Abstract
Genetic diversity can affect population viability and can be reduced by both acute and chronic mechanisms. Using the history of the establishment and management of two invasive rat species on Tetiaroa atoll, French Polynesia, we investigated the intensity and longevity of contrasting population bottleneck mechanisms on genetic diversity and bottleneck signal. Using microsatellite loci we show how both a chronic reduction over approximately 50 years of a Rattus exulans population caused by the arrival of its competitor R. rattus, and an acute reduction in a R. rattus population caused by a failed eradication approximately 10 years ago, caused similar magnitudes of genetic diversity loss. Furthermore, these strong bottleneck signals were in addition to the lasting signal from initial colonisation by each species many decades to centuries earlier, characterising a genetic paradox of biological invasion. These findings have implications for the study of population genetics of invasive species, and underscore how important historical context of population dynamics is when interpreting snapshots of genetic diversity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genetic diversity is considered important for, and representative of, the long-term potential for a population or species to confront and survive environmental change (Frankham et al. 2010; Songer 2018). Populations founded from larger numbers of individuals or by multiple introductions tend to have higher genetic diversity (Uller and Leimu 2011), while a relationship has consistently been demonstrated between low genetic diversity and extinction risk (e.g. Spielman et al. 2004; Frankham 2005a, b; Blomqvist et al. 2010; Menzies et al. 2012). Loss of genetic diversity is primarily mediated by fluctuations in population size, especially where these are reductions associated with population bottlenecks that are either short (acute) or long (chronic) in their duration (Frankham et al. 2010). Population bottlenecks can cause fixation of alleles and as long as the population size remains reduced, dominance of genetic drift over natural selection and elevated inbreeding rates can ultimately lead to reduced heterozygosity and inbreeding depression (Frankham et al. 2010). However, if population size is restored, genetic variability begins to increase due to low inbreeding and new mutations (Nei et al. 1975).
Biological invasions pose a genetic paradox with respect to population bottlenecks (Estoup et al. 2016). Introduced species typically colonise in small numbers, thus founding populations with low genetic diversity, and thereby assumed to have low evolutionary potential (Frankham 2005a, b). Nonetheless, they are still able to establish, spread, and adapt (Dlugosch and Parker 2008). The elements of this paradox have been addressed in several taxonomic groups, and the most common outcome of these studies is concluding that loss of genetic diversity in introduced populations is less frequent and intense than initially thought (e.g. Kolbe et al. 2004; Bossdorf et al. 2005; Roman and Darling 2007; Uller and Leimu 2011; Estoup et al. 2016). However, the amount of genetic diversity lost may depend on the nature and extent of any bottleneck. Understanding the outcomes of different types of population bottlenecks can provide new insights about invasive species’ population biology and behaviour and their potential management (Allendorf and Lundquist 2003).
Rats are one of the most widely introduced vertebrates to accompany humans: notably the Pacific rat (Rattus exulans), black rat (R. rattus) and brown rat (R. norvegicus) (Drake and Hunt 2009). Leveraging the history of invasive rat arrival and management on a remote coral atoll we investigated two different temporal intensities of bottlenecks, chronic and acute, on genetic diversity, and benchmarked them against natural variation. R. exulans colonised the entire atoll at least 100 years ago, while R. rattus only arrived in the early 1970s on a subset of the motu (Sachet and Fosberg 1983). Where R. rattus colonised, as the much larger animal and superior competitor this caused predictable suppression and changes in R. exulans body size and behaviour, although not diet (Russell et al. 2015). We evaluated (i) the chronic bottleneck impact from suppression by a superior competitor, approximately 50 years ago, on R. exulans; (ii) the acute bottleneck impact of a failed rat eradication attempt, approximately 10 years ago, on R. rattus. We hypothesise that the sustained reduction in population size from a chronic bottleneck will result in significantly greater loss of genetic diversity compared to a transient reduction in population size from an acute bottleneck.
Material and methods
Study site
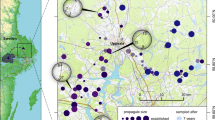
Tetiaroa (17°07′15″S 149°29′30″W) (Fig. 1) is a South Pacific atoll located 50 km north of Tahiti in the Society Islands, French Polynesia. It comprises 12 islets, locally known as “motu” (total land area 530.9 ha), arranged around a ring-shaped coral reef. The vegetation of most motu has been altered by humans, and they are now dominated by abandoned coconut plantations, with the exception of Reiono that retains a significant amount of native vegetation (Sachet and Fosberg 1983; Russell et al. 2011). Rattus exulans colonised all large motu while R. rattus only colonised the northern motu chain (i.e. Onetahi, Honuea, Tiaraunu, Tauini, Auroa, Hiraanae and Oroatera) (Russell et al. 2011).
