Abstract
After its bloom in the Black and Caspian Sea in the late 1980s and early 90s, there has been an increased interest in understanding the ecology of the invasive zooplanktivorous comb jellyfish Mnemiopsis leidyi and its potentially severe impacts on the functioning of marine systems. In the last decade, M. leidyi has colonized most of the Mediterranean Sea, including the Adriatic Sea, and in 2016 it was recorded in the Venice Lagoon (Malej et al. J Sea Res 124:10–16, 2017). The impact M. leidyi could have on a semi-enclosed ecosystem like the Venice Lagoon is of concern as it is an important nursery and foraging area for several fish species as well as an area of mussel, clam, and crab fishery and aquaculture. Historically, the feeding preference of M. leidyi was determined by morphological identification of gut contents. This is the first study investigating the in-situ gut contents of this species using DNA metabarcoding, which overcomes the limit in identifying partially digested prey. In this study, M. leidyi’s gut contents collected in the Venice Lagoon were evaluated by metabarcoding and compared to the in-situ mesozooplankton community. The results indicate that its blooming period is in the late summer and that it feeds on a variety of prey, mostly coinciding with the zooplankton assemblage. Notably, some groups, like decapod larvae and the slow-swimming larvae of gastropods and bivalves, appear to be favored. Conversely, the relative abundance of copepods was higher in-situ than in the gut contents.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The progressive global spread of the highly invasive zooplanktivorous comb jelly Mnemiopsis leidyi (A. Agassiz 1865), together with the increasing awareness of its potential impacts on the functioning of marine systems, has led to a rising interest in their ecology (Shiganova et al. 2019a). The ability of this ctenophore to adapt and colonize new areas is partly due to its broad temperature and salinity tolerance, being hermaphroditic, having the capability to self-fertilize, and its capability to regenerate (Purcell et al. 2001).
The bloom of M. leidyi in the Black and Caspian Sea ecosystems in the late 1980s and early 90s likely became possible due to a shortage of predators and competitors by overfishing (Shiganova et al. 2001). Subsequently, these blooms of M. leidyi have been associated with severe declines in fish stocks and affected the ecosystem production (Shiganova and Bulgakova 2000; Lynam et al. 2006) as M. leidyi outbreaks can exert a top-down control and induce trophic cascades (McNamara et al. 2013; Roohi et al. 2010 Schneider and Behrends 1994). The pressures on the ichthyoplankton community can act mainly in two ways: first, through direct predation on fish eggs and larvae, and second, through intense competition for food with zooplanktivorous fishes species and larvae (Purcell et al. 2001), indirectly affecting the abundance of ichthyoplankton. Mnemiopsis leidyi is known to feed on a variety of prey, depending on food availability and its life stage (Shiganova and Bulgakova 2000; Sullivan and Gifford 2004).
Most of the Mediterranean Sea, from the eastern basin to the western basin, has been colonized by M. leidyi (Shiganova et al. 2019b). In 2016, it was first recorded in the Venice Lagoon (VL) after likely being introduced via ballast water (Malej et al. 2017). Ballast water is a global vector in human-mediated invasions providing a fast dispersal mechanism for many marine taxa and therefore massively increasing the risk of NIS introduction (Marchini et al. 2015; Vidjak et al. 2019). In fact, the VL is highly impacted by human activities (Lotze et al. 2006; Solidoro et al. 2010) and is a known hotspot of NIS introduction (Marchini et al. 2015; Vidjak et al. 2019; Pansera et al. 2021), due to its heavy maritime traffic. This makes it both starting point as a source of new introductions as well as continuous re-introduction via ballast water.
The concerns regarding the ecological and economic impacts M. leidyi could have in the Northern Adriatic Sea ecosystem are enormous as the sea is an important nursery and foraging area for sardines and anchovies, which together account for approximately 41% of total Adriatic marine catches (Morello and Arneri 2009; Shokralla et al. 2012). The Northern Adriatic coast, together with the VL, is not only a vital nursery area for fishes, but is also an important production area for mussels (Mytilus) and clams (Ruditapes and Chamalea), and is an essential area for crab fisheries and aquaculture. These economically important species are part of the zooplankton community during their larval stages and are likely consumed by M. leidyi, increasing the pressure on this economic branch, both for small local businesses as well as for the industrial production. Moreover, high densities of M. leidyi clog fishermen’s nets and cooling systems of power plants incurring further economic costs on coastal communities (Purcell et al. 2007; Palmieri et al. 2014).
In the past, to study feeding preferences the gut content of M. leidyi was analyzed through morphological identification. However, this approach has its limits, as it only allows identification of undigested or partially digested prey. In addition, identification based on morphological features for some groups like larval stages or cryptic species is difficult. DNA metabarcoding, a molecular approach based on sequencing a short DNA fragment that is unique to each species and can therefore be used for species discrimination, has been previously used for gut content analyses, e.g., on fishes (Albaina et al. 2016), on the jellyfish Chrysaora (Meredith et al. 2016) or on the anemone Metridium (Wells et al. 2022). To the best of our knowledge, this is the first study using DNA metabarcoding for gut content analyses of M. leidyi.
This study aims to identify the feeding preferences of the comb jellyfish M. leidyi in the VL and to speculate on its potential impact on zooplankton abundances and biodiversity through the use of DNA metabarcoding. This study will increase the knowledge about factors driving a possible decline in fish stocks, indicating if it is due to competition for zooplankton or to direct feeding of M. leidyi on fish eggs or larvae. Considering the importance of this ecosystem for several meroplanktonic species, many of which are exploited commercially, the threat the feeding pressure of M. leidyi could have on this zooplanktonic compartment is an additional concern.
