Abstract
Due to novel or dynamic fluctuations in environmental conditions and resources, host and parasite relationships can be subject to diverse selection pressures that may lead to significant changes during and after invasion of a parasite. Genomic analyses are useful for elucidating evolutionary processes in invasive parasites following their arrival to a new area and host. Philornis downsi (Diptera: Muscidae), the avian vampire fly, was introduced to the Galápagos Islands circa 1964 and has since spread across the archipelago, feeding on the blood of developing nestlings of endemic land birds. Since its discovery, there have been significant changes to the dynamics of P. downsi and its novel hosts, such as shifting mortality rates and changing oviposition behaviour, however no temporal genetic studies have been conducted. We collected P. downsi from nests and traps from a single island population over a 14-year period, and genotyped flies at 469 single nucleotide polymorphisms (SNPs) using restriction-site associated DNA sequencing (RADSeq). Despite significant genetic differentiation (FST) between years, there was no evidence for genetic clustering within or across four sampling years between 2006 and 2020, suggesting a lack of population isolation. Sibship reconstructions from P. downsi collected from 10 Darwin’s finch nests sampled in 2020 showed evidence for shifts in reproductive behaviour compared to a similar genetic analysis conducted in 2004–2006. Compared with this previous study, females mated with fewer males, individual females oviposited fewer offspring per nest, but more unique females oviposited per nest. These findings are important to consider within reproductive control techniques, and have fitness implications for both parasite evolution and host fitness.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biological invasions by pathogenic and parasitic species are an increasing threat to the health of humans, wildlife, and ecosystems (Fisher et al. 2012; Early et al. 2016; Ricciardi et al. 2017; Jactel et al. 2020). Island populations are especially vulnerable to the impacts of invasive pathogens and parasites, given their isolation, susceptibility to disease and lack of evolved resistance (Wikelski et al. 2004; Russell et al. 2017; Brettell et al. 2021). Invasions can exert extreme selection pressures on both introduced and native species, as well as parasites and hosts (Sakai et al. 2001; Keller and Taylor 2008; Whitney and Gabler 2008; Le Roux 2021), which can influence both the success of invasions and the effectiveness of control methods (Leger and Espeland 2010; Chown et al. 2016; Mayer et al. 2021). This is particularly important for invasive parasites, which are affected by both novel environmental processes and novel hosts. Environmental variables within a parasite’s invasive range may impact its growth and life cycle, and thereby influence how parasites affect their novel hosts (e.g., Cline et al. 2014; Liu et al. 2021). Novel hosts exert unique selection pressures on invasive parasites that may lead to altered host-parasite dynamics (Telfer and Bown 2012; McIntire and Juliano 2021) via shifts in parasite strategy and population genetic structure (Brown et al. 2009; Emde et al. 2014; Beaurepaire et al. 2019). Long-term studies of parasite invasions are critical for identifying evolutionary changes within introduced populations, which may be consequential for the conservation and survival of vulnerable host species.
Long-term studies of invasion dynamics are rare, particularly for invasive species with complex life cycles, such as pathogens and parasites (Miura et al. 2006; Feis et al. 2016). Genetic analyses are useful for elucidating the invasion history, contemporary dispersal, and evolutionary shifts within introduced parasite populations (Lawson Handley et al. 2011; Cristescu 2015; Kamenova et al. 2017). Understanding these genetic processes within invasive species can inform management by identifying dispersal routes between populations, the risk of subsequent invasions, or the evolution of resistance to control measures (Sakai et al. 2001; Gaskin et al. 2011). The efficacy of biological control methods often requires rigorous and contemporary information on mating behaviour, selection patterns and demography of the target population (Roderick and Navajas 2003; Lance and McInnis 2005). For example, the Sterile Insect Technique (SIT) is a method used to suppress and eradicate insect pests (e.g., Diptera: Enkerlin et al. 2017; Lepidoptera: Dyck et al. 2021; Gato et al. 2021; Coleoptera: Himuro et al. 2022). SIT involves the release of large numbers of sterile adult males, and relies on low female remating frequency (i.e., polyandry) and low dispersal (Hendrichs et al. 2005; Lance and McInnis 2005). Dispersal rates may increase significantly during insect invasions and range expansions as selection can favour individuals with greater dispersal capacity (Travis and Dytham 2002; Lombaert et al. 2014; Dudaniec et al. 2022). Rates of female remating have been found to increase during range expansion in insects (Laugier et al. 2013; Crowther et al. 2019), potentially as a strategy to increase female fecundity and genetic diversity at colonised sites. Therefore, changes to parasite dispersal and reproductive behaviours can affect the feasibility of costly and time-consuming control measures. Changes to parasite reproductive behaviour may also affect parasite relatedness, and therefore host fitness (Buckling and Brockhurst 2008; Gleichsner et al. 2018). Kin selection predicts that high parasite relatedness leads to decreased competition between parasites and more prudent exploitation of the host, therefore higher host fitness and lower host mortality (Frank 1992; Buckling and Brockhurst 2008). Thus, understanding these changes in invasive systems is critical for both effective management, and measuring impacts on affected native host species.
The avian vampire fly, P. downsi (Diptera: Muscidae) (Dodge and Aitken, 1968), was discovered in the nests of Darwin’s finches on the Galápagos Islands in 1997 (Fessl et al. 2001). It was accidentally introduced to the Galápagos archipelago circa 1964 from its native distribution on the South American mainland (Fessl et al. 2001, 2018; Causton et al. 2006). The free-living larvae of P. downsi reside in nests of many land bird species, feeding on the blood and tissue of developing nestlings (Fessl et al. 2006b). This novel parasitism has significant effects on avian hosts on the Galápagos, causing anaemia, mortality, and permanent physical deformations (Fessl et al. 2006a; Kleindorfer and Dudaniec 2016; Katsis et al. 2021). Philornis downsi has been detected on 15 of the 17 islands across the archipelago, infesting nearly all studied passerine species (Wiedenfeld et al. 2007; Fessl et al. 2018; McNew and Clayton 2018). Due to its severe mortality effects on hosts, particularly critically endangered Darwin’s finch species (Lawson et al. 2017; Kleindorfer et al. 2021), control and eradication of P. downsi on the Galápagos is a high priority.
Previous studies have found that P. downsi shows little evidence for genetic differentiation across five islands in the Galápagos archipelago (Santa Cruz, Floreana, Isabela, Santiago and San Cristobal) and across the lowland and highland habitats (Dudaniec et al. 2008a; Koop et al. 2020), indicative of moderate to high genetic dispersal. However, given strong selection pressures associated with invasion (Le Roux 2021), genetic drift in small founding populations (Polechová 2018), and interactions with multiple novel hosts (Telfer and Bown 2012; McIntire and Juliano 2021), P. downsi is expected to show evolutionary shifts since its introduction. Further, previous research detected morphological changes in P. downsi, with an ~ 11% decrease in female body length and a ~ 26% decrease in female abdomen size between 2004–2016, the latter trait being strongly correlated with fecundity (Common et al. 2020). Similarly, host mortality rates and parasite intensity (number of parasites per nest) shifted between 2000 and 2014, increasing in some species while decreasing in others (Dudaniec et al. 2007; Cimadom et al. 2014; Kleindorfer and Dudaniec 2016). The oviposition behaviour of P. downsi also appears to be changing over recent decades. Previously, females oviposited primarily during late incubation, so their eggs hatched simultaneously with eggs of their host (O’Connor et al. 2010). In later years, larvae were detected in incubating nests of several host species, suggesting females are ovipositing earlier in the nesting cycle (Common et al. 2019). These changes in body size, intensity, and oviposition behaviour lend support that shifts in dispersal and reproductive behaviour may be occurring across time and could be measurable at the genetic level.
The mating and oviposition behaviour of P. downsi has not been measured genetically since Dudaniec et al. (2010), which used microsatellite data to explore P. downsi remating frequency and oviposition behaviour with data from 2004 to 2006. Via microsatellite-based sibship reconstructions (i.e., inference of full- and half-sibling relationships between individuals within a nest) and genetic relatedness, Dudaniec et al. (2010) found that one to six females infested a single nest, each contributing an average of ~ 5 offspring per nest. Multiple mating was common, with each female P. downsi mating with an average of 1.9 males (Dudaniec et al. 2010). In this study, we analyse P. downsi collected from traps and the nests of the small tree finch (Camarhynchus parvulus), medium tree finch (C. pauper) and small ground finch (Geospiza fuliginosa) on Floreana Island (Galápagos) with four sampling years that span a 14-year period. This time period represents 64% of the period in which the fly has been documented in finch nests (Fessl et al. 2001). With genomic dataset with higher resolution than the previous microsatellite study, derived from restriction site-associated DNA sequencing (RADSeq), we aim to examine temporal shifts in genetic structure and reproductive behaviour in P. downsi. Specifically, we examine inter-annual variation in (i) genetic divergence, (ii) effective population size, and (iii) evidence for population bottlenecks. Within infra-populations (i.e., the parasites present within each host nest), we use sibship reconstructions to examine for temporal shifts in (iv) genetic relatedness, (v) the number of female flies ovipositing per nest, (vi) the number of offspring assigned per female, (vii) the number of male flies contributing to the offspring in each nest, and (viii) the number of males assigned to the offspring of each female, (i.e., an estimate of female remating frequency). We anticipate that this information will offer an updated and temporal insight into P. downsi evolution within its invasive Galápagos range, with implications for host fitness, and future control programs.
Materials and methods
Study species
Philornis downsi is a Dipteran ectoparasite that feeds on developing nestlings (Fessl and Tebbich 2002; Kleindorfer and Dudaniec 2016; Common et al. 2019). Adult P. downsi are vegetarian and non-parasitic, feeding on decaying vegetable matter and fruit (Fessl et al. 2018). Adult females lay their eggs in incubating or brooding nests of 150 known host bird species across 10 Orders in their native distribution across mainland South America, and their invasive range in the Galápagos Islands (Fessl et al. 2018; McNew and Clayton 2018). The eggs of P. downsi usually hatch concurrently with host species hatching, and first instar larvae move to the nares of the nestlings where they feed on blood and tissues of the developing birds (Fessl et al. 2006b). Second and third instar larvae also feed within the nares, but commonly move to the base of the nest during the day, feeding both internally, within the nares and ear canals, and externally, piercing the skin of the nestling, at night (Fessl et al. 2006b; O’Connor et al. 2010). Larvae pupate in the bottom of the nest after 4–7 days of feeding, emerging as adults after 7–14 days (Kleindorfer et al. 2014; Lahuatte et al. 2016; Bulgarella et al. 2017).