We specifically compared populations across three motu (Table 1): Honuea (28.2 ha), historically occupied by R. exulans and R. rattus, and Reiono (21.4 ha) and Rimatuu (88.3 ha), both only occupied by R. exulans (Russell et al. 2011). Rimatuu is larger than Reiono; this provides a contrast to compare the effect of population size on the genetic diversity of R. exulans. Honuea and Reiono are similar in size, which provides a contrast to compare the genetic diversity of R. exulans in the presence of its dominant competitor R. rattus (Honuea) versus in its absence (Reiono). We also compared R. rattus populations on Honuea before and after a severe bottleneck due to an unsuccessful eradication attempt in January 2010 that left approximately 13–23 survivors from both species (Russell et al. 2015).
Rat trapping
We studied rats over five expeditions from 2009 to 2018. In July 2009 rats were kill-trapped across all motu on the atoll to obtain tissue samples (see Russell et al. 2011). In January 2010 rats were kill-trapped on Honuea over 7 nights as the first stage of an eradication attempt in a 25 m grid of 117 traps across the entire motu, and tissue samples were taken (see Russell et al. 2015). In December 2017 rats were live-trapped over 4 nights on Rimatuu and 5 nights on Reiono (50 live traps in a 10 × 5 trap-grid at 10 m spacing) but no tissue samples were taken. In August 2018 rats were kill-trapped across Reiono to obtain tissue samples prior to their successful eradication, but no density estimation was undertaken (see Samaniego et al. 2020). In November 2018 rats were live-trapped over 4 nights on Honuea (40 traps in a 10 × 4 trap-grid at 12.5 m spacing) and tissue samples were taken (Table 2). All trap locations were geolocated, and during live-trapping rats were marked with an individually numbered ear tag and released. Rat tails were collected from kill-trapped rats in July 2009 and January 2010, and from euthanised live-trapped rats in August 2018, while ear snips were collected from live-trapped rats in November 2018.
Population size analysis
Spatially explicit capture-recapture analysis using the ‘SECR’ package in R software 4.0.3 (R Core Team 2017) was used to estimate the density of each rat species on each motu (Borchers and Efford 2008). Spatially explicit capture-recapture can be used for differing trap layouts, device types, and number of trapping nights, ensuring these aspects are accounted for in the analysis. We estimated density using a full likelihood model where density and capture probability (g0) varied with session (i.e. motu or time) but sigma (σ) was constant (in particular to allow estimation of density on Honuea in January 2010 when only kill trapping was used). We assumed a half-normal detection curve and a buffer of 40 m around each trapping grid. Because data were sparse, we used initial values of density of 10, capture probability of 0.1 and sigma of 10 m. Derived density estimates were multiplied by motu area to give an indication of the population size of each rat species on each motu. We note our estimate of rat density on Reiono in December 2017 differs to that of Samaniego et al. (2020) as we excluded the effects of age and sex in our models, here preferring to estimate density directly with the full likelihood rather than deriving it from a conditional likelihood.
DNA extraction, genotyping and sequencing
Genomic DNA was extracted from tail and ear tissue samples preserved in ethanol using Qiagen DNeasy Blood and Tissue Kit following the manufacturer’s protocol.
We used 14 microsatellite markers to genotype the populations. Eleven markers were developed from R. norvegicus by Jacob et al. (1995) and have been successfully used for R. rattus (e.g. Brouat et al. 2014; Pichlmueller and Russell 2018; Gatto-Almeida et al. 2020). Some were previously also used for R. exulans (e.g. Russell et al. 2011; Amos et al. 2016): D2Rat234, D11Mgh5, D5Rat83, D7Rat13, D10Rat2, D12Rat76, D15Rat77, D16Rat81, D18Rat96, D19Mit2 and D20Rat46. Three other markers specifically designed for R. rattus characterised by Loiseau et al. (2008) (Rr14, Rr17, Rr114) were used. Markers were chosen from different chromosomes to avoid physical linkage. Forward primers were labelled with fluorescent dyes: 6-FAM, HEX, VIC and PET (Applied Biosystems).
The polymerase chain reactions (PCR) were performed as multiplex reactions in 10 µl volumes containing 1 µl of extracted DNA (> 40 ng DNA/µl), 1 × QIAGEN Multiplex PCR master mix and 0.2 µM of each primer used in the reaction. Loci were multiplexed in sets of two to seven. The thermocycler conditions were: initial heat activation of 95 °C for 15 min, 30 cycles of 94 °C for 30 s, 55 °C for 90 s and 72 °C for 60 s, and final extension of 72 °C for 20 min.