Material and methods
Study site and sampling
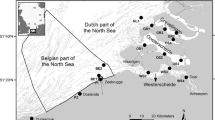
The study was conducted in the VL, a Mediterranean microtidal lagoon with a surface of about 550 km2, a mean depth of the tidal flats of −1.2 m above mean sea level (AMSL) and it reaches −10/−15 m AMSL in the natural tidal channels. It is connected through three inlets to the Northern Adriatic Sea, a shallow coastal area (mean depth of 35 m) strongly influenced by the inputs of large rivers and characterized by mesotrophic conditions and by a notable spatial and temporal variability of physico-chemical and trophic gradients (Bernardi-Aubry et al. 2006, 2020). The VL is a heterogeneous system characterized by a number of environmental gradients and a mosaic of habitats (e.g., intertidal marshes and mudflats, and natural and navigation channels) that are the result of complex natural and man-induced drivers (Tagliapietra et al. 2009). With each tidal cycle, about one-third of the total volume of the lagoon is exchanged (Gačić et al. 2004), and the residence times range from a few days, in the vicinity of the inlets, to over 60 days in the inner areas (Cucco and Umgiesser 2006).
Based on morphological identification, the zooplankton community in the VL is composed of about 80% copepods (with Acartia as the most abundant genus) and about 10% chordates (mostly composed of Appendicularia, Ascidiacea larvae, and Actinopterygii larvae or eggs), followed by echinoderms and mollusks (Camatti et al. 2008; Schroeder et al. 2020). In the areas nearby the inlets it shows higher abundances of marine taxa, like cladocerans and appendicularians (Solidoro et al. 2010).
The sampling was performed as part of a study with monthly samplings of 16 stations within the VL, representative of different environmental conditions, from April 2018 to March 2019 (Fig. 1). Both in-situ zooplankton community and Mnemiopsis leidyi individuals were sampled using an HydroBios Apstein net with a 0.4 m diameter opening and 200 μm mesh. Environmental data, such as temperature, salinity, oxygen, turbidity and Chl-a were measured using a multiparametric CTD probe (SBE 19plus) at the sampling sites. Mnemiopsis leidyi individuals larger than 1.5 cm in length were measured (total biovolume) and immediately frozen at −20 °C, while zooplankton samples were preserved in 96% ethanol for genetic analyses. For the gut content analyses, M. leidyi individuals were thawed, gut contents were extracted with a Pasteur pipette under a stereomicroscope (Zeiss, Discovery V8), and then all gut contents within one station were pooled.
Molecular analyses
For the zooplankton taxonomic composition assay, from the in-situ samples, a representative subsample (about one-third of the total sample) was taken, the ethanol removed by centrifugation, and afterwards, the samples were rinsed with PBS (1x), while the extracted gut contents of M. leidyi were centrifuged to remove excess liquids. All samples were successively homogenized by bead-beating for one minute. Genomic DNA was extracted using the E.Z.N.A.® Mollusc DNA kit (Omega Bio-Tek) following the manufacturer’s instructions and increasing the initial volume of reagents (lysis and binding buffer) provided by the kit proportionally to the sample volume. The quality and quantity of the extracted DNA were assessed with a NanoDrop 2000 Spectrophotometer (ThermoScientific). The amplification was performed using a degenerated forward primer jdgLCO1490 (5’-TCAACAAAYCAYAARGAYATYGG-3’) (Schroeder et al. 2021) in combination with the reverse internal primer mlCOIintR proposed by Leray et al. (2013) with a target length of 319 bp. The reverse primer mlCOIintR was slightly modified compared to the original in order to match the forward internal primer mlCOIintF by interchanging the “S” with “W” nucleotides: 5’-GGRGGRTAWACWGTTCAWCCWGTWCC-3’ instead of 5’-GGRGGRTASACSGTTCASCCSGTSCC-3’. As shown by Schroeder et al. (2021), this primer pair performed well for zooplankton biodiversity assessments and was chosen for this study due to its impediments in amplifying ctenophores. In this way, it could be avoided to primarily amplify the host DNA, but rather the actual gut content.
The amplification was performed as by Schroeder et al. (2021). Briefly, a two-step polymerase chain reaction (PCR) was performed, amplifying the target regions in the first PCR and bounding the sample-tags in the secondary PCR. Afterwards, the library was purified, quantified, and prepared for high-throughput sequencing by pooling an equimolar amount of amplicon products. Emulsion PCR was conducted using the Ion One Touch System (Life Technologies) following the manufacturer’s recommendations, and DNA was bound to Ion Sphere particles (Life Technologies) for clonal amplification automatically enriched with the Ion OneTouch ES system (Life Technologies). For sequencing, the library was loaded on a 316™ chip with 650 flows in an Ion Personal Genome Machine (PGM, Life Technologies).
Bioinformatics and statistics
The bioinformatic workflow was conducted following Schroeder et al. (2021), including sequence preparation (filtering and trimming), error corrections, chimera removal, and several steps of taxonomic assignment. Those steps included assignments against a COI reference database of marine metazoan sequences deposited in GenBank following the query used by Schroeder et al. (2020). As similarity thresholds 97% and 94% were chosen and an additional recovery of putative metazoan sequences at 85% similarity was performed. Those 85% hits were then clustered de-novo at 97% (q2-vsearch (Rognes et al. 2016)), and compared against the GenBank database with BlastN + (Camacho et al. 2009). Metazoan clusters with a BLASTn p-identity of at least 94% were joined to the final dataset and those with a BLASTn p-identity of < 94% and > 90% were considered as a “best match”, hence putative metazoan OTUs with low taxonomic confidentiality (see Schroeder et al. 2021).