Field sample collection
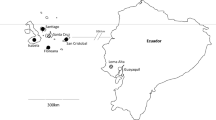
Philornis downsi adults, larvae and pupae were collected during the 2006, 2008, 2014 and 2020 Darwin’s finch (Passeriformes: Thraupidae) breeding seasons (January-April) from nests of small ground finches (G. fuliginosa), small tree finches (C. parvulus) and medium tree finches (C. pauper) on Floreana Island. The study site was located in the highlands on Floreana (01°17’S, 090°27’W, 300–400 m asl), a humid Scalesia forest at the base of Cerro Pajas volcano. Nests were located and monitored from incubation to nest termination (fledging or nestling death) following well-established field protocols (Kleindorfer et al. 2014). Nests were monitored every three days during the egg phase and every two days during the feeding phase, with nest activity determined using a borescope. Within 24 h of nest termination, nests were dismantled to collect all P. downsi specimens residing within the base of the nest. All collected specimens were identified to age class (i.e., first—third instar larvae, pupae, adult, Common et al. 2019) under a Leica MS5 dissecting microscope, preserved in 90% ethanol and stored in a −20 ℃ freezer. The number of nests per year and the number of specimens collected from each nest are presented in Table S1.
To sample adult P. downsi, McPhail traps (BioQuip Products, California, USA) were deployed in two of the four sampling years, 2014 (February 18th–April 15th) and 2020 (January 19th–March 5th). McPhail traps were baited with 150 mL of fermented papaya sugar mixture (600 g ripe papaya, 75 g white sugar, 4 L water, blended and fermented for three days; Lincango and Causton 2009). In 2014, 28 traps were placed every 15 m along four 90 m transects at two to seven metres high (m above ground). Bait lure was replaced, and specimens were collected every seven days, for a total of seven trapping events. In 2020, 32 traps were placed every 50 m in a 200 m × 100 m lattice in two study plots, and along two 200 m transects in two separate plots (Common et al. 2022). Traps were hung at four and seven metres high, to capture both male and female P. downsi, as previous research found a difference in capture height between the sexes (Kleindorfer et al. 2016). Bait lure was replaced, and specimens were collected every five days, for a total of nine trapping events.
DNA extraction, sequencing, and filtering
DNA extraction was undertaken using whole specimens of 285 P. downsi individuals (larvae, pupae and adults) by Eurofins BioDiagnostics Inc. (Wisconsin, USA). Sample sizes for each sampling year were: 2006 = 27; 2008 = 40; 2014 = 43; 2020 = 175 (Table S2). The whole specimen was extracted for larvae (n = 26) and pupae (n = 199). For adult specimens, the head, thorax, and several legs were used for extraction (n = 60). Extracted DNA concentrations were standardised to 10 ng/μl and prepared in to paired-end RAD libraries with the Sbf I restriction enzyme, similar to the method of Baird et al. (2008) and following a protocol performed by Floragenex, Inc. (Oregon, USA), as described in Text S1. Samples were sequenced on the Illumina HiSeq4000 platform.
A bioinformatics pipeline implemented by Floragenex Inc. (Oregon, USA) was used to process the raw sequencing data, call variants and filtering, as described in Text S1. A locus was retained if it was present in a minimum of 60% of individuals and had a minor allele frequency (MAF) of 0.02 and a minimum read depth > 8. Individual missingness (i.e., the percentage of missing data per individual specimen) was calculated in vcftools (Danecek et al. 2011). Due to poor DNA quality, missing data was of concern, therefore we subsampled and analysed the data for 10% and 30% missingness (Figure S1). These values for missing data were chosen to explore the effects of low and high missing data on our results, and to examine these patterns with a stringent (10%) data set and a dataset with more individuals (30%). Further filtering was conducted in Plink 1.9 (Chang et al. 2015) for each dataset to identify and remove loci deviating from Hardy–Weinberg equilibrium (HWE) at P < 0.01. To minimise the effects of linkage disequilibrium (LD), we excluded one marker from each pair with R2 > 0.5 using the window-based method.
Genetic variation and structure
To assess changes in genetic structure across the sampling period, all specimens collected from nests and traps were analysed (1) for all years pooled, and (2) separately per year (2006, 2008, 2014, 2020), and for each of the 10% and 30% missingness datasets. Samples from a given year are independent generations as previous studies found that P. downsi only survives to the next year’s breeding season, but not subsequent breeding seasons, and our sampling periods are 2–6 years apart (Causton et al. 2019; Bulgarella et al. 2022).
To avoid potential bias in allele frequencies due to highly related individuals within nests, we calculated pairwise relatedness in vcftools (Danecek et al. 2011), which calculates relatedness using the method developed by Manichaikul et al. (2010). We subsequently removed one individual from each pair that had a relatedness value > 0.25 (i.e., full siblings) from the dataset. Trapping data represents a random sample of the adult fly population and therefore all trapping samples were included in the analysis, together with those collected from nests. The sample sizes for analysing genetic structure per year were: 10% missingness: 2006 = 15, 2008 = 14, 2014 = 19, 2020 = 73; total N = 121; 30% missingness: 2006 = 20, 2008 = 22, 2014 = 28, 2020 = 101; total N = 171.
Genetic diversity parameters were calculated per year using the R package hierfstat (Goudet 2005) including observed heterozygosity (Ho), expected heterozygosity (He), and the inbreeding coefficient (FIS). Allelic richness statistics were calculated per year using the R package PopGenReport (Adamack and Gruber 2014). Pairwise FST between years was calculated using the method developed by Weir, Cockerham (1984) implemented in hierfstat, with bootstrapped 95% confidence intervals and 10,000 permutations to determine significance. An analysis of molecular variance (AMOVA) was calculated to determine the variance between years in GenoDive 3.05 (Meirmans 2020).
We examined for genetic structure using two approaches: (1) pooling all samples across the four years, and (2) separating the samples by year to determine within-year substructure. Two methods to detect genetic structure were applied. We used discriminant analysis of principal components (DAPC; Jombart et al. 2010) implemented in the package adegenet (Jombart 2008). DAPC is a model-free approach that transforms genotypes into principal components (PC), applies a discriminant analysis to the number of PCs retained to optimize among-group variation and minimize within-group variation, calculating the optimum number of clusters using Bayesian Information Criterion. Finally, we used the software STRUCTURE 2.3.4 (Pritchard et al. 2000), which utilises an iterative Bayesian clustering method, to calculate allele frequencies and individual assignments to genetic clusters. In STRUCTURE, 10 runs with 20,000 Markov Chain Monte Carlo (MCMC) iterations after a 10,000-iteration burn-in period was conducted for each K value (all years combined: K = 1–10; per year analysis: K = 1–5). The number of distinct genetic clusters, K, was selected using STRUCTURESELECTOR (Li and Liu 2018), which implements two methods for K selection. The change in K (ΔK) approach compares the rate of change of the log probability to predict the optimal K (Evanno et al. 2005). STRUCTURESELECTOR also reports four estimators MEDMEDK, MEDMEAK, MAXMEDK, and MAXMEAK to determine the optimal K (Puechmaille 2016). These methods assign subpopulations to a cluster if the mean or median individual membership coefficient was above the threshold of 0.5.
Effective population size
The effective population size (Ne) of P. downsi on Floreana Island was estimated using the program COLONY 2.0.6.7 (Jones and Wang 2010). COLONY uses a maximum likelihood method on the frequency of full- and half-sibling assignments within pairs of randomly selected individuals to determine effective population size (Wang 2009). This method was selected as it is more flexible than other Ne estimation methods, and therefore more robust in handling violations of assumptions, such as non-random mating (Wang 2009). As COLONY assumes all individuals are from the same population within a single generation, the samples were split per year and analysed separately. Highly related individuals (r > 0.25) were removed from the dataset for the purpose of Ne estimation, and the 10% and 30% missingness datasets were further analysed separately. To explore the effect of genotyping error rate on Ne estimates (e.g., allelic dropouts and missing data), all datasets were analysed using three error rates: 1%, 5% and 10% (Wang 2019; Guppy et al. 2020).
Bottleneck detection
We used the program BOTTLENECK 1.2.02 (Piry et al. 1999) to assess whether a recent population bottleneck has occurred. BOTTLENECK tests for heterozygosity excess compared to expectations under mutation-drift equilibrium (Cornuet and Luikart 1996) by comparing expected and observed heterozygosity and allele frequency per locus to determine if a locus is in heterozygosity excess or deficit, and whether this difference is significant. Data were analysed for each year (and for each of the 10% and 30% datasets) under two mutation models, the infinite allele model (IAM) and the stepwise mutation model (SMM) as these models are most suitable for biallelic SNP data (Cornuet and Luikart 1996; Kogura et al. 2011). The sign test with 1000 permutations was used to determine the significance of heterozygosity excess across loci. To detect a mode-shift in allele frequencies that may be indicative of a population bottleneck (Luikart et al. 1998), we examined the allele frequency distribution. An approximately L-shaped distribution is expected under mutation-drift equilibrium (Luikart et al. 1998).
Within-nest relatedness and sibship reconstruction
To estimate genetic relatedness and reconstruct sibships within each sampled nest, P. downsi were analysed from seven small ground finch (G. fuliginosa) and three small tree finch (C. parvulus) nests collected in 2020. Parasite intensity in sampled nests was 41.7 ± 7.6 (N = 10). Due to the small body size of first and second instar larvae, only third instar, pupae and adult specimens collected from nests were sequenced. Dudaniec et al. (2010) found relatedness did not vary with the number of individuals sampled from a nest and concluded a subsample of 10% of the infra-population was deemed sufficient to estimate in-nest relatedness. Thus, at least 10% of each infrapopulation was analysed (average 24%, range 13.3–36.0%) for a total of 98 specimens sampled from 10 nests.
