoceros beetle in Solomon Islands: a tale of two invasions. Biol Invasions 25, 2659–2678 (2023). https://doi.org/10.1007/s10530-023-03063-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-023-03063-x