Species richness per sample was quantified according to the measure of the first Hill number—MOTU/taxa richness (q = 0) using the R package iNEXT (Hsieh et al. 2019). Spatial and temporal patterns of the environmental factors based on Euclidean distances of normalized data were assessed using repeated-measure permutational analysis of variance (PERMANOVA) with the sampling months as a fixed factor and the stations as a random factor (PRIMER 6 + and PERMANOVA software package; PRIMER-E, Ltd., UK). To visualize the similarities between the samples in terms of environmental conditions, a principal coordinates analysis (PCoA) was performed. With the R software (R Core Team 2018), differences between months and stations were tested using the Kruskal–Wallis test, while Spearman’s correlations were calculated between biovolume (ml/m3) and environmental parameters. The Spearman’s correlations between the in-situ zooplankton community and M. leidyi’s gut content was also calculated for groups at different taxonomic levels (square-root transformed percentages) as well as for the most abundant species. Where confidence intervals were calculated, 95% bias-corrected bootstrap confidence intervals with 5000 replicates were used with the R packages boot (Canty and Ripley 2022). Beta-diversity was calculated from dissimilarity matrices built according to Bray–Curtis distances using the metaMDS script with the autotransform function (R package vegan, Oksanen et al. 2019) and plotted as NMDS plots by month and by location, where the stations were grouped by location (“inner”, “med” and “inlet”), based on residence time and salinity as a proxy of connectivity. Specifically, the “inlet” stations are those located within the three inlets (station 4, 11 and 15; mean salinity: 34.4 ± 1.5), the “med” stations are those with intermediate connectivity to the sea (station 2, 7, 9, 10, 12 and 13; mean salinity: 32.1 ± 3.6), while the “inner” stations are those with reduced connectivity to the sea (station 1, 5, 6, 8, 14 and 16; mean salinity: 29.6 ± 4.6). Differences between the gut content and the zooplankton community were evaluated by one-way PERMANOVA and the species contribution to those differences was evaluated by a similarity percentage analysis (SIMPER), based on a Bray–Curtis distance matrix of square-root transformed percentages (adonis2 and simper functions in R package vegan, Oksanen et al. 2019).
Results
Environmental characteristics
The environmental parameters differed significantly both temporally and spatially (Fig. 2a, Table S1). The temporal pattern followed a temperature gradient, a parameter that showed especially high variability owed to the shallow nature of the VL. Temperatures ranged from 3.0 to 30.5 °C (mean: 18.3 °C ± 8.2) and exhibited the typical seasonal trend (KW: χ2 = 180.63, df = 11, p < 2.2e−16). In the months with lower temperature values, Chl-a was also lower, ranging from 0.7 to 49.3 ug/l (5.1 ug/l ± 6.9). In contrast, turbidity and salinity were more related to the location, with higher salinities (KW: χ2 = 122.54, df = 15, p < 2.2e−16) and lower turbidity values (KW: χ2 = 112.7, df = 15, p < 2.2e−16) in the inlet stations (4, 11 and 15) and the nearby areas (Fig. 2b). Overall, the salinity values ranged from 9.0 to 36.3 (30.9 ± 4.2), the turbidity from 0.8 to 38.5 NTU (6.3 NTU ± 5.6) and the oxygen from 56.9 to 188.2% (102.3% ± 17.4).
Biovolume of Mnemiopsis leidyi
During the study period (April 2018 to March 2019), the first individuals of M. leidyi, including larval stages (~ 0.5 cm length), were detected in June. Individuals longer than 1.5 cm were found in 44 samples, from June to February, with variable total biovolume ranging from 1.3 to 78 ml/m3. Still, the highest biovolumes of M. leidyi were found during late summer, especially from July to October (Fig. 3a). After November, the presence was significantly reduced, and only single individuals were detected in the samples. Concurrently, the presence of larval stages increased again. In fact, temperature showed a weak, but significant positive correlation to the biovolume of M. leidyi (p < 0.001, rho = 0.37, N = 44). However, although the abundances differed between stations (Fig. 3b), none of the other environmental parameters showed significant correlations.
Diet of Mnemiopsis leidyi
The number of raw sequences was 2.3 × 106 reads for the 44 samples of M. leidyi gut content and 3.2 × 106 reads for the 44 samples of in-situ mesozooplankton samples. After taxonomic assignments, the final number of sequences of the gut contents of M. leidyi was 768,611 with 122 OTUs. Of these sequences, 71.1% were detected at 97% similarity threshold, 14.9% at 94%, and 14% by the recovery of putative metazoans (see Material and Methods for more detail). The mesozooplankton community resulted in 233 OTUs representing 1,486,969 sequences (assignments at 97%: 87.5%; at 94%: 9.3%; by the recovery of putative metazoans: 3.2%). For the following analyses except for the beta-diversity estimates, the most stringent dataset was used: excluding the “best match” assignments, thus, those putative metazoan OTUs with low taxonomic confidentiality due to low similarity assignment. This stringent dataset was composed of 107 OTUs with 672,956 sequences of M. leidyi gut content and 213 OTUs with 1,464,823 sequences of in-situ mesozooplankton (see rarefaction curves in Supplementary Material Fig. S1).