Mean pairwise genetic relatedness was calculated between individuals within each nest in vcftools using the method of Manichaikul et al. (2010). The program COLONY 2.0.6.7 (Jones and Wang 2010) was used to reconstruct sibship and parentage within each infrapopulation. COLONY uses a maximum likelihood method to infer parentage and sibship structure from multi-locus genotype data (Jones and Wang 2010). We ran COLONY across the two datasets: 10% and 30% missingness. Each nest was run using an error rate of 1%, 5% and 10% to explore the effect of genotyping error on number of putative maternal and paternal genotypes identified in each nest (Wang 2019; Guppy et al. 2020). Polyandry was selected for females, as studies on other Dipterans (Arnqvist and Nilsson 2000; Dunn et al. 2005), and on P. downsi specifically (Dudaniec et al. 2010), found evidence for multiple mating. As in Dudaniec et al. (2010), monogamy was selected for males because it is highly unlikely that females that mated with the same male are ovipositing in the same nest due to random mating and a large population size.
We explored the effects of individual missingness (10% and 30%) and error rate on the number of putative maternal and paternal genotypes inferred from COLONY using ANOVA in R 4.1.0 (R Core Development Team 2020). Furthermore, to test the assumption that 10% of the infrapopulation sampled is sufficient to assess relatedness, we used multiple linear regression in R to explore the effect of the percentage of the total infrapopulation genotyped and total infrapopulation size (parasite intensity) on the number of putative female and male genotypes, and on mean pairwise individual relatedness within nests.
Results
Data filtering
A total of 127,871 SNP variants were obtained from RAD sequencing. After removing SNPs due to low quality or missing data, 7021 SNPs remained. The total number of reads and RAD clusters per dataset are presented in Table S3. After removing SNPs with a MAF < 0.02, those in linkage disequilibrium and deviating from HWE, a total of 469 SNPs remained in the 10% missingness dataset (N = 138) and 462 SNPs remained in the 30% missingness dataset (N = 188) (Table S4). For analyses that required only unrelated individuals (i.e., genetic structure), individuals with pairwise relatedness greater than 0.25 were removed, giving final sample sizes of: 10% = 121, 30% = 171.
Genetic diversity and structure
Across years combined, observed heterozygosity was lower than the expected heterozygosity (Ho = 0.290, He = 0.383, Table 1; Table S5 for results at 30% missingness) and the inbreeding coefficient (FIS) was = 0. 243 (Table 1). Within years, Ho ranged from 0.255 to 0.323 and FIS from 0.112 to 0.352 (Table 1). Mean allelic richness increased consistently from 2006 (1.91) to 2020 (2.00, Table 1). The study year 2020 was the only year with private alleles (i.e., unique alleles found only within this sampling year), with four alleles being unique to the 2020 specimens (Table 1). Pairwise FST between years ranged from 0.003 to 0.010. All pairwise FST values between years were significant to P < 0.05, suggesting some genetic differentiation between study years (Table S6a). However, analysis of molecular variance (AMOVA) comparing years showed that only 0.7% of the molecular variance was explained by year (mean FST = 0.007 ± 0.001 standard deviation (SD), P < 0.001), with 12.2% of the variation explained among individuals (mean FIS = 0.123 ± 0.017 SD, P < 0.001), and 87.1% explained within individuals (mean FIT = 0.129 ± 0.017).
For the analysis of genetic structure pooling all years using DAPC, the lowest BIC value suggested an optimal K of 1 at 10% missingness, and K = 2 at 30% missingness (Figure S2). The result of K = 2 was not explained by year, sex or age and therefore is not biologically meaningful. The most likely number of clusters of all years pooled determined by the maximum likelihood method of STRUCTURE was K = 2 for 10% missingness and K = 1 for 30% missingness, and the ΔK method of cluster selection suggested K = 3 for both datasets. However, the assignment probabilities of each individual to the second or third clusters were low (e.g., 10%: cluster 1 = 0.46 ± 0.009, cluster 2 = 0.46 ± 0.009; cluster 3 = 0.08 ± 0.01), suggesting little to no support for genetic clustering across years. DAPC analysis on data separated by year suggested an optimal K of 1 for each year, with the exception of 2020 using the 30% missingness dataset, which had an optimal K of 2 (Figure S3). The mean percentage of individual missingness was high in individuals assigned to cluster 2 compared to cluster 1 (e.g., 2020; cluster 1: 3.78% ± 0.40, N = 82; cluster 2: 21.15% ± 1.14, N = 18), which may partially explain the detection of two genetic clusters. Furthermore, the clusters did not follow any pattern of clustering by age, sex or host species. When analysing each year separately using STRUCTURE, K = 1 was also deduced with the MedK method for each year. Across all analyses, the individuals assigned to the second or third clusters were not supported by assignment probabilities and did not show a pattern across year, host species, age or sex. Therefore, the P. downsi population on Floreana shows little to no evidence for restricted gene flow or genetic divergence across the sampling period.
Effective population size and bottlenecks
Estimates of the effective population size of P. downsi on Floreana Island were low and varied across years (Table S7). Estimated Ne using COLONY was highest in 2006 (10% missingness: Ne = 420, 95% jack-knifed CI = 115–∞, Table S7) and 2014 (10% missingness: Ne = 342, 95% jack-knifed CI = 133–∞, Table S7). Estimated Ne was lowest in 2020 (10% missingness: Ne = 64, 95% jack-knifed CI = 45–93, Table S7). However, many of the upper 95% jack-knifed confidence intervals were undefined, and so we interpret the Ne values as relative estimates across years and with significant caution. Evidence for population bottlenecks was supported by a mode-shift distortion of allele frequencies from the expected L-shaped distribution in all study years (Figure S4), detected by BOTTLENECK. There was a significant deviation from expected equilibrium heterozygosity using both the IAM and the SMM (Table S8, sign test: P < 0.001) in all study years.
Infrapopulation genetic relatedness and sibship reconstruction
Mean individual pairwise relatedness varied between P. downsi infrapopulations, ranging from -0.089 to 0.205 (10%: 0.079 ± 0.016; 30%: 0.027 ± 0.030). Relatedness did not vary with the percentage of the infrapopulation genotyped (Table S9, 10% range: 13.3–36.0% genotyped per nest; 0.001 ± 0.003, P = 0.709) or infrapopulation size in either dataset (Table S9, 10%: −0.001 ± 0.001, P = 0.344). Therefore, our genetic relatedness estimates were not affected by genotyping effort per infrapopulation.
The percentage of individual missingness (i.e., 10% or 30% dataset) did not significantly affect the number of putative parental genotypes identified in each nest (Table S10, putative maternal genotypes: F1,50 = 1.63, P = 0.405; putative paternal genotypes: F1,50 = 10.21, P = 0.182). There was also no effect of error rate on number of parental genotypes (Table S10, putative maternal genotypes: F1,50 = 2.11, P = 0.636; putative males: F1,50 = 13.78, P = 0.299). The number of putative maternal or paternal genotypes did not differ with percentage of the infrapopulation genotyped or infrapopulation size (Table S11- S12). Thus, our genotyping effort was sufficient to characterise the sibship relationships within infrapopulations.
All results below are presented for the 10% missingness dataset at 5% error rate (Table 2, see Table S13 for results of 30% missingness dataset, which gave similar results). Multiple female infestations per nest were common, with an estimate of 4.88 ± 0.48 (range = 3–7) maternal genotypes estimated from each nest. Each of these maternal genotypes were associated with 14.3–33.3% of the infrapopulation. The number of P. downsi offspring assigned to each maternal genotype ranged between 1.00 and 3.00 (Table 2, mean = 1.89 ± 0.28). The number of paternal genotypes contributing to each infrapopulation was 6.13 ± 0.61 (range = 3–7). There was evidence for female multiple mating, with a mean of 1.27 ± 0.09 (range = 1.00–1. 75) paternal genotypes per maternal genotype.
Discussion
Understanding how demographic and reproductive processes change throughout parasite invasion is critical for tracking novel host-parasite interactions and the success of control techniques, especially when native host species are at risk. We found evidence that female reproductive behaviour has shifted since the 2004–2006 genetic study conducted by Dudaniec et al. (2010). Compared to 2004–2006, the number of female infestations per nest increased by ~ 70% from 2004 to 2020 (Fig. 1). The number of paternal genotypes per nest remained stable (5.44 ± 0.45 in Dudaniec et al. 2010, compared to 6.13 ± 0.61, Fig. 1), but the number of reported males per female (i.e., rate of multiple mating) decreased ~ 35%. Concordantly, the genetically assigned females in 2020 had fewer offspring assigned to each of them per nest, with a smaller percentage of the infrapopulation assigned to each maternal genotype comparing time periods (2004–2006: 43.6% ± 4.38, from Dudaniec et al. 2010; 2020: 22.0% ± 2.22). Female P. downsi are investing fewer offspring per nest, but more females are ovipositing in each nest. These changed have occurred with no significant change to parasite intensity within nests over the sampling period (2004–2006: 30.8 ± 16.5, Dudaniec et al. 2010; 2021: 43.4 ± 8.3; Common et al. 2021). Our study did not detect any genetic clustering of P. downsi on Floreana Island across our four sampling years between 2006 and 2020, however FST indicated some genetic differentiation between years. Although there was no compelling evidence for neutral genetic structure, the changes in reproductive behaviour we report suggest that P. downsi may be under selection pressures that could lead to adaptation to its novel environment and hosts.