PCR products were diluted tenfold, then 1 µl of the PCR solution was mixed with 0.4 µL of GeneScan™ 600 LIZ® (Thermo Fisher Scientific) and 10 µL of Hi-Di™ Formamide (Applied Biosystems). This mixture was submitted to heat shock treatment: 95 °C for 5 min and 4 °C for 5 min. Genotyping runs were performed on an ABI3130XL (Applied Biosystems) automated sequencer and analysed using GENEIOUS prime version 2019.0.4 (https://www.geneious.com) including the Microsatellite Analysis External Plugin version 1.4.6 (Biomatters Ltd.)
Genetic analysis
For both species we used the software Micro-Checker version 2.2.3 (Van Oosterhout et al. 2004) to test for the presence of null alleles, using Bonferroni correction and 3000 randomisations. The number of alleles (Na), allelic richness (Ar), observed heterozygosity (Ho), and gene diversity within population (Hs) were calculated using FSTAT 2.9.4 (Goudet 1995). FIS and FST values were calculated according to Weir and Cockerham (1984) for each locus and population.
To determine the strength and impact of the chronic and acute bottlenecks, we compared genetic diversity between populations. To measure genetic diversity we estimated allelic richness, but due to bias caused by uneven sample sizes we used the rarefaction method with the software ADZE 1.0 for comparing allelic richness across populations while correcting for sample sizes differences (Szpiech et al. 2008). To check for bottleneck signals we calculated the M-ratio described by Garza and Williamson (2001) using Arlequin 3.5 (Excoffier et al. 2005). This index can be interpreted as the average percentage of intermediate allelic states that are occupied, considering that when a population is reduced in size, genetic drift is enhanced and allelic states will eventually be lost (Garza and Williamson 2001). Following the criteria established by Garza and Williamson (2001), populations with M < 0.68 can be assumed to have gone through a population bottleneck, with lower values indicating stronger bottlenecks. As recent bottlenecked populations generally lose rare alleles first, and those alleles contribute relatively little to expected heterozygosity, an excess of heterozygosity is also expected to occur (Cornuet and Luikart 1996). We used the software Bottleneck v.1.2.02 (Piry et al. 1999) to perform a one-tailed Wilcoxon sign rank test of heterozygosity excess under the two-phase model of mutation with 5000 iterations. As suggested by Piry et al. (1999), we used 95% single-step mutations and 5% multiple-step mutations, and a variance of 12. This test computes the expected heterozygosity under the assumption of mutation-drift equilibrium and compares it to the observed heterozygosity to establish if there is a significant heterozygosity excess (Cornuet and Luikart 1996).
Results
Population size and chronic bottleneck: Rattus exulans
Two hundred and twenty-two R. exulans were captured across the three motu and 109 individuals were genotyped: (Tables 2 and 3). The estimated population densities for R. exulans ranged from 2.2 individuals ha−1 (i.e. 62 individuals) in sympatry with R. rattus (Honuea) to 65.5 ha−1 (i.e. 1402 individuals) where the species was allopatric (Reiono) (Table 3).
Loci D11Mgh5 and D16Rat81 did not amplify in this species and were excluded from the final analysis. The genotyping success of the 12 remaining loci was 96.2% and mean number of alleles per locus across the entire atoll was 9.2. Uncorrected allelic richness and gene diversity indicated markedly lower diversity in Honuea (Ar = 3.91, Hs = 0.444) when compared to Rimatuu (Ar = 5.40, Hs = 0.536) and Reiono (Ar = 5.40, Hs = 0.637).
Micro-Checker detected an excess of homozygotes only for locus D20Rat46 in the Rimatuu population. While this can indicate the presence of null alleles, no loci presented significant FIS values (Online Resource 1). Omission of this marker had no effect on the results so we still present data based on inclusion of all 12 amplified markers. M-ratios for all three populations indicated genetic bottleneck signal, with Honuea (M = 0.289) presenting a lower index than Rimatuu (M = 0.304) and Reiono (M = 0.362). The Wilcoxon sign rank test showed no signal of heterozygosity excess for any of the three populations (p > 0.05) (Table 3).
Despite the different sizes of the motu, allelic richness accumulation curves from Rimatuu and Reiono had the same profile and these two populations accumulated distinct alleles with sample size at a higher rate than on Honuea (Fig. 2a). Furthermore, while Honuea is of a similar area to Reiono, the lower accumulation of alleles with sample size indicates that the genetic diversity of this population is indeed lower regardless of sample size artefacts. This pattern also held for 10 out of the 12 individual loci (Online Resource 2).