The taxonomic assignment of the gut content of M. leidyi indicated that it feeds on a variety of prey. The most abundant phylum of prey was Arthropoda with a mean of 62% (95% CI [52, 71]), with Copepoda as the most represented class (26%; 95% CI [20, 34])), followed by the classes of Decapoda (20%; 95% CI [12, 30]) and Branchiopoda (composed of cladocerans only) (12%; 95% CI [6, 23]). The second most abundant phylum was Mollusca (21; 95% CI [14, 29]), composed mainly of Gastropoda (15%; 95% CI [10, 23]) and Bivalvia (5%; 95% CI [3, 10]); the third was Annelida (composed of Polychaeta only) (12%; 95% CI [7, 21]), and the fourth was Nemertea (3%; 95% CI [1, 5]). However, the high values of standard deviation indicated a high variability between the samples (Fig. 4, S2, Table 1).
The in-situ mesozooplankton community showed similar compositions to the gut contents. Several groups showed significant correlations between the gut content and the in-situ mesozooplankton community: Annelida, Cladocera, Calanoida, Cyclopoida, Poecilostomatoida, Decapoda, Hydrozoa and Nemertea (p < 0.01; Table 1). However, there were some differences: the relative abundance of arthropods was higher in-situ than in the gut contents (89% compared to 62%), with higher proportions of calanoids (59% vs. 25%) and cladocerans (21% vs. 12%). Decapods, however, showed higher relative abundances in the gut contents (5% in-situ vs. 20% in gut contents). Similarly, mollusks (3% vs. 21%), Nemertea (0.02% vs. 3%) and Polychaeta (1% vs. 12%) are accumulated by M. leidyi (Table 1).
Regarding the relative abundances of the four most abundant copepod genera in the gut content of M. leidyi (relative abundance calculated in relation to copepods), the genus Acartia made up 71.9% (95% CI [60.2, 81.2]) of the copepod sequences, followed by Centropages with 7.1% (95% CI [3, 16.4]), Oithona with 6.6% (95% CI [3.4, 12.7]) and Paracalanus with 3.4% (95% CI [1.7, 6.8]). Within the in-situ zooplankton community, Acartia was again at the first rank with 76.9% (95% CI [66.9, 85.1]) of the copepod community, followed by Paracalanus with 8.2% (95% CI [4.9, 12.8]), by Centropages with 5.6% (95% CI [2.8, 11.2]) and by Temora with 3.5% (95% CI [1.3, 8.3]). The composition of the in-situ zooplankton community and the gut content differed significantly (one-way PERMANOVA: SS = 2.4, R2 = 0.1, F = 8.8, p = 0.001). The species mostly contributing to the difference between the gut content and the zooplankton community was the highly abundant copepod Acartia tonsa with 13% contribution, which was more abundant in-situ, indicating a reduced capture by M. leidyi. Similarly, the cladocerans Pleopis polyphemoides, Penilia avirostris and Pseudevadne tergestina contributed with 6%, 4% and 4%, (respectively) to the difference, and the copepods Centropages ponticus, Paracalanus spp. with 3% and 2%, respectively (SIMPER analysis, Table S2, Fig. 5a, b, d). On the other hand, specially several meroplanktonic taxa seemed to accumulate in M. leidyi’s gut, such as the crabs, Dyspanopeus sayi and Carcinus aestuarii (contribution to difference of 7% and 3%), as well as the bivalve Ruditapes philippinarum (2% contribution), a species of immense commercial interest in the VL. Likewise, polychaete larvae, the gastropods Bittium reticulatum and Haminoea orteai, and the nemertean Cephalothrix sp. were more abundant in the gut content and contributed with 5%, 5%, 2% and 2%, respectively, to the difference between the gut content and the zooplankton community, indicating an accumulation by M. leidyi (Fig. 5c, d, Table S2). The beta-diversity analyses, based on the most inclusive dataset, hence including also the “best match” assignments in order to incorporate as many putative metazoan OTUs as possible, resulted in a clear temporal differentiation by month as well as a spatial one by location for both the zooplankton community and the gut contents (Fig. 6). The differences between the gut content samples were greater than between the in-situ zooplankton samples. However, it emerged that the two datasets were almost overlapping rather than creating two different clusters. This overlap indicated that the feeding of M. leidyi depends mainly on the food available at that specific moment and location (Fig. 6).
Correlation plot (% based on square-rooted data) of the most abundant copepod species A, B, mollusk species C and other taxa D. Shaded areas indicate 95% confidence intervals and a 1:1 line was added to highlight preference/avoidance. Spearman’s correlations between the two datasets are given in the corresponding color
Beta-diversity estimates based on Bray–Curtis similarities plotted on NMDS of Mnemiopsis leidyi’s gut content and the in-situ mesozooplankton community. Numbers plotted on datapoint refer to the sampling station and colors of points refer to the sampling month (left) or location (right) of each sample. Those groups are highlighted plotting convex hulls, the distances to their centroid and the standard deviation of the points with the respective colors (M. leidyi gut content in grey and in-situ community in black)
Discussion
Understanding the characteristics of blooms of the zooplanktivorous invasive predator Mnemiopsis leidyi is increasingly important, due to its ongoing successful invasion of new regions and its potential impact on zooplankton densities and ecosystem production (Shiganova et al 2019a). The top-down effect of the predation pressure on zooplankton, which is especially significant during intense blooms of M. leidyi, can favor a substantial decrease in zooplankton and a correlated increase in phytoplankton (Shiganova 1998; Finenko et al. 2006; Tiselius and Møller 2017), accompanied by a decline in fish stocks, as already experienced in the Black and Caspian Seas (Shiganova and Bulgakova 2000). Considering the importance of the Venice Lagoon as a nursery area, the massive blooms experienced in the last years in this habitat raise concerns regarding its already ongoing and future effects on the ecosystem production and ecosystem services. Hence, within the "blue economy" with various business categories falling under this definition, such as environmental regulation, fish farming and fishing, providing additional insights into the potential impact of invasive species on the ecosystem, is crucial to satisfy both economic demands and environmental protection.