Summary of COLONY results, comparing the results found by Dudaniec et al. (2010) from P. downsi collected in Darwin’s finch nests between 2004 and 2006 (microsatellite data, with typing errors (5%); N nests = 57; N individuals = 1020), to P. downsi collected from Darwin’s finch nests in 2020 (SNP data, 10% missingness, 5% error dataset; N nests = 10; N individuals = 77). Data are presented as mean ± standard error
Shifts in reproductive behaviour
Sibship reconstructions of P. downsi offspring collected from nests revealed temporal shifts in female reproductive behaviour over a 14-year period. Between 2004 and 2006, Dudaniec et al. (2010) found that on average, approximately three females oviposited in a single nest, each mating with an approximate average of two males and contributing an average of five offspring per nest. Compared to the previous microsatellite data that used the same analytical approach (Dudaniec et al. 2010), we found evidence that P. downsi females are mating with fewer males on average, more females are ovipositing per nest, and each female is contributing fewer offspring per nest (Fig. 1). Although the number of loci used in this study was low (469 loci at 10% missingness and 462 loci at 30% missingness), it is sufficient for reconstructing family structure as ~ 10 SNPs is equivalent to one microsatellite (Wang and Santure 2009). Thus, this genetic dataset has much higher resolution than the previous 2010 study (Dudaniec et al. 2010), which used eight microsatellites (Dudaniec et al. 2008b). The two different datasets may have differences in data resolution and therefore this may account for some of the differences in the results. However, studies evaluating the performance of SNPs versus microsatellites found high power to resolve relatedness and parentage when using 100–500 SNPs, and results between the two types of data were highly congruent (Flanagan and Jones 2019; Premachandra et al. 2019; Weng et al. 2021). Despite the congruence of the two datasets, we note that there may be inherent variation in their results due to the differences in genetic approach, thus care should be taken when comparing them. The maximum likelihood method implemented in COLONY (Jones and Wang 2010) is robust even with high inbreeding coefficients (~ 0.4; Wang and Santure 2009). It is also important to note that we were unable to genotype smaller first and second instar larvae collected from the nests, and therefore we may have missed offspring from females ovipositing later in the nesting cycle. This methodology was also used by Dudaniec et al. (2010), with only a small proportion (1%) of genotyped specimens being second instar. Hence, our estimates of the number of females per nest may be underestimated but are comparable to Dudaniec et al. (2010).
Mating of females to multiple males (i.e., polyandry) is common in insects and generally beneficial to female fecundity (Arnqvist and Nilsson 2000; Dunn et al. 2005; Abraham et al. 2011). However, increased multiple mating can decrease female longevity in insects, due to the increased cost of both mating and egg production (Arnqvist and Nilsson 2000; Kawagoe et al. 2001; Gotoh and Tsuchiya 2008). Given that females must survive during the arid, non-breeding period on the Galápagos (Causton et al. 2019; Bulgarella et al. 2022), selection for increased female longevity may be driving the observed decrease in multiple mating. The costs and benefits of polyandry in P. downsi are currently not understood. Research into the fitness of invasive species under different rates of multiple mating may further our understanding of the functional significance of changes to mating behaviour.
There are several potential explanations for, and fitness consequences of, changing oviposition behaviour in P. downsi. Female body size, in particular abdomen size, has decreased on Floreana Island between 2004 and 2016 (Common et al. 2020). Female body and abdomen size are correlated with fecundity (i.e., number of eggs laid: Pincheira-Donoso and Hunt 2017; Common et al. 2020). Smaller-bodied females may have fewer eggs to lay, and therefore may lay fewer eggs overall or per nest. Because of this, more females may be able to oviposit per nest, without increasing intensity. Female insects and parasites have been known to adjust their clutch size depending on conspecific density, competition, or host quality (Van Alphen and Visser 1990; Damman 1991; Visser and Rosenheim 1998; Díaz-Fleischer and Aluja 2003; Aluja et al. 2019). A decrease in the number of eggs laid per nest may also be due to smaller host size (Díaz-Fleischer and Aluja 2003; Dudaniec et al. 2007). Nestlings are dying younger, and younger nestlings are smaller than older nestlings (G. fuliginosa and C. parvulus nestling body mass increases approximately 1 g per day: Kleindorfer et al. unpublished data) meaning P. downsi larvae are feeding on smaller hosts than in earlier years (Kleindorfer et al. 2014). Earlier age of host death also creates an unreliable resource, as host death, often due to P. downsi parasitism, could occur before larvae are ready to pupate. The unreliability of host resources could contribute to declining parasite clutch sizes, as females oviposit fewer eggs in more nests as a form of spatial bet hedging (Hopper 1999; McLaughlin and Wasserberg 2021). Due to small sample sizes of our two host species, we were unable to analyse P. downsi reproductive behaviour between species, which is important for continued monitoring of host-parasite dynamics. Exploration of P. downsi clutch size on larger-bodied hosts, such as the Galápagos Mockingbird (Mimus parvulus), which is better able to tolerate P. downsi parasitism (Knutie et al. 2016), may reveal differences in oviposition behaviour between different host species.
The changes in oviposition behaviour we report may also have implications for avian host fitness, as their nests are forced to sustain increasingly unrelated cohorts of parasites in their nest. When the most optimal parasite strategy requires prudent exploitation of the host (i.e., when parasite transmission requires a living host or requires host resources to reach maturity), increasing parasite relatedness is predicted to decrease virulence (i.e., damage to the host, Buckling and Brockhurst 2008). This is because cooperation between highly related individuals results in indirect fitness benefits for the parasite, i.e., lowered virulence ensures the host survives, allowing for parasites to reach maturity at greater rates, therefore maximising inclusive fitness (Buckling and Brockhurst 2008). Alternatively, where cohorts are highly unrelated, more virulent parasites have a competitive advantage, selecting for increased host exploitation to increase individual parasite development and fitness (Frank 1992; Buckling and Brockhurst 2008). Increased parasite relatedness has further been found to increase parasite transmission, decrease host reproduction and survival (Davies et al. 2002; Gleichsner et al. 2018). Darwin’s finches may therefore be facing increased exploitation by P. downsi as infrapopulation relatedness decreases, potentially increasing nestling mortality and driving decreasing host age at death (Kleindorfer et al. 2014).
Genetic diversity and bottlenecks
Genetic drift or bottleneck events after species’ introductions can decrease genetic variation, with a strong negative effect on allelic richness, particularly shortly after the event occurs (Dlugosch and Parker 2008; Santos et al. 2012). Allelic richness increased in P. downsi across years from 2.06 to 2.38. Allelic richness and heterozygosity in invasive populations increase and stabilize over large time scales, with high gene flow, and with multiple introductions (Dlugosch and Parker 2008; Greenbaum et al. 2014). Observed heterozygosity in P. downsi was lower compared to Dudaniec et al. (2008a, b) from 2004 to 2006, but consistent with Koop et al. (2020) from 2015 to 2017. The decrease in heterozygosity we detect across time, particularly in earlier years (2006–2014) may be due to genetic drift after introduction or bottleneck effects. Multiple introductions of P. downsi to the archipelago from the mainland have not been investigated, so historical introductions may have occurred. As no P. downsi have been detected on the Galápagos prior to 1964, there may have been enough generations after invasion to recover from founder effects.
We found evidence for a recent bottleneck in all years, consistent with past studies (Dudaniec et al. 2008a). With low rainfall and hence a paucity of active hosts, coupled with shorter male survivorship (188 days) compared with females (265 days; Causton et al. 2019), the annual bottleneck on Floreana Island may be associated with high male (and likely female) mortality at the end of the year, prior to the onset of the next fly breeding season. Therefore, large variation in annual P. downsi populations are expected, not only because of the initial colonisation bottleneck, but also given annual reductions in population size after the host breeding season ends (Causton et al. 2019). Only relatively small numbers of adults survive to the next breeding season (Causton et al. 2019; Bulgarella et al. 2022), and therefore only a small proportion of the census population contributes to the next generation.
We report a high inbreeding coefficient (FIS) in P. downsi across years of 0.243. Studies have found that FIS can be correlated with the proportion of missing data, and when data were not missing at random (Marandel et al. 2020). Therefore, our results may be biased towards a higher FIS; although values of our 10% dataset are in line with those found on Floreana by Koop et al. (2020; FIS = 0.19). Although high, our reported FIS was also consistent with estimates for P. downsi on other islands, on the South American mainland (Koop et al. 2020), and with other invasive insect populations (Karsten et al. 2013; Kirk et al. 2013; Andersen and Mills 2018; Do et al. 2022).
Lack of genetic structure
Our study found no evidence for genetic clustering in the P. downsi Floreana Island population within and across years, suggesting significant gene flow that is consistent across time, and thus, suggests the island is not genetically isolated. Genetic differentiation (FST) was lower in this study than previously found in P. downsi populations (Dudaniec et al. 2008a; Koop et al. 2020). However, this is not unexpected as previous studies compared genetic differentiation across islands and habitats, whereas this study compared within a single island and habitat across time. Despite low FST values, pairwise comparisons between years were all significant, indicating subtle genetic differentiation across the years examined. This differentiation may be affected by the annual population bottlenecks we detected and as suggested by other studies of P. downsi (Dudaniec et al. 2008a; Causton et al. 2019). Philornis downsi shows high dispersal capability between habitats and islands (Dudaniec et al. 2008a; Fessl et al. 2018; Koop et al. 2020), and combined with frequent tourist movement (Toral-Granda et al. 2017) and strong air currents (Peck 1994) between islands, it is likely that the island populations of P. downsi are well-connected. The lack of genetic structure detected in this study could support this, however more study is required to fully understand P. downsi movement and gene flow across the entire archipelago, and how these drive or restrict genetic differentiation of separate island populations.
High gene flow may swamp selection and therefore local adaptation to specific islands or host species (Tigano and Friesen 2016; Jacob et al. 2017), but testing for selection was beyond the scope of our data resolution. It is possible that P. downsi may still be adapting to the habitat and hosts of the Galápagos Islands, given high genetic differentiation and low gene flow reported between the archipelago and mainland source populations in Ecuador (Koop et al. 2020). Evidence for gene flow between islands in more recent studies (Koop et al. 2020), and the lack of genetic structure we report on Floreana suggests that P. downsi on this island is no longer showing signs of genetic isolation from other islands, as concluded by Dudaniec et al. (2008a) who examined gene flow between three islands.