Acute bottleneck: Rattus rattus
One hundred and eight R. rattus individuals were captured on Honuea pre and post the eradication attempt, and 85 individuals were genotyped: 67 from Honuea before the eradication attempt (Honuea Pre) and 18 from Honuea 8 years after the population bottleneck (Honuea Post). The estimated population size of R. rattus was 3.0 individuals ha−1 (i.e. 84 individuals) in 2010 and 21.8 individuals ha−1 (i.e. 610 individuals) in 2018. A total of 80 R. rattus were captured in 2010 during the eradication attempt, which given the total estimated population of 84 individuals suggests a very small number of R. rattus survived, indicating a very strong demographic bottleneck (Table 4).
The genotyping success was 90.1% and mean number of alleles per locus (both pre and post) was 7.9. As expected, Honuea Pre showed higher indexes of diversity (i.e. number of alleles, gene diversity) compared to Honuea Post. Uncorrected allelic richness also indicated higher diversity in the population pre-bottleneck (Ar = 5.32 for Honuea Pre, Ar = 4.01 for Honuea Post). Honuea Post showed a significant and positive FIS value (Table 4) indicating an inbred population.
Micro-Checker detected an excess of homozygotes for four markers in Honuea Pre (D11Mgh5, D12Rat76, D19Mit2 and Rr14) and for six markers in Honuea Post (D12Rat76, D15Rat77, D5Rat83, Rr14, D20Rat46 and D11Mgh5). Exclusion of these loci did not affect allele accumulation curve shapes so presented curves are based on all 14 markers, and indicate significantly lower mean allelic richness in the population post bottleneck (Fig. 2b). Individual loci held this pattern for all but two loci (Online Resource 3). FIS values for D11Mgh5, D12Rat76, D15, D19Mit2 and Rr14 markers were also significant (Table S1) indicating departure from Hardy–Weinberg equilibrium (HWE) in these loci.
M-ratios for both populations indicated genetic bottleneck signal, with Honuea Post (M = 0.287) being stronger than Honuea Pre (M = 0.324). The Wilcoxon sign rank test, as observed for R. exulans, showed no signal of heterozygosity excess (p > 0.05) (Table 4).
Discussion
Understanding how small populations, especially those on islands, are able to persist despite low population sizes and concomitant low genetic diversity is central to the small population paradigm of conservation biology (Boyce 2002). Recent introductions of non-native species to islands present natural experiments to investigate this question. Several studies report that introduced species have not suffered genetic bottlenecks during the invasion process, most likely due to multiple invasion events and/or large propagule pools (Kolbe et al. 2004; Roman and Darling 2007; Uller and Leimu 2011). However, we found reductions in genetic diversity and lasting evidence of genetic bottlenecks in invasive rats following population size reductions. Contradicting our hypothesis, both a chronic reduction due to asymmetrical competition, and an acute reduction due to an eradication attempt, similarly reduced allelic diversity by about 40%.
Population size and chronic bottleneck: R. exulans
On Honuea, R. exulans experienced a chronic suppression of its population for over 50 years due to over-invasion by the larger, dominant competitor species R. rattus (Russell et al. 2015). This reduced, and likely held, the R. exulans population at around 60 individuals (2.2 individuals ha-1) (at the time of estimation in January 2010 just prior to an eradication attempt), an order of magnitude lower than that of the population size in the absence of competition estimated on nearby similar sized Reiono. However, carrying capacity may have differed slightly between motu due to habitat differences; Reiono being more forested and likely more more favourable for rats. As we predicted, the allelic richness accumulation curves showed significant differences between Honuea and Reiono due to the chronic bottleneck on the former. This conclusion is strengthened given the genetic diversity on Reiono was similar to that on Rimatuu which had also not gone through a population bottleneck. It is interesting that although Rimatuu is four times larger than Reiono, both motu showed similar genetic diversity indices. This may indicate that genetic diversity on Rimatuu was not fully enumerated, but the allelic accumulation curve for Rimatuu follows the same profile of stabilization as Reiono. Furthermore, we reaffirmed that the similarity in the accumulation curves observed between neighbouring Reiono and Rimatuu was not due to gene flow occurring between those islands (Online Resource 4) also found by Russell et al. (2011).
The M-ratio indicated strong bottleneck signals for all three R. exulans populations. Considering that the M-ratio retains information about past demographic history (Garza and Williamson 2001), we can attribute those results to the original foundation bottleneck endured by the likely small number of founders arriving on Tetiaroa atoll at least a century ago. However, the Honuea population experienced the strongest bottleneck of the three, likely from the additional chronic population suppression caused by the more recent arrival of R. rattus 50 years ago. The reduced genetic diversity of R. exulans on Honuea corresponds with other trait differences observed on the same population prior to the eradication attempt (Russell et al. 2015). These include reductions in body-size and access to optimal habitat due to being the inferior competitor, and are common observations for R. exulans throughout its invaded range (Wilmshurst et al. 2021). Changes in R. exulans body size after this period of over-invasion (Russell et al. 2015) are likely to be phenotypic and generated by suboptimal habitat selection and/or individual diet limitation and energy expenditure, although at the population level diet did not appear to change in the presence of competition (Russell et al. 2015). However, these short-term changes in phenotypic and genotypic diversity will accelerate microevolution in an island rodent population (Pergrams and Ashley 2001; van der Geer 2020).