In this study, M. leidyi was found to be ubiquitous in the VL and showed a seasonal persistence (at different life stages), hence tolerating the measured temperature range of 3–30.5 °C. These findings confirm its high ecological tolerance which makes it a successful invader (Shiganova et al 2019a, b; Shiganova 2020), and highlights the need to improve our knowledge on this species, including its feeding preference. Spatial differences in abundance found within the lagoon may be driven not only by prey availability and environmental preference, but also by hydrodynamic processes that accumulate M. leidyi in specific areas. Seasonal differences were evident with highest abundances in terms of biovolume [ml/m3] detected during (late-)summer (July–October) with temperature as the main abiotic driver, likewise stated by many authors, e.g., Kremer (1979), who mentioned temperature and prey abundance as key factors affecting its seasonal patterns. Other factors that make semi-enclosed lagoons especially vulnerable are potential low oxygen levels that can occur especially during summer (Bernardi-Aubry et al. 2020). However, M. leidyi, as other gelatinous species, can potentially benefit from it as they are generally more tolerant to hypoxia compared to their preys. Decker et al. (2004) showed a reduced jumping frequency of the copepod A. tonsa under hypoxic conditions. This indicates that in hypoxic waters, less-tolerant prey might be more vulnerable to predation, therefore favoriting capture rates by M. leidyi.
Several authors have studied the feeding preferences of M. leidyi in the past. However, to our knowledge, this is the first study applying DNA metabarcoding based on high-throughput sequencing technologies to investigate its dietary composition. The primary benefit of this method compared to morphology-based identification in analyzing the feeding preference is the detection of partially digested prey and cryptic species. However, the (relative) quantification of prey items that are more effortlessly digestible, e.g., soft organisms like fish larvae, or that have been ingested beforehand, may be underestimated. Other studies used DNA metabarcoding for gut content analysis by comparing it with the zooplankton community. For example, Wells et al. (2022) who found that the gut contents of the anemone Metridium farcimen were primarily made up of arthropods (52% of sequences), especially crab larvae, barnacles (larvae or molts), copepods, and insects, as well as Meredith et al. (2016), who found that the gut contents of the jelly fish Chrysaora quinquecirrha were composed of copepods, fish, ctenophores, anemones, amphipods, barnacles, shrimp, polychaetes, flukes, flatworms, echinoderms, gastropods, bivalves and hemichordates.
The literature, based on morphological identification, indicates that M. leidyi’s diet often reflects the composition of ambient prey (e.g., Javidpour et al. 2009; Madsen and Riisgård 2010; Granhag et al. 2011). Copepods often dominated the diet of M. leidyi, but also meroplanktonic larvae of polychaetes, mollusks, decapods and barnacles are consumed (e.g., Kremer 1979; Purcell et al. 2001; Colin et al. 2015). In our study, the diet of M. leidyi was very variable, but mainly included copepods, decapods, cladocerans, gastropods, bivalves and polychaetes, but also echinoderms, nemerteans and cnidarians, hence a composition that characterizes a typical lagoon community. During winter, the dietary composition showed a peak in polychaete larvae, in consistency with Larson (1987) and McNamara et al. (2010), which reported the ingestion of polychaetes larvae by M. leidyi. However, this noticeable difference of the winter samples may also be a result of higher uncertainty due to the smaller sample size (see biovolume during winter).
Similarly to Decker et al. (2004) and Roohi et al. (2010), also in our study A. tonsa was the most abundant copepod species, both in-situ and in the gut content. However, in general, copepods and cladocerans were less represented in the gut content than in-situ, while meroplanktonic groups such as decapod and mollusk larvae were more abundant in the gut content, indicating a preferential feeding on the latter ones. In fact, due to the capture mechanisms of M. leidyi, less mobile organisms such as mollusks seemed to be a vulnerable prey which is consistent with the literature (e.g., Madsen and Riisgård 2010; Marchessaux et al. 2021). Depending on prey mobility the capture mechanism varies: Slow-moving or immobile organisms, like mollusk and barnacle larvae or eggs are captured by creating an undetectable current by the cilia within the auricles which together with the mucus gets the prey to be trapped in their tentila (Waggett and Costello 1999; Haddock 2007; Colin et al. 2015), while highly mobile preys, like fast swimming copepods, are captured by collision with the inside of the lobes (Mutlu 1999; Purcell et al. 2001; Javidpour et al. 2009). For example, within copepods, smaller, and therefore probably less mobile, species like Oithona nana and O. davisae or Euterpina acutifrons seemed to be captured preferentially. In comparison, the larger species Temora stylifera and Paracalanus spp., being potentially faster, are less abundant in the gut content as they may escape from M. leidyi more easily. It has to be kept in mind, especially regarding the holoplanktonic copepods, that DNA metabarcoding does not allow to differentiate between life stages. Therefore, more than size differences between copepods species, the actual life stage of each species at that specific moment may have a more significant effect on the vulnerability of species to the feeding pressure of M. leidyi.