Effective population size
It is important to note the infinite values and upper bounds in our estimates of effective population size, which indicate that values of Ne could be much higher than reported (Wang 2009; Do et al. 2014). We emphasize that these results should be interpreted with caution. The small Ne values may be an artefact of our sampling design or poor data resolution. Particularly in the 2020 study year, a higher proportion of specimens were collected from nests. Although highly related individuals were removed before analysis, it is possible that this sampling design artificially inflated sibship frequencies and hence underestimated the effective population size. Larger numbers of individuals collected from traps with increased number of loci analysed could refine the estimation of effective population size.
The ‘relative’ effective population size of P. downsi on Floreana Island showed considerable variation between years. Ne was highest in 2006 and 2014 (but values for 2008 could not be obtained for the 10% missingness dataset), and lowest in 2020. Notably, this temporal decrease matches a decrease in mean in-nest P. downsi intensities (Common et al. 2021) and daily catch rates of adult P. downsi across the sampling years (2014: 0.35 ± 0.03; Kleindorfer et al. unpublished data; 2020: 0.28 ± 0.02; Common et al. 2022). The estimated values for effective population size were significantly smaller than expected, given the widespread distribution and high in-nest intensity of P. downsi. However, there are many reasons why Ne may be lower than expected. Positive selection (Charlesworth 2009), individual variation in reproductive success (Hedrick 2005), and temporal variation in population size (Hedrick 2009) can all result in smaller Ne in relation to census size, with evidence for all these factors present in P. downsi.
Missing data
Due to high rates of missing data in our dataset, we considered the effects of individual missingness on our results. To maximise the number of individuals included in the dataset, we analysed the data with a maximum of 30% missingness, and to maximise the confidence in our data, we analysed the data with a maximum of 10% missingness. Overall, we found results between the 10% and 30% individual missingness data sets were congruent. As stated previously, the inbreeding coefficient, FIS, was also higher in the 30% missingness dataset, likely because FIS is correlated with missingness where missing data is non-random (Marandel et al. 2020). Although there were differences in cluster assignments between the two datasets, the number of genetic clusters remained at one for both rates of missingness. We anticipate that missingness in the data likely drove the detection of K = 2 or 3, and therefore it is important to understand how missingness is distributed across individuals within each cluster, as this may bias results and lead to falsely identifying genetic clusters (Yi and Latch 2022). For the in-nest relatedness and sib-ship reconstruction analyses, we did not find an effect of missingness on the number of putative maternal or paternal genotypes. Therefore, although 30% missingness was sufficient in our study to explore genetic structure and relatedness, researchers must be aware of the biases associated with high missingness datasets. Where missing data is a problem, we recommend a more conservative value of 10% individual missingness to ensure high confidence of results.”
Management implications
The lack of genetic structure and divergence in P. downsi between years suggests a lack of isolation and frequent gene flow from other islands to Floreana, and possibly throughout the archipelago. Therefore, a targeted, island-specific control method for P. downsi may not be effective in the face of ongoing colonisations. Archipelago-wide strategies, such as increasing quarantine restrictions on boats moving between islands, may be more effective in the longer term. More information on gene flow and migration between human inhabited and uninhabited islands, and Galápagos and the mainland, is needed to fully determine the efficacy of targeted control methods. Changing reproductive behaviours may affect some control methods as female remating is a key factor in the success of SIT. The decrease in remating frequency reported in this study may decrease the risk of females mating with a viable male, and therefore increase SIT success (Hendrichs et al. 2005; Lance and McInnis 2005). Another potential control technique is the utilisation of synthetic gene drives; selfish transgenic elements that bias heredity with the aim to suppress or eradicate populations, such as pests or invasive species (Sinkins and Gould 2006; Bunting et al. 2022). Determining the efficacy of synthetic gene drives requires detailed information of both the genetic structure and life history of target populations (James et al. 2020). The lack of population isolation on Floreana suggests that use of a self-sustaining gene drive on this island could spread throughout the archipelago. Our study, and other recent genetic and ecological studies on P. downsi provide the knowledge base upon which potential control methods can be explored.
Conclusions
While some previous studies have examined spatial genetic variation in P. downsi (Dudaniec et al. 2008a; Koop et al. 2020), here we investigated temporal changes in genetic variation and reproductive behaviours and provided insight into the evolutionary dynamics of this invasive host-parasite system. For the Floreana Island population of P. downsi sampled between 2006 and 2020, we characterised genetic processes and changes approximately 25 years after the fly’s discovery in Darwin’s finch nests. There was evidence for subtle genetic differentiation between years, however there was limited to no evidence for genetic clustering. If gene flow was restricted to Floreana and the P. downsi population was isolated, we would expect to see genetic differentiation across the 14-year sampling period, particularly given the population bottlenecks we observe, the low estimated effective population size and previous studies indicating host-parasite co-evolution over this period. Changes to reproductive behaviour are concordant with previous morphological shifts in P. downsi, and may suggest host-parasite co-evolutionary interactions or environment adaptations that require further study. The inclusion of multiple locations and comparison to the mainland for exploring signatures of selection would provide a more comprehensive idea of how P. downsi is responding to novel habitats and hosts during its invasion. Decreased relatedness among parasites in nests could be leading to increased host exploitation, and therefore increasing rates of mortality in threatened Darwin’s finch populations. Historical and contemporary estimates of genetic migration rates could further resolve the parasite’s colonisation history and gene flow across the archipelago. This information could then be used to predict the success of targeted management strategies and how control will affect gene flow and adaptation. Continued research into temporal genetic changes in parasitic populations is imperative for planning adaptive and effective control strategies, and hence the conservation of vulnerable host species.
Data availability
Genetic datasets (filtered vcf files) and sample information used within this study can be accessed from Figshare: https://figshare.com/s/fd7a7d058daf20a498fa.
References
Abraham S, Goane L, Rull J, Cladera J, Willink E, Vera MT (2011) Multiple mating in Anastrepha fraterculus females and its relationship with fecundity and fertility. Entomol Exp Appl 141:15–24. https://doi.org/10.1111/j.1570-7458.2011.01160.x
Adamack AT, Gruber B (2014) PopGenReport: simplifying basic population genetic analyses in R. Methods Ecol Evol 5:384–387. https://doi.org/10.1111/2041-210X.12158
Aluja M, Birke A, Díaz-Fleischer F, Rull J (2019) Phenotypic plasticity in clutch size regulation among populations of a potential invasive fruit fly from environments that vary in host heterogeneity and isolation. Bull Entomol 109:169–177. https://doi.org/10.1017/S0007485318000329
Andersen JC, Mills NJ (2018) Comparative genetics of invasive populations of walnut aphid, Chromaphis juglandicola, and its introduced parasitoid, Trioxys pallidus, in California. Ecol Evol 8:801–811. https://doi.org/10.1002/ece3.3667
Arnqvist G, Nilsson T (2000) The evolution of polyandry: multiple mating and female fitness in insects. Anim Behav 60:145–164. https://doi.org/10.1006/anbe.2000.1446
Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA (2008) Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3:e3376. https://doi.org/10.1371/journal.pone.0003376
Beaurepaire AL, Moro A, Mondet F, Le Conte Y, Neumann P, Locke B (2019) Population genetics of ectoparasitic mites suggest arms race with honeybee hosts. Sci Rep 9:11355. https://doi.org/10.1038/s41598-019-47801-5
Brettell LE, Martin SJ, Riegler M, Cook JM (2021) Vulnerability of island insect pollinator communities to pathogens. J Invertebr Pathol 186:107670. https://doi.org/10.1016/j.jip.2021.107670
Brown CR, Padhi A, Moore AT, Brown MB, Foster JE, Pfeffer M, O’Brien VA, Komar N (2009) Ecological divergence of two sympatric lineages of Buggy Creek virus, an arbovirus associated with birds. Ecology 90:3168–3179. https://doi.org/10.1890/08-1731.1
Buckling A, Brockhurst MA (2008) Kin selection and the evolution of virulence. Heredity 100:484–488. https://doi.org/10.1038/sj.hdy.6801093