We detected no heterozygosity excess in any of the R. exulans populations. The Honuea population was expected to show heterozygosity excess as it had been suppressed for 50 years. However, detection of this bottleneck signal is strongly dependent on the number of generations since the bottleneck happened, the severity of the bottleneck and the mutation rate of the loci being studied (Cornuet and Luikart 1996; Piry et al. 1999). Therefore, it is possible that any signal that occurred was lost during this population’s extended suppression. It therefore probably had enough time to achieve equilibrium between rare alleles and heterozygosity by the time we collected the first samples in 2010, even though the overall allelic richness remained lower from the bottleneck.
Acute bottleneck: R. rattus
For R. rattus, we predicted that the acute reduction in population size would cause a lower long-term loss of genetic diversity. According to Nei et al. (1975), after a bottleneck the loss of alleles in the first generation is drastic if the population size is small. However, if the population size reaches the original or a higher level (as occurred here), the mean number of alleles created by mutation is expected to increase. Considering that invasive rat populations typically recover to carrying capacity on islands within two years (Russell et al. 2008; Kappes et al. 2019), and the population on Honuea apparently recovered similarly based on follow-up monitoring 20 months later (J. Russell unpubl. data), we expected, this population would have recovered some of the lost genetic diversity. However, in contrast to what was expected, instead we found a significant reduction in allelic diversity in the post-eradication population, still present nearly 10 years after the bottleneck. This reduction was almost identical in size to that recorded for the chronically suppressed R. exulans population. Even though the population size after the eradication attempt was now higher than before the eradication attempt occurred, the genetic diversity remained lower than before this intervention, with indications of an inbred population (significant FIS values). The low diversity maintained suggests that the only source of new alleles for this population is through mutation rather than immigration (by swimming), and apparently this takes longer than expected to recover diversity.
The M-ratio indicated strong bottleneck signal for both R. rattus populations and, as for R. exulans, this probably also reflects the original foundation bottleneck endured in the first colonisation of Tetiaroa atoll. When the population was severely reduced by the eradication attempt in 2010, the M-ratio still would not have recovered from the original bottleneck, and then endured a new decline. This new decline might not have been as significant as the first one, once the population was already depleted of alleles. This probably explains why the M-ratio value observed on Honuea after the eradication attempt differed little from that observed before the eradication, instead reflecting the original introduction to Tetiaroa.
We also detected no heterozygosity excess in either of the R. rattus populations. However, because heterozygosity excess tests detect bottlenecks for only a given window of time after a bottleneck has been endured, and R. rattus reproduce throughout the year in the tropics (Russell and Holmes 2015), the decade that had passed since the bottleneck was induced may have caused any signal to be lost, as also found for R. exulans.
Our study also sheds further light on the important topic of what occurs following a failed rat eradication (Kappes et al. 2019). The pre-eradication total rat population size on Honuea in 2010 (both species combined) was estimated by spatially explicit capture-recapture in this study at 146 individuals (3.0 individuals ha-1), which falls within the range of the earlier removal-trapping estimate of 138–148 by Russell et al. (2015). However, from our breakdown of the number of survivors by each species, most of the Honuea eradication survivors appear to have been R. exulans, which was the more trap-averse species during the eradication trapping campaign, and is now known to be more likely to survive a multi-rat species tropical eradication (Griffiths et al. 2014). Thus, although many rats survived the eradication attempt, most of these were R. exulans, and the R. rattus on Honuea indeed experienced a very strong demographic bottleneck, one as strong as that for a chronically suppressed population. In contrast to R. exulans on Tetiaroa atoll, R. rattus exist as a meta-population (Russell et al. 2011), and after the failed eradication, the re-establishing rat population on Honuea may have constituted both survivors and reinvading R. rattus from nearby motu, as has been found elsewhere (Pichlmueller and Russell 2018). However, the post eradication sub-island trapping grid we used (in contrast to the island-wide grid for pre-eradication) was in the more amenable forest habitat in the south-west of Honuea which may have inflated our post eradication density estimate (see Fig. 3. in Russell et al. 2015).