In this study, a standard sampling net with a mesh size of 200 µm was used to collect the in-situ zooplankton community. The ingested prey, however, may include zooplankton smaller than 200 µm, like nauplii or small bivalve larvae, which might be underestimated in the sampled zooplankton community. Therefore, the selectivity of the 200 µm sampling net could be another explanation for the higher relative abundance of small sized organisms in the gut content compared to the in-situ zooplankton assemblages. Hence, the additional use of e.g., an 80 µm plankton net to better describe the smaller size fraction of the community could be beneficial (Pansera et al. 2014). In fact, Wells et al. (2022) found that the diet of another planktivorous animal (an anemone) was more closely related to an 80 µm plankton sample than a 330 µm. The diet of M. leidyi is known to differ at different life stages. While larvae and post-larvae of M. leidyi consume primarily microphyto- and microzooplankton prey like dinoflagellates or ciliates (Sullivan and Gifford 2004), adults feed on a variety of holo- and meroplankton organisms (Shiganova and Bulgakova 2000). Therefore, as in this study only adult M. leidyi individuals above 1.5 cm (therefore possibly feeding on larger prey compared to smaller individuals) were included in the gut content analysis, the use of a standard mesozooplankton net with a mesh size of 200 µm should have a limited bias.
Differences between the collected zooplankton community and the gut contents can appear also due to the fact that the zooplankton samples are just a snapshot in time, whereas gut content samples are a summation of prey encountered over the time it takes to digest them (Wells et al. 2022).
As previously mentioned, the VL represents an ecosystem of huge ecological but especially socio-economic importance. It is not only a vital nursery area for fishes, but it is also an area for mussel, clam, and crab aquaculture. Hence, the impact of M. leidyi on the ecosystem functioning is of increasing interest. Given the severe impact on the fish stocks in the Black and Caspian Seas in the late 80s and early 90s, the investigation on this zooplanktonic compartment (fish larvae and eggs) is of special interest. However, this study indicated no significant direct predation on fish larvae or eggs with few fish sequences especially in the gut contents. This is probably explained by the dominance of benthic fish species in the VL, like Zosterisessor ophiocephalus, and the fact that the spawning times may not coincide with the major blooming period of M. leidyi (Franzoi et al. 2010). Moreover, the reproductive strategy of lagoon resident fish species is adapted to prevent seaward flushing of eggs and larvae by spawning demersal eggs attached to the aquatic vegetation or other substrates, while the planktonic stage is reduced or lacking (Dando 1984). Therefore, rather than direct predation on fish eggs and larvae, competition for zooplankton may have an impact on the fish stock in the VL. Given the relatively high abundances of meroplanktonic taxa in the gut content, the impact M. leidyi seems to have on the meroplanktonic compartment of the zooplankton community may be more significant than expected and could increase the pressure on the local economy and industrial production of crabs and clams. As a conclusion, while in other geographic areas the major concern regarding the arrival and large blooming of M. leidyi mainly refers to the fish stocks and its associated economy, in the VL and the Northern Adriatic coasts, M. leidyi’s impact may be greater on the meroplanktonic compartment, and consequently on the mussel, clam, and crab fishery and aquaculture.
Data availability and materials
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Albaina A, Aguirre M, Abad D et al (2016) 18S rRNA V9 metabarcoding for diet characterization: a critical evaluation with two sympatric zooplanktivorous fish species. Ecol Evol 6:1809–1824. https://doi.org/10.1002/ece3.1986
Bernardi-Aubry F, Acri F, Bastianini M et al (2006) Seasonal and interannual variations of phytoplankton in the Gulf of Venice (Northern Adriatic Sea). Chem Ecol 22:S71–S91. https://doi.org/10.1080/02757540600687962
Bernardi-Aubry F, Acri F, Scarpa GM, Braga F (2020) Phytoplankton—macrophyte Interaction in the Lagoon. Water 12:2810
Camacho C, Coulouris G, Avagyan V et al (2009) BLAST+: Architecture and applications. BMC Bioinform 10:1–9. https://doi.org/10.1186/1471-2105-10-421
Camatti E, Comaschi A, De Olazabal A, Fonda Umani S (2008) Annual dynamics of the mesozooplankton communities in a highly variable ecosystem (North Adriatic Sea, Italy). Mar Ecol 29:387–398. https://doi.org/10.1111/j.1439-0485.2008.00256.x
Canty A, Ripley BD (2022) boot: Bootstrap R (S-Plus) functions. R Package Version 1(3–28):1
Colin SP, MacPherson R, Gemmell B et al (2015) Elevating the predatory effect: Sensory-scanning foraging strategy by the lobate ctenophore Mnemiopsis leidyi. Limnol Oceanogr 60:100–109. https://doi.org/10.1002/lno.10007
Cucco A, Umgiesser G (2006) Modeling the Venice Lagoon residence time. Ecol Modell 193:34–51. https://doi.org/10.1016/j.ecolmodel.2005.07.043
Dando PR (1984) Reproduction in estuarine fish. In: Potts GW, Wootton RJ (eds) Fish Reproduction: Strategies and Tactics. Academic Press, London, pp 155–170