Bulgarella M, Quiroga MA, Boulton RA, Ramírez IE, Moon RD, Causton CE, Heimpel GE (2017) Life cycle and host specificity of the parasitoid Conura annulifera (Hymenoptera: Chalcididae), a potential biological control agent of Philornis downsi (Diptera: Muscidae) in the Galápagos islands. Ann Entomol Soc Am 110:317–328. https://doi.org/10.1093/aesa/saw102
Bulgarella M, Lincango MP, Lahuatte PF, Oliver JD, Cahuana A, Ramírez IE, Sage R, Colwitz AJ, Freund DA, Miksanek JR, Moon RD, Causton CE, Heimpel GE (2022) Persistence of the invasive bird-parasitic fly Philornis downsi over the host interbreeding period in the Galapagos islands. Sci Rep 12:2325. https://doi.org/10.1038/s41598-022-06208-5
Bunting MD, Pfitzner C, Gierus L, White M, Piltz S, Thomas PQ (2022) Generation of gene drive mice for invasive pest population suppression. Methods Mol Biol 2495:203–230. https://doi.org/10.1007/978-1-0716-2301-5_11
Causton CE, Peck SB, Sinclair BJ, Roque-Albelo L, Hodgson CJ, Landry B (2006) Alien insects: threats and implications for conservation of Galápagos islands. Ann Entomol Soc Am 99:121–143. https://doi.org/10.1603/0013-8746(2006)099[0121:AITAIF]2.0.CO;2
Causton CE, Moon RD, Cimadom A, Boulton RA, Cedeño D, Lincango MP, Tebbich S, Ulloa A (2019) Population dynamics of an invasive bird parasite, Philornis downsi (Diptera: Muscidae), in the Galapagos Islands. PLoS ONE 14:e0224125. https://doi.org/10.1371/journal.pone.0224125
Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell SM, Lee JJ (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. https://doi.org/10.1186/s13742-015-0047-8
Charlesworth B (2009) Effective population size and patterns of molecular evolution and variation. Nat Rev Genet 10:195–205. https://doi.org/10.1038/nrg2526
Chown SL, Hodgins KA, Griffin PC (2016) Biological invasions, climate change, and genomics. Crop Breeding. Apple Academic Press, 59–114
Cimadom A, Ulloa A, Meidl P, Zöttl M, Zöttl E, Fessl B, Nemeth E, Dvorak M, Cunninghame F, Tebbich S (2014) Invasive parasites, habitat change and heavy rainfall reduce breeding success in Darwin’s finches. PLoS ONE 9:e107518. https://doi.org/10.1371/journal.pone.0107518
Cline TJ, Kitchell JF, Bennington V, McKinley GA, Moody EK, Weidel BC (2014) Climate impacts on landlocked sea lamprey: implications for host-parasite interactions and invasive species management. Ecosphere. https://doi.org/10.1890/ES14-00059.1
Common LK, Dudaniec RY, Colombelli-Négrel D, Kleindorfer S (2019) Taxonomic shifts in Philornis larval behaviour and rapid changes in Philornis downsi Dodge & Aitken (Diptera: Muscidae): An invasive avian parasite on the Galápagos Islands. In: Sarwar M (ed) Life cycle and development of Diptera. IntechOpen, England
Common LK, O’Connor JA, Dudaniec RY, Peters KJ, Kleindorfer S (2020) Evidence for rapid downward fecundity selection in an ectoparasite (Philornis downsi) with earlier host mortality in Darwin’s finches. J Evol Biol 33:524–533. https://doi.org/10.1111/jeb.13588
Common LK, Sumasgutner P, Dudaniec RY, Colombelli-Négrel D, Kleindorfer S (2021) Avian vampire fly (Philornis downsi) mortality differs across Darwin’s finch host species. Sci Rep 11:15832. https://doi.org/10.1038/s41598-021-94996-7
Common LK, Sumasgutner P, Sumasgutner SC, Colombelli-Négrel D, Dudaniec RY, Kleindorfer S (2022) Temporal and spatial variation in sex-specific abundance of the avian vampire fly (Philornis downsi). Parasitol Res 121:63–74. https://doi.org/10.1007/s00436-021-07350-1
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014. https://doi.org/10.1093/genetics/144.4.2001
Cristescu ME (2015) Genetic reconstructions of invasion history. Mol Ecol 24:2212–2225. https://doi.org/10.1111/mec.13117
Crowther LP, Wright DJ, Richardson DS, Carvell C, Bourke AFG (2019) Spatial ecology of a range-expanding bumble bee pollinator. Ecol Evol 9:986–997. https://doi.org/10.1002/ece3.4722
Damman H (1991) Oviposition behaviour and clutch size in a group-feeding Pyralid moth, Omphalocera munroei. J Anim Ecol 60:193–204. https://doi.org/10.2307/5454
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, Depristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, Genomes Project Analysis G (2011) The variant call format and VCFtools. Bioinformatics 27:2156-2158. https://doi.org/10.1093/bioinformatics/btr330
Davies CM, Fairbrother E, Webster JP (2002) Mixed strain schistosome infections of snails and the evolution of parasite virulence. Parasitology 124:31–38. https://doi.org/10.1017/S0031182001008873
Díaz-Fleischer F, Aluja M (2003) Influence of conspecific presence, experience, and host quality on oviposition behavior and clutch size determination in Anastrepha ludens (Diptera: Tephritidae). J Insect Behav 16:537–554. https://doi.org/10.1023/A:1027307424150
Dlugosch KM, Parker IM (2008) Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol 17:431–449. https://doi.org/10.1111/j.1365-294X.2007.03538.x
Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR (2014) NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour 14:209–214. https://doi.org/10.1111/1755-0998.12157
Do Y, Park W-B, Park J-K, Kim CJ, Choi MB (2022) Genetic and morphological variation of Vespa velutina nigrithorax which is an invasive species in a mountainous area. Sci Rep 12:4737. https://doi.org/10.1038/s41598-022-08756-2
Dudaniec RY, Fessl B, Kleindorfer S (2007) Interannual and interspecific variation in intensity of the parasitic fly, Philornis downsi, in Darwin’s finches. Biol Conserv 139:325–332. https://doi.org/10.1016/j.biocon.2007.07.006
Dudaniec RY, Gardner MG, Donnellan S, Kleindorfer S (2008a) Genetic variation in the invasive avian parasite, Philornis downsi (Diptera, Muscidae) on the Galápagos archipelago. BMC Ecol 8:13. https://doi.org/10.1186/1472-6785-8-13
Dudaniec RY, Gardner MG, Kleindorfer S (2008b) Isolation, characterization and multiplex polymerase chain reaction of novel microsatellite loci for the avian parasite Philornis downsi (Diptera: Muscidae). Mol Ecol Resour 8:142–144. https://doi.org/10.1111/j.1471-8286.2007.01900.x
Dudaniec RY, Gardner MG, Kleindorfer S (2010) Offspring genetic structure reveals mating and nest infestation behaviour of an invasive parasitic fly (Philornis downsi) of Galápagos birds. Biol Invasions 12:581–592. https://doi.org/10.1007/s10530-009-9464-x
Dudaniec RY, Carey AR, Svensson EI, Hansson B, Yong CJ, Lancaster LT (2022) Latitudinal clines in sexual selection, sexual size dimorphism, and sex-specific genetic dispersal during a poleward range expansion. J Anim Ecol 91:1104–1118. https://doi.org/10.1111/1365-2656.13488
Dunn DW, Sumner JP, Goulson D (2005) The benefits of multiple mating to female seaweed flies, Coelopa frigida (Diptera: Coelpidae). Behav Ecol Sociobiol 58:128–135. https://doi.org/10.1007/s00265-005-0922-x
Dyck VA, Hendrichs J, Robinson AS (2021) Sterile insect technique: principles and practice in area-wide integrated pest management. Taylor & Francis, Oxfordshire United Kingdom
Early R, Bradley BA, Dukes JS, Lawler JJ, Olden JD, Blumenthal DM, Gonzalez P, Grosholz ED, Ibañez I, Miller LP, Sorte CJB, Tatem AJ (2016) Global threats from invasive alien species in the twenty-first century and national response capacities. Nat Commun 7:12485. https://doi.org/10.1038/ncomms12485
Emde S, Rueckert S, Kochmann J, Knopf K, Sures B, Klimpel S (2014) Nematode eel parasite found inside acanthocephalan cysts - a “Trojan horse” strategy? Parasit Vectors 7:504. https://doi.org/10.1186/s13071-014-0504-8
Enkerlin WR, Gutiérrez Ruelas JM, Pantaleon R, Soto Litera C, Villaseñor Cortés A, Zavala López JL, Orozco Dávila D, Montoya Gerardo P, Silva Villarreal L, Cotoc Roldán E, Hernández López F, Arenas Castillo A, Castellanos Dominguez D, Valle Mora A, Rendón Arana P, Cáceres Barrios C, Midgarden D, Villatoro Villatoro C, Lira Prera E, Zelaya Estradé O, Castañeda Aldana R, López Culajay J, Ramírez y Ramírez F, Liedo Fernández P, Ortíz Moreno G, Reyes Flores J, Hendrichs J (2017) The Moscamed regional programme: review of a success story of area-wide sterile insect technique application. Entomol Exp Appl 164:188–203. https://doi.org/10.1111/eea.12611
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Feis ME, Goedknegt MA, Thieltges DW, Buschbaum C, Wegner KM (2016) Biological invasions and host–parasite coevolution: different coevolutionary trajectories along separate parasite invasion fronts. Zoology 119:366–374. https://doi.org/10.1016/j.zool.2016.05.012
Fessl B, Tebbich S (2002) Philornis downsi– a recently discovered parasite on the Galápagos archipelago – a threat for Darwin’s finches? Ibis 144:445–451. https://doi.org/10.1046/j.1474-919X.2002.00076.x
Fessl B, Couri MS, Tebbich S (2001) Philornis downsi Dodge & Aitken, new to the Galapagos islands (Diptera, Muscidae). Stud Dipterol 8:317–322
Fessl B, Kleindorfer S, Tebbich S (2006a) An experimental study on the effects of an introduced parasite in Darwin’s finches. Biol Conserv 127:55–61. https://doi.org/10.1016/j.biocon.2005.07.013
Fessl B, Sinclair BJ, Kleindorfer S (2006b) The life-cycle of Philornis downsi (Diptera: Muscidae) parasitizing Darwin’s finches and its impacts on nestling survival. Parasitology 133:739–747. https://doi.org/10.1017/S0031182006001089
Fessl B, Heimpel GE, Causton CE (2018) Invasion of an avian nest parasite, Philornis downsi, to the Galápagos Islands: Colonization history, adaptations to novel ecosystems, and conservation challenges. In: Parker PG (ed) Disease ecology: galapagos birds and their parasites. Springer International Publishing, Cham, pp 213–266
Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ (2012) Emerging fungal threats to animal, plant and ecosystem health. Nature 484:186–194. https://doi.org/10.1038/nature10947
Flanagan SP, Jones AG (2019) The future of parentage analysis: from microsatellites to SNPs and beyond. Mol Ecol 28:544–567. https://doi.org/10.1111/mec.14988
Frank SA (1992) A kin selection model for the evolution of virulence. Proc R Soc Lond Ser B: Biol Sci 250:195–197. https://doi.org/10.1098/rspb.1992.0149
Gaskin JF, Bon M-C, Cock MJW, Cristofaro M, Biase AD, De Clerck-Floate R, Ellison CA, Hinz HL, Hufbauer RA, Julien MH, Sforza R (2011) Applying molecular-based approaches to classical biological control of weeds. Biol Control 58:1–21. https://doi.org/10.1016/j.biocontrol.2011.03.015
Gato R, Menéndez Z, Prieto E, Argilés R, Rodríguez M, Baldoquín W, Hernández Y, Pérez D, Anaya J, Fuentes I, Lorenzo C (2021) Sterile insect technique: successful suppression of an Aedes aegypti field population in Cuba. Insects. 12(5):469
Gleichsner AM, Reinhart K, Minchella DJ (2018) The influence of related and unrelated co-infections on parasite dynamics and virulence. Oecologia 186:555–564. https://doi.org/10.1007/s00442-017-4035-9
Gotoh T, Tsuchiya A (2008) Effect of multiple mating on reproduction and longevity of the phytoseiid mite Neoseiulus californicus. Exp Appl Acarol 44:185. https://doi.org/10.1007/s10493-008-9143-0
Goudet J (2005) hierfstat, a package for r to compute and test hierarchical F-statistics. Mol Ecol Notes 5:184–186. https://doi.org/10.1111/j.1471-8286.2004.00828.x
Greenbaum G, Templeton AR, Zarmi Y, Bar-David S (2014) Allelic richness following population founding events—a stochastic modeling framework incorporating gene flow and genetic drift. PLoS ONE 9:e115203. https://doi.org/10.1371/journal.pone.0115203
Guppy JL, Jones DB, Kjeldsen SR, Le Port A, Khatkar MS, Wade NM, Sellars MJ, Steinig EJ, Raadsma HW, Jerry DR, Zenger KR (2020) Development and validation of a RAD-Seq target-capture based genotyping assay for routine application in advanced black tiger shrimp (Penaeus monodon) breeding programs. BMC Genom 21:541. https://doi.org/10.1186/s12864-020-06960-w
Hedrick P (2005) Large variance in reproductive success and the Ne/N ratio. Evolution 59:1596–1599. https://doi.org/10.1111/j.0014-3820.2005.tb01809.x
Hedrick PW (2009) Genetics of populations. Jones & Bartlett Publishers, Massachusetts
Hendrichs J, Vreysen M, Enkerlin W, Cayol J, Dyck V, Robinson A (2005) Strategic options in using sterile insects for area-wide integrated pest management. In: Dyck VA, Hendrichs J, Robinson AS (eds) Sterile insect technique. Springer, Dordrecht, p 841
Himuro C, Kohama T, Matsuyama T, Sadoyama Y, Kawamura F, Honma A, Ikegawa Y, Haraguchi D (2022) First case of successful eradication of the sweet potato weevil, Cylas formicarius (Fabricius), using the sterile insect technique. PLoS ONE 17:e0267728. https://doi.org/10.1371/journal.pone.0267728
Hopper KR (1999) Risk-spreading and bet-hedging in insect population biology. Annu Rev Entomol 44:535–560. https://doi.org/10.1146/annurev.ento.44.1.535
Jacob S, Legrand D, Chaine AS, Bonte D, Schtickzelle N, Huet M, Clobert J (2017) Gene flow favours local adaptation under habitat choice in ciliate microcosms. Nat Ecol Evol 1:1407–1410. https://doi.org/10.1038/s41559-017-0269-5
Jactel H, Desprez-Loustau M-L, Battisti A, Brockerhoff E, Santini A, Stenlid J, Björkman C, Branco M, Dehnen-Schmutz K, Douma JC (2020) Pathologists and entomologists must join forces against forest pest and pathogen invasions. NeoBiota 58:107. https://doi.org/10.3897/neobiota.58.54389
James SL, Marshall JM, Christophides GK, Okumu FO, Nolan T (2020) Toward the definition of efficacy and safety criteria for advancing gene drive-modified mosquitoes to field testing. Vector-Borne and Zoonotic Diseases 20:237–251. https://doi.org/10.1089/vbz.2019.2606
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. https://doi.org/10.1093/bioinformatics/btn129
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94. https://doi.org/10.1186/1471-2156-11-94
Jones OR, Wang J (2010) COLONY: A program for parentage and sibship inference from multilocus genotype data. Mol Ecol Resour 10:551–555. https://doi.org/10.1111/j.1755-0998.2009.02787.x
Kamenova S, Bartley TJ, Bohan DA, Boutain JR, Colautti RI, Domaizon I, Fontaine C, Lemainque A, Le Viol I, Mollot G, Perga ME, Ravigné V, Massol F (2017) Chapter three—invasions toolkit: current methods for tracking the spread and impact of invasive species. In: Bohan DA, Dumbrell AJ, Massol F (eds) Advances in ecological research. Academic Press, pp 85–182
Karsten M, van Vuuren BJ, Barnaud A, Terblanche JS (2013) Population genetics of Ceratitis capitata in South Africa: implications for dispersal and pest management. PLoS ONE 8:e54281. https://doi.org/10.1371/journal.pone.0054281
Katsis AC, Colombelli-Négrel D, Common LK, O’connor JA, Dudaniec RY, García-Loor J, Kleindorfer S (2021) Nestling behaviour predicts naris deformation in Darwin’s finches parasitized by the avian vampire fly. Biol J Linn Soc. https://doi.org/10.1093/biolinnean/blab092
Kawagoe T, Suzuki N, Matsumoto K (2001) Multiple mating reduces longevity of females of the windmill butterfly Atrophaneura alcinous. Ecol Entomol 26:258–262. https://doi.org/10.1046/j.1365-2311.2001.00326.x
Keller SR, Taylor DR (2008) History, chance and adaptation during biological invasion: separating stochastic phenotypic evolution from response to selection. Ecol Lett 11:852–866. https://doi.org/10.1111/j.1461-0248.2008.01188.x
Kirk H, Dorn S, Mazzi D (2013) Worldwide population genetic structure of the oriental fruit moth (Grapholita molesta), a globally invasive pest. BMC Ecol 13:12. https://doi.org/10.1186/1472-6785-13-12
Kleindorfer S, Dudaniec RY (2016) Host-parasite ecology, behavior and genetics: a review of the introduced fly parasite Philornis downsi and its Darwin’s finch hosts. BMC Zool 1:1. https://doi.org/10.1186/s40850-016-0003-9
Kleindorfer S, Peters KJ, Custance G, Dudaniec RY, O’Connor JA (2014) Changes in Philornis infestation behavior threaten Darwin’s finch survival. Curr Zool 60:542–550. https://doi.org/10.1093/czoolo/60.4.542
Kleindorfer S, Peters KJ, Hohl L, Sulloway FJ (2016) Flight behaviour of an introduced parasite affects its Galapagos island hosts: Philornis downsi and Darwin’s finches. In: Weis JS, Sol D (eds) Biological invasions and animal behaviour. Cambridge University Press, Cambridge, pp 158–79
Kleindorfer S, Common LK, Sumasgutner P (2021) Nesting success and nesting height in the critically endangered medium tree finch (Camarhynchus pauper). Birds 2(4):427–444
Knutie SA, Owen JP, McNew SM, Bartlow AW, Arriero E, Herman JM, DiBlasi E, Thompson M, Koop JA, Clayton DH (2016) Galápagos mockingbirds tolerate introduced parasites that affect Darwin’s finches. Ecology. 97(4):940–50
Kogura Y, Seeb JE, Azuma N, Kudo H, Abe S, Kaeriyama M (2011) The genetic population structure of lacustrine sockeye salmon, Oncorhynchus nerka, in Japan as the endangered species. Environ Biol Fishes 92:539–550. https://doi.org/10.1007/s10641-011-9876-1
Koop JAH, Causton CE, Bulgarella M, Cooper E, Heimpel GE (2020) Population structure of a nest parasite of Darwin’s finches within its native and invasive ranges. Conserv Genet. https://doi.org/10.1007/s10592-020-01315-0
Lahuatte PF, Lincango MP, Heimpel GE, Causton CE (2016) Rearing larvae of the avian nest parasite, Philornis downsi (Diptera: Muscidae), on chicken blood-based diets. J Insect Sci 16(1):84
Lance D, McInnis D (2005) Biological basis of the sterile insect technique. Sterile Insect Technique. Springer, pp. 69–94
Laugier GJM, Le Moguédec G, Tayeh A, Loiseau A, Osawa N, Estoup A, Facon B (2013) Increase in male reproductive success and female reproductive investment in invasive populations of the Harlequin Ladybird Harmonia axyridis. PLoS ONE 8:e77083. https://doi.org/10.1371/journal.pone.0077083
Lawson Handley LJ, Estoup A, Evans DM, Thomas CE, Lombaert E, Facon B, Aebi A, Roy HE (2011) Ecological genetics of invasive alien species. Biocontrol 56:409–428. https://doi.org/10.1007/s10526-011-9386-2
Lawson LP, Fessl B, Hernán Vargas F, Farrington HL, Francesca Cunninghame H, Mueller JC, Nemeth E, Christian Sevilla P, Petren K (2017) Slow motion extinction: inbreeding, introgression, and loss in the critically endangered mangrove finch (Camarhynchus heliobates). Conserv Genet 18:159–170. https://doi.org/10.1007/s10592-016-0890-x
Le Roux J (2021) The Evolutionary Ecology of Invasive Species. Elsevier Science, Amsterdam
Leger EA, Espeland EK (2010) PERSPECTIVE: coevolution between native and invasive plant competitors: implications for invasive species management. Evol Appl 3:169–178. https://doi.org/10.1111/j.1752-4571.2009.00105.x
Li Y-L, Liu J-X (2018) StructureSelector: a web-based software to select and visualize the optimal number of clusters using multiple methods. Mol Ecol Resour 18:176–177. https://doi.org/10.1111/1755-0998.12719
Lincango P, Causton C (2009) Ensayos de atrayentes para la captura de la mosca parásito, Philornis downsi (Diptera: Muscidae) en las Islas Galápagos. Puerto Ayora, Galapagos, Ecuador: Charles Darwin Foundation
Liu Y, Henkel J, Beaurepaire A, Evans JD, Neumann P, Huang Q (2021) Comparative genomics suggests local adaptations in the invasive small hive beetle. Ecol Evol 11:15780–15791. https://doi.org/10.1002/ece3.8242
Lombaert E, Estoup A, Facon B, Joubard B, Grégoire JC, Jannin A, Blin A, Guillemaud T (2014) Rapid increase in dispersal during range expansion in the invasive ladybird Harmonia axyridis. J Evol Biol 27:508–517. https://doi.org/10.1111/jeb.12316