The tail’s end
In contrast to earlier findings indicating loss of genetic diversity in introduced populations is uncommon (Kolbe et al. 2004; Bossdorf et al. 2005; Roman and Darling 2007; Uller and Leimu 2011; Estoup et al. 2016), we found bottleneck signals in both species of invasive rats on Tetiaroa atoll resulted from the original foundation events. Despite reduced genetic diversity, both populations successfully established and spread. Signals from the more recent acute and chronic bottleneck events were not detected. The heterozygosity excess test was potentially inappropriate to our time frame and/or species because, although the test did not indicate any recent bottlenecks, notable loss of allelic richness was detected. Although this can be due to the reduced genetic diversity caused by the original bottleneck, these tests have failed to detect well-known population collapses (Peery et al. 2012). Further investigations that use the heterozygosity test should consider the possibility of this index not being as sensitive as previously thought.
Our study was on neutral markers, though we can reasonably expect that loss of genetic diversity would also have occurred in markers under selection, likely leading to an overall reduction in population fitness (e.g. Chapman et al. 2009; Da Silva et al. 2009; Forstmeier et al. 2012; Ruiz-López et al. 2012).
Our study focused on invasive species, for which reducing long-term population viability through suppression is the goal, but also has important ramifications for reintroduction projects of rodents for conservation purposes, e.g. common hamsters in Europe (La Haye et al. 2017), but especially to islands. e.g. Anacapa deer mice (Ozer et al. 2011) and R. exulans to island cultural reserves in New Zealand (Ricardo et al. 2020).
Lastly, in our study the 50/500 rule seems to be supported as an appropriate yardstick for determining long-term population vulnerability or viability when considering genetic diversity (Franklin 1980). The rule states that (Ne) should not be smaller than 50 in the short term and not smaller than 500 in the long term (Jamieson and Allendorf 2012). This rule has been used as a guide to assess minimum viable Ne, aiming to avoid extreme inbreeding levels or genetic drift. On Tetiaroa atoll, R. exulans on Honuea was close to fifty individuals after the arrival of its competitor, which brought the population to the Ne limit, increasing its inbreeding levels and/or genetic drift. In addition, R. rattus on Honuea displayed higher inbreeding and genetic drift after the bottleneck when the population had recovered its demographic population size but not its allelic diversity. These are in good agreement with the idea that rat populations on Tetiaroa atoll represent a real example of genetic paradox, even though it remains unclear how long these species might naturally persist on the atoll.
Availability of data and material
The microsatellite data file is available in the Figshare digital repository and can be accessed at: https://doi.org/10.17608/k6.auckland.13661024.v1
References
Allendorf FW, Lundquist LL (2003) Introduction: population biology, evolution, and control of invasive species. Conserv Biol 17:24–30. https://doi.org/10.1046/j.1523-1739.2003.02365.x
Amos W, Nichols HJ, Churchyard T, Brooke MDL (2016) Rat eradication comes within a whisker! a case study of a failed project from the South Pacific. R Soc Open Sci 3:160110. https://doi.org/10.1098/rsos.160110
Blomqvist D, Pauliny A, Larsson M, Flodin LÅ (2010) Trapped in the extinction vortex? Strong genetic effects in a declining vertebrate population. BMC Evol Biol 10:33. https://doi.org/10.1186/1471-2148-10-33
Borchers DL, Efford MG (2008) Spatially explicit maximum likelihood methods for capture-recapture studies. Biometrics 64:377–385. https://doi.org/10.1111/j.1541-0420.2007.00927.x
Bossdorf O, Auge H, Lafuma L et al (2005) Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia 144:1–11. https://doi.org/10.1007/s00442-005-0070-z
Boyce MS (2002) Reconciling the small-population and declining-population paradigms. In: Beissinger SR, McCullough DR (eds) Population viability analysis. University of Chicago Press, Chicago, pp 41–49
Brouat C, Tollenaere C, Estoup A et al (2014) Invasion genetics of a human commensal rodent: the black rat Rattus rattus in Madagascar. Mol Ecol 23:4153–4167. https://doi.org/10.1111/mec.12848
Chapman JR, Nakagawa S, Coltman DW et al (2009) A quantitative review of heterozygosity-fitness correlations in animal populations. Mol Ecol 18:2746–2765. https://doi.org/10.1111/j.1365-294X.2009.04247.x