Decker MB, Breitburg DL, Purcell JE (2004) Effects of low dissolved oxygen on zooplankton predation by the ctenophore Mnemiopsis leidyi. Mar Ecol Prog Ser 280:163–172. https://doi.org/10.3354/meps280163
Finenko GA, Kideys AE, Anninsky BE et al (2006) Invasive ctenophore Mnemiopsis leidyi in the Caspian Sea: feeding, respiration, reproduction and predatory impact on the zooplankton community. Mar Ecol Prog Ser 314:171–185. https://doi.org/10.3354/meps314171
Franzoi P, Franco A, Torricelli P (2010) Fish assemblage diversity and dynamics in the Venice lagoon. Rend Lincei 21:269–281. https://doi.org/10.1007/s12210-010-0079-z
Gačić M, Mancero Mosquera I, Kovačević V et al (2004) Temporal variations of water flow between the Venetian lagoon and the open sea. J Mar Syst 51:33–47. https://doi.org/10.1016/j.jmarsys.2004.05.025
Granhag L, Møller LF, Hansson LJ (2011) Size-specific clearance rates of the ctenophore Mnemiopsis leidyi based on in situ gut content analyses. J Plankton Res 33:1043–1052. https://doi.org/10.1093/plankt/fbr010
Haddock SHD (2007) Comparative feeding behavior of planktonic ctenophores. Integr Comp Biol 47:847–853. https://doi.org/10.1093/icb/icm088
Hsieh TC, Ma KH, Chao A (2019) iNEXT: iNterpolation and EXTrapolation for species diversity. R Package Version 2:19
Javidpour J, Molinero JC, Lehmann A et al (2009) Annual assessment of the predation of Mnemiopsis leidyi in a new invaded environment, the Kiel Fjord (Western Baltic Sea): a matter of concern? J Plankton Res 31:729–738. https://doi.org/10.1093/plankt/fbp021
Kremer P (1979) Predation by the ctenophore Mnemiopsis leidyi in Narragansett Bay, Rhode Island. Estuaries 2:97–105. https://doi.org/10.2307/1351633
Larson RJ (1987) In situ feeding rates of the ctenophore Mnemiopsis mccrady. Estuaries Coasts 10:87–91
Lotze HK, Lenihan HS, Bourque BJ et al (2006) Depletion, degradation, and recovery potential of estuaries and coastal seas. Science 312(5781):1806–1809
Lynam CP, Gibbons MJ, Axelsen BE, Sparks CA, Coetzee J, Heywood BG, Brierley AS (2006) Jellyfish overtake fish in a heavily fished ecosystem. Curr Biol 16(13):R492–R493
Madsen CV, Riisgård HU (2010) Ingestion-rate method for measurement of clearance rates of the ctenophore Mnemiopsis leidyi. Aquat Invasions 5:357–361. https://doi.org/10.3391/ai.2010.5.4.04
Malej A, Tirelli V, Lučić D et al (2017) Mnemiopsis leidyi in the northern Adriatic: here to stay? J Sea Res 124:10–16. https://doi.org/10.1016/j.seares.2017.04.010
Marchessaux G, Belloni B, Gadreaud J, Thibault D (2021) Predation assessment of the invasive ctenophore Mnemiopsis leidyi in a French Mediterranean lagoon. J Plankton Res 43:161–179. https://doi.org/10.1093/plankt/fbab002
Marchini A, Ferrario J, Sfriso A, Occhipinti-Ambrogi A (2015) Current status and trends of biological invasions in the Lagoon of Venice, a hotspot of marine NIS introductions in the Mediterranean Sea. Biol Invasions 17:2943–2962. https://doi.org/10.1007/s10530-015-0922-3
McNamara ME, Lonsdale DJ, Cerrato RM (2010) Shifting abundance of the ctenophore Mnemiopsis leidyi and the implications for larval bivalve mortality. Mar Biol 157:401–412. https://doi.org/10.1007/s00227-009-1327-6
McNamara ME, Lonsdale DJ, Cerrato RM (2013) Top-down control of mesozooplankton by adult Mnemiopsis leidyi influences microplankton abundance and composition enhancing prey conditions for larval ctenophores. Estuar Coast Shelf Sci 133:2–10. https://doi.org/10.1016/j.ecss.2013.04.019
Meredith RW, Gaynor JJ, Bologna PAX (2016) Diet assessment of the Atlantic Sea Nettle Chrysaora quinquecirrha in Barnegat Bay, New Jersey, using next-generation sequencing. Mol Ecol 25:6248–6266. https://doi.org/10.1111/mec.13918
Morello EB, Arneri E (2009) Anchovy and sardine in the Adriatic Sea—an ecological review. In: Gibson RN, Atkinson RJA, Gordon JDM (eds) Oceanography and Marine Biology An Annual Review, vol 47. Taylor & Francis, Milton Park, pp 209–256
Mutlu E (1999) Distribution and abundance of ctenophores and their zooplankton food in the Black Sea. II Mnemiopsis Leidyi Mar Biol 135:603–613. https://doi.org/10.1007/s002270050661
Oksanen J, Blanchet FG, Friendly M, et al (2019) Package “vegan”- Community ecology package. CRAN
Palmieri MG, Barausse A, Luisetti T, Turner K (2014) Jellyfish blooms in the Northern Adriatic Sea: Fishermen’s perceptions and economic impacts on fisheries. Fish Res 155:51–58. https://doi.org/10.1016/j.fishres.2014.02.021
Pansera M, Granata A, Guglielmo L, Minutoli R, Zagami G, Brugnano C (2014) How does mesh-size selection reshape the description of zooplankton community structure in coastal lakes? Estuar Coast Shelf Sci 151:221–235
Pansera M, Camatti E, Schroeder A et al (2021) The non-indigenous Oithona davisae in a Mediterranean transitional environment: coexistence patterns with competing species. Sci Rep 11:1–14. https://doi.org/10.1038/s41598-021-87662-5
Purcell JE, Shiganova TA, Decker MB, Houde ED (2001) The ctenophore Mnemiopsis in native and exotic habitats: U.S. estuaries versus the Black Sea basin. Hydrobiologia 451:145–176. https://doi.org/10.1023/A:1011826618539
Purcell JE, Uye SI, Lo WT (2007) Anthropogenic causes of jellyfish blooms and their direct consequences for humans: a review. Mar Ecol Prog Ser 350:153–174. https://doi.org/10.3354/meps07093