Luikart G, Allendorf FW, Cornuet JM, Sherwin WB (1998) Distortion of allele frequency distributions provides a test for recent population bottlenecks. J Hered 89:238–247. https://doi.org/10.1093/jhered/89.3.238
Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen W-M (2010) Robust relationship inference in genome-wide association studies. Bioinformatics 26:2867–2873. https://doi.org/10.1093/bioinformatics/btq559
Marandel F, Charrier G, Lamy J-B, Le Cam S, Lorance P, Trenkel VM (2020) Estimating effective population size using RADseq: effects of SNP selection and sample size. Ecol Evol 10:1929–1937. https://doi.org/10.1002/ece3.6016
Mayer M, Shine R, Brown GP (2021) Rapid divergence of parasite infectivity and host resistance during a biological invasion. Biol J Linn Soc 132:861–871. https://doi.org/10.1093/biolinnean/blaa229
McIntire KM, Juliano SA (2021) Detrimental effects of a failed infection by a co-invasive parasite on a native congeneric parasite and its native host. Biol Invasions 23:1637–1648. https://doi.org/10.1007/s10530-021-02464-0
McLaughlin LG, Wasserberg G (2021) Spatial bet hedging in sand fly oviposition: factors affecting skip oviposition in Phlebotomus papatasi sand flies. Vector-Borne Zoonotic Dis 21:280–288. https://doi.org/10.1089/vbz.2020.2737
McNew SM, Clayton DH (2018) Alien invasion: biology of Philornis flies highlighting Philornis downsi, an introduced parasite of Galápagos birds. Annu Rev Entomol 63:369–387. https://doi.org/10.1146/annurev-ento-020117-043103
Meirmans PG (2020) genodive version 3.0: easy-to-use software for the analysis of genetic data of diploids and polyploids. Mol Ecol Resour 20:1126–1131. https://doi.org/10.1111/1755-0998.13145
Miura O, Torchin ME, Kuris AM, Hechinger RF, Chiba S (2006) Introduced cryptic species of parasites exhibit different invasion pathways. Proc Natl Acad Sci USA 103:19818. https://doi.org/10.1073/pnas.0609603103
O’Connor JA, Robertson J, Kleindorfer S (2010) Video analysis of host–parasite interactions in nests of Darwin’s finches. Oryx 44:588–594. https://doi.org/10.1017/S0030605310000086
Peck SB (1994) Aerial dispersal of insects between and to islands in the Galápagos Archipelago. Ecuador Ann Entomol Soc Am 87:218–224. https://doi.org/10.1093/aesa/87.2.218
Pincheira-Donoso D, Hunt J (2017) Fecundity selection theory: concepts and evidence. Biol Rev 92:341–356. https://doi.org/10.1111/brv.12232
Piry S, Luikart G, Cornuet JM (1999) Computer note. BOTTLENECK: a computer program for detecting recent reductions in the effective size using allele frequency data. J Hered 90:502–503. https://doi.org/10.1093/jhered/90.4.502
Polechová J (2018) Is the sky the limit? On the expansion threshold of a species’ range. PLoS Biol 16:e2005372. https://doi.org/10.1371/journal.pbio.2005372
Premachandra HKA, Nguyen NH, Knibb W (2019) Effectiveness of SNPs for parentage and sibship assessment in polygamous yellowtail kingfish Seriola lalandi. Aquaculture 499:24–31. https://doi.org/10.1016/j.aquaculture.2018.09.022
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945. https://doi.org/10.1093/genetics/155.2.945
Puechmaille SJ (2016) The program structure does not reliably recover the correct population structure when sampling is uneven: subsampling and new estimators alleviate the problem. Mol Ecol Resour 16:608–627. https://doi.org/10.1111/1755-0998.12512
R Core Development Team (2020) R: A language and environment for statistical computing. R version 4.0.0. R Foundation for Statistical Computing, Vienna, Austria
Ricciardi A, Blackburn TM, Carlton JT, Dick JTA, Hulme PE, Iacarella JC, Jeschke JM, Liebhold AM, Lockwood JL, MacIsaac HJ, Pyšek P, Richardson DM, Ruiz GM, Simberloff D, Sutherland WJ, Wardle DA, Aldridge DC (2017) Invasion science: a horizon scan of emerging challenges and opportunities. Trends Ecol Evol 32:464–474. https://doi.org/10.1016/j.tree.2017.03.007
Roderick GK, Navajas M (2003) Genes in new environments: genetics and evolution in biological control. Nat Rev Genet 4:889–899. https://doi.org/10.1038/nrg1201
Russell JC, Meyer J-Y, Holmes ND, Pagad S (2017) Invasive alien species on islands: impacts, distribution, interactions and management. Environ Conserv 44:359–370. https://doi.org/10.1017/S0376892917000297
Sakai AK, Allendorf FW, Holt JS, Lodge DM, Molofsky J, With KA, Baughman S, Cabin RJ, Cohen JE, Ellstrand NC, McCauley DE, O’Neil P, Parker IM, Thompson JN, Weller SG (2001) The population biology of invasive species. Annu Rev Ecol Syst 32:305–332. https://doi.org/10.1146/annurev.ecolsys.32.081501.114037
Santos J, Pascual M, Simões P, Fragata I, Lima M, Kellen B, Santos M, Marques A, Rose MR, Matos M (2012) From nature to the laboratory: the impact of founder effects on adaptation. J Evol Biol 25:2607–2622. https://doi.org/10.1111/jeb.12008
Sinkins SP, Gould F (2006) Gene drive systems for insect disease vectors. Nat Rev Genet 7:427–435. https://doi.org/10.1038/nrg1870
Telfer S, Bown K (2012) The effects of invasion on parasite dynamics and communities. Funct Ecol 26:1288–1299. https://doi.org/10.1111/j.1365-2435.2012.02049.x
Tigano A, Friesen VL (2016) Genomics of local adaptation with gene flow. Mol Ecol 25:2144–2164. https://doi.org/10.1111/mec.13606
Toral-Granda MV, Causton CE, Jäger H, Trueman M, Izurieta JC, Araujo E, Cruz M, Zander KK, Izurieta A, Garnett ST (2017) Alien species pathways to the Galapagos islands. Ecuador PLOS One 12:e0184379. https://doi.org/10.1371/journal.pone.0184379
Travis JM, Dytham C (2002) Dispersal evolution during invasions. Evol Ecol Res 4:1119–1129
Van Alphen JJ, Visser ME (1990) Superparasitism as an adaptive strategy for insect parasitoids. Annu Rev Entomol 35:59–79. https://doi.org/10.1146/annurev.en.35.010190.000423
Visser ME, Rosenheim JA (1998) The influence of competition between foragers on clutch size decisions in insect Parasitoids. Biol Control 11:169–174. https://doi.org/10.1006/bcon.1997.0589
Wang J (2009) A new method for estimating effective population sizes from a single sample of multilocus genotypes. Mol Ecol 18:2148–2164. https://doi.org/10.1111/j.1365-294X.2009.04175.x
Wang J (2019) Pedigree reconstruction from poor quality genotype data. Heredity 122:719–728. https://doi.org/10.1038/s41437-018-0178-7
Wang J, Santure AW (2009) Parentage and sibship inference from multilocus genotype data under polygamy. Genetics 181:1579–1594. https://doi.org/10.1534/genetics.108.100214
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370. https://doi.org/10.2307/2408641
Weng Z, Yang Y, Wang X, Wu L, Hua S, Zhang H, Meng Z (2021) Parentage analysis in giant grouper (Epinephelus lanceolatus) using microsatellite and SNP markers from genotyping-by-sequencing data. Genes 12(7):1042
Whitney KD, Gabler CA (2008) Rapid evolution in introduced species, ‘invasive traits’ and recipient communities: challenges for predicting invasive potential. Divers Distrib 14:569–580. https://doi.org/10.1111/j.1472-4642.2008.00473.x
Wiedenfeld DA, Jiménez GU, Fessl B, Kleindorfer S, Carlos Valarezo J (2007) Distribution of the introduced parasitic fly Philornis downsi (Diptera, Muscidae) in the Galápagos islands. Pac Conserv Biol 13:14–19. https://doi.org/10.1071/PC070014
Wikelski M, Foufopoulos J, Vargas H, Snell H (2004) Galápagos Birds and Diseases: Invasive Pathogens as Threats for Island Species. Ecology and Society 9. http://www.jstor.org/stable/26267654
Yi X, Latch EK (2022) Nonrandom missing data can bias principal component analysis inference of population genetic structure. Mol Ecol Resour 22:602–611. https://doi.org/10.1111/1755-0998.13498
Acknowledgements
We thank the Galápagos National Park for permission to conduct research and the Charles Darwin Foundation for logistical support. We thank the community of Floreana Island for their invaluable support. We thank all volunteers, students and researchers who helped collect samples for this project across the years, with special thanks to David Arango Roldán, Çağlar Akçay, Jefferson García Loor, Mario Gallego-Abenza, Alena Hohl, Leon Hohl, Andrew Katsis, Jody O’Connor, Petra Pesak, Katharina Peters, and Verena Puehringer-Sturmayr. We thank Eurofins BioDiagnostics USA for DNA extraction and Jason Boone and Jessica Walsh at Floragenex, Inc., for DNA sequencing and technical support. This publication is contribution number 2466 of the Charles Darwin Foundation of the Galápagos Islands.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. This study was funded by the Australian Federation of University Women (SA), Australian Research Council, Earthwatch Institute, Flinders University, Galápagos Conservation Fund, Macquarie University, Max Planck Institute for Ornithology, Mohamed bin Zayed Species Conservation Fund, Royal Society for the Protection of Birds/Birdfair, Rufford Small Grant Foundation and the University of Vienna.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study design and collected the samples. Analysis and the first draft of the manuscript was completed by L.K.C. All authors commented on and edited previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no financial or non-financial competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Common, L.K., Kleindorfer, S., Colombelli-Négrel, D. et al. Genetics reveals shifts in reproductive behaviour of the invasive bird parasite Philornis downsi collected from Darwin’s finch nests. Biol Invasions 25, 563–581 (2023). https://doi.org/10.1007/s10530-022-02935-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-022-02935-y