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014. https://doi.org/10.12968/vetn.2014.5.7.372
Da Silva A, Gaillard JM, Yoccoz NG et al (2009) Heterozygosity-fitness correlations revealed by neutral and candidate gene markers in roe deer from a long-term study. Evolution 63:403–417. https://doi.org/10.1111/j.1558-5646.2008.00542.x
Dlugosch KM, Parker IM (2008) Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol 17:431–449. https://doi.org/10.1111/j.1365-294X.2007.03538.x
Drake DR, Hunt TL (2009) Invasive rodents on islands: integrating historical and contemporary ecology. Biol Invasions 11:1483–1487. https://doi.org/10.1007/s10530-008-9392-1
Estoup A, Ravigné V, Hufbauer R et al (2016) Is there a genetic paradox of biological invasion? Annu Rev Ecol Evol Syst 47:51–72. https://doi.org/10.1146/annurev-ecolsys-121415
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform 1:47–50. https://doi.org/10.1177/117693430500100003
Forstmeier W, Schielzeth H, Mueller JC et al (2012) Heterozygosity-fitness correlations in zebra finches: microsatellite markers can be better than their reputation. Mol Ecol 21:3237–3249. https://doi.org/10.1111/j.1365-294X.2012.05593.x
Frankham R (2005a) Genetics and extinction. Biol Conserv 126:131–140. https://doi.org/10.1016/j.biocon.2005.05.002
Frankham R (2005b) Resolving the genetic paradox in invasive species. Heredity 94:385. https://doi.org/10.1038/sj.hdy.6800634
Frankham R, Ballou JD, Briscoe DA (2010) Introduction to conservation genetics, 2nd edn. Cambridge Univeristy Press, New York
Frankham R (2018) Conservation genetics. In: Fath B (ed) Encyclopedia of ecology, 2nd edn. Elsevier Publishing, pp 382–390
Franklin IR (1980) Evolutionary change in small populations. In: Conservation biology—an evolutionary-ecological perspective. Sinauer Associates, U.S.A., Sunderland, Massachusetts, pp 135–150
Garza JC, Williamson EG (2001) Detection of reduction in population size using data from microsatellite loci. Mol Ecol 10:305–318. https://doi.org/10.1046/j.1365-294x.2001.01190.x
Gatto-Almeida F, Pichlmueller F, Micheletti T et al (2020) Using genetics to plan black rat (Rattus rattus) management in Fernando de Noronha archipelago, Brazil. Perspect Ecol Conserv 18:44–50. https://doi.org/10.1016/j.pecon.2020.01.001
Goudet J (1995) FSTAT (version 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Griffiths R, Wegmann A, Hanson C et al (2014) The Wake Island rodent eradication—part success, part failure, but wholly instructive. Proc Vertebr Pest Conf 26:101–111. https://doi.org/10.5070/v426110487
Jacob HJ, Brown DM, Bunker RK et al (1995) A genetic linkage map of the laboratory rat, Rattus norvegicus. Nat Genet 9:63–69. https://doi.org/10.1038/ng0195-63
Jamieson IG, Allendorf FW (2012) How does the 50/500 rule apply to MVPs? Trends Ecol Evol 27:578–584. https://doi.org/10.1016/j.tree.2012.07.001
Kappes PJ, Bond AL, Russell JC, Wanless RM (2019) Diagnosing and responding to causes of failure to eradicate invasive rodents. Biol Invasions 21:2247–2254. https://doi.org/10.1007/s10530-019-01976-0
Kolbe JJ, Glor RE, Schettino LRG et al (2004) Genetic variation increases during a biological invasion. Nature 431:177–181. https://doi.org/10.1038/nature02807
La Haye MJJ, Reiners TE, Raedts R et al (2017) Genetic monitoring to evaluate reintroduction attempts of a highly endangered rodent. Conserv Genet 18:877–892. https://doi.org/10.1007/s10592-017-0940-z
Loiseau A, Rahelinirina S, Rahalison L et al (2008) Isolation and characterization of microsatellites in Rattus rattus. Mol Ecol Resour 8:916–918. https://doi.org/10.1111/j.1755-0998.2008.02115.x
Menzies BR, Renfree MB, Heider T et al (2012) Limited genetic diversity preceded extinction of the Tasmanian tiger. PLoS One 7:e35433. https://doi.org/10.1371/journal.pone.0035433
Nei M, Maruyama T, Chakraborty R (1975) The bottleneck effect and genetic variability in populations. Evolution 29:1–10. https://doi.org/10.1111/j.1558-5646.1975.tb00807.x
Ozer F, Gellerman H, Ashley MV (2011) Genetic impacts of Anacapa deer mice reintroductions following rat eradication. Mol Ecol 20:3525–3539. https://doi.org/10.1111/j.1365-294X.2011.05165.x
Peery ZM, Kirby R, Reid BN et al (2012) Reliability of genetic bottleneck tests for detecting recent population declines. Mol Ecol 21:3403–3418. https://doi.org/10.1111/j.1365-294X.2012.05635.x
Pergrams OWR, Ashley MV (2001) Microevolution in rodents. Genetica 112:245–246. https://doi.org/10.1023/A:1013343923155