R Core Team (2018). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Rognes T, Flouri T, Nichols B et al (2016) VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016:1–22. https://doi.org/10.7717/peerj.2584
Roohi A, Kideys AE, Sajjadi A et al (2010) Changes in biodiversity of phytoplankton, zooplankton, fishes and macrobenthos in the Southern Caspian Sea after the invasion of the ctenophore Mnemiopsis leidyi. Biol Invasions 12:2343–2361. https://doi.org/10.1007/s10530-009-9648-4
Schneider G, Behrends G (1994) Population dynamics and the trophic role of Aurelia aurita medusae in the Kiel Bight and western Baltic. ICES J Mar Sci 51:359–367
Schroeder A, Stanković D, Pallavicini A et al (2020) DNA metabarcoding and morphological analysis—assessment of zooplankton biodiversity in transitional waters. Mar Environ Res. https://doi.org/10.1016/j.marenvres.2020.104946
Schroeder A, Pallavicini A, Edomi P et al (2021) Suitability of a dual COI marker for marine zooplankton DNA metabarcoding. Mar Environ Res 170:105444. https://doi.org/10.1016/j.marenvres.2021.105444
Shiganova TA (1998) Invasion of the Black Sea by the ctenophore Mnemiopsis leidyi and recent changes in pelagic community structure. Fish Oceanogr 7:305–310. https://doi.org/10.1046/j.1365-2419.1998.00080.x
Shiganova TA (2020) Adaptive strategies of Mnemiopsis leidyi A. Agassiz in different environments of the Eurasian seas. Marine Pollut Bull 161:111737
Shiganova TA, Bulgakova YV (2000) Effects of gelatinous plankton on Black Sea and Sea of Azov fish and their food resources. ICES J Mar Sci 57:641–648. https://doi.org/10.1006/jmsc.2000.0736
Shiganova TA, Mirzoyan ZA, Studenikina EA et al (2001) Population development of the invader ctenophore Mnemiopsis leidyi, in the Black Sea and in other seas of the Mediterranean basin. Mar Biol 139:431–445. https://doi.org/10.1007/s002270100554
Shiganova TA, Alekseenko E, Kazmin AS (2019a) Predicting range expansion of invasive ctenophore Mnemiopsis leidyi A. agassiz 1865 under current environmental conditions and future climate change scenarios. Estuar Coast Shelf Sci 227:106347
Shiganova TA, Sommer U, Javidpour J et al (2019b) Patterns of invasive ctenophore Mnemiopsis leidyi distribution and variability in different recipient environments of the Eurasian seas: a review. Mar Environ Res 152:104791
Shokralla S, Spall JL, Gibson JF, Hajibabaei M (2012) Next-generation sequencing technologies for environmental DNA research. Mol Ecol 21:1794–1805. https://doi.org/10.1111/j.1365-294X.2012.05538.x
Solidoro C, Bandelj V, Bernardi FA et al (2010) Response of the Venice Lagoon ecosystem to natural and anthropogenic pressures over the last 50 years. Coast Lagoons Crit Habitats Environ Chang. https://doi.org/10.1201/EBK1420088304
Sullivan LJ, Gifford DJ (2004) Diet of the larval ctenophore Mnemiopsis leidyi A. Agassiz (Ctenophora, Lobata). J Plankton Res 26:417–431. https://doi.org/10.1093/plankt/fbh033
Tagliapietra D, Sigovini M, Ghirardini AV (2009) A review of terms and definitions to categorise estuaries, lagoons and associated environments. Mar Freshw Res 60:497–509. https://doi.org/10.1071/MF08088
Tiselius P, Møller LF (2017) Community cascades in a marine pelagic food web controlled by the non-visual apex predator Mnemiopsis leidyi. J Plankton Res 39:271–279. https://doi.org/10.1093/plankt/fbw096
Vidjak O, Bojanić N, de Olazabal A et al (2019) Zooplankton in Adriatic port environments: indigenous communities and non-indigenous species. Mar Pollut Bull 147:133–149. https://doi.org/10.1016/j.marpolbul.2018.06.055
Waggett R, Costello JH (1999) Capture mechanisms used by the lobate ctenophore, Mnemiopsis leidyi, preying on the copepod Acartia tonsa. J Plankton Res 21:2037–2052. https://doi.org/10.1093/plankt/21.11.2037
Wells CD, Paulay G, Nguyen BN, Leray M (2022) DNA metabarcoding provides insights into the diverse diet of a dominant suspension feeder, the giant plumose anemone Metridium farcimen. Environ DNA 4:147–156
Acknowledgements
This study is based on a monitoring program within LTER-ITALY (Italian Long-Term Ecological Research Network; http://www.lteritalia.it/) that we wish to acknowledge. A.S. was supported by the joint PhD Program “Environmental Life Sciences” of the University of Trieste and the University of Udine (https://sites.google.com/view/phd-envlifesci/home) co-financed by the National Research Council, Institute of Marine Sciences (CNR ISMAR). The authors wish to thank F. Bernardi Aubry, G. Zennaro and L. Dametto for the technical support during sampling, D. Stanković for its bioinformatics support and F. Gionechetti and the whole team of the Laboratory of Applied and Comparative Genomics at the University of Trieste.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization: CE, PA, SA, PM; Formal analysis and investigation: SA, PM; Writing—original draft preparation: SA; Writing—review and editing: CE, PA, PM, SA; Funding acquisition: CE, PA.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Schroeder, A., Camatti, E., Pansera, M. et al. Feeding pressure on meroplankton by the invasive ctenophore Mnemiopsis leidyi. Biol Invasions 25, 2007–2021 (2023). https://doi.org/10.1007/s10530-023-03023-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-023-03023-5