Pichlmueller F, Russell JC (2018) Survivors or reinvaders? Intraspecific priority effect masks reinvasion potential. Biol Conserv 227:213–218. https://doi.org/10.1016/j.biocon.2018.09.020
Piry S, Luikart G, Cornuet J-M (1999) BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered 90:502–503. https://doi.org/10.1093/jhered/90.4.502
Ricardo H, Wilson DJ, Wehi PM (2020) Kiore (Rattus exulans) distribution and relative abundance on a small highly modified island. NZ J Zool 47:350–359. https://doi.org/10.1080/03014223.2020.1785515
Roman J, Darling JA (2007) Paradox lost: genetic diversity and the success of aquatic invasions. Trends Ecol Evol 22:454–464. https://doi.org/10.1016/j.tree.2007.07.002
Ruiz-López MJ, Gañan N, Godoy JA et al (2012) Heterozygosity-fitness correlations and inbreeding depression in two critically endangered mammals. Conserv Biol 26:1121–1129. https://doi.org/10.1111/j.1523-1739.2012.01916.x
Russell JC, Holmes ND (2015) Tropical island conservation: rat eradication for species recovery. Biol Conserv 185:1–7. https://doi.org/10.1016/j.biocon.2015.01.009
Russell JC, Towns DR, Clout MN (2008) Review of rat invasion biology—implications for island biosecurity. Science for Conservaton 286. Department of Conservation, Wellington, NZ
Russell JC, Caut S, Anderson SH, Lee M (2015) Invasive rat interactions and over-invasion on a coral atoll. Biol Conserv 185:59–65. https://doi.org/10.1016/j.biocon.2014.10.001
Russell JC, Faulquier L, Tonione MA (2011) Rat invasion of Tetiaroa Atoll, French Polynesia. In: Veitch CR, Clout MN, Towns DR (eds) Island invasives: eradication and management. IUCN, Gland, Switzerland, pp 118–123
Sachet M, Fosberg FR (1983) An ecological reconnaissance of Tetiaroa Atoll, Society Islands. Atoll Research Bulletin No. 275. The Smithsonian Institution, Washington, DC
Samaniego A, Griffiths R, Gronwald M et al (2020) A successful Pacific rat Rattus exulans eradication on tropical Reiono Island (Tetiaroa atoll, french polynesia) despite low baiting rates. Conserv Evid 17:12–14
Songer M (2018) Wildlife ecology. In: Fath B (ed) Encyclopedia of ecology, 2nd edn. Elsevier Publishing, pp 509–516
Spielman D, Brook BW, Frankham R (2004) Most species are not driven to extinction before genetic factors impact them. Proc Natl Acad Sci USA 101:15261–15264. https://doi.org/10.1073/pnas.0403809101
Szpiech ZA, Jakobsson M, Rosenberg NA (2008) ADZE: A rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 24:2498–2504. https://doi.org/10.1093/bioinformatics/btn478
Uller T, Leimu R (2011) Founder events predict changes in genetic diversity during human-mediated range expansions. Glob Chang Biol 17:3478–3485. https://doi.org/10.1111/j.1365-2486.2011.02509.x
van der Geer AAE (2020) Size matters: micro-evolution in Polynesian rats highlights body size changes as initial stage in evolution. PeerJ 8:e9076. https://doi.org/10.7717/peerj.9076
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538. https://doi.org/10.1111/j.1471-8286.2004.00684.x
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370. https://doi.org/10.2307/2408641
Wilmshurst JM, Ruscoe WA, Russell JC et al (2021) Family Muridae. In: King CM, Forsyth DM (eds) The handbook of New Zealand mammals, 3rd edn. Otago University Press, pp 161–240
Acknowledgements
Thanks to the Tetiaroa Society and The Brando resort for facilitating research on Tetiaroa atoll for the past decade, especially Neil Davies, Frank Murphy, Nicolas Leclerc and Moana Le Rohellec.
Funding
Open Access funding enabled and organized by CAUL and its institutions. FGA was funded by Brazilian Ministry of Science and Technology CAPES (Grant No. 88881.189218/2018-01), FP was funded by NZ BioHeritage National Science Challenge (Grant No. 1617-44-033), TWB was funded by European Union's Horizon 2020 research and innovation programme Marie Skłodowska-Curie fellowship (Grant No. 747120), AS was funded by Island Conservation and RSPB, JCR was funded by FRST NZ S&T postdoctoral fellowship (Grant No. UCAL 0801).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Ethical approval
Research was conducted in 2009 and 2010 under UC Berkeley Animal Care and Use Committee protocol R332-0111, and in 2017 and 2018 under University of Auckland Animal Ethics Committee approval R1677.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gatto-Almeida, F., Pichlmueller, F., Bodey, T.W. et al. The tails of two invasive species: genetic responses to acute and chronic bottlenecks. Biol Invasions 24, 3263–3273 (2022). https://doi.org/10.1007/s10530-022-02844-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-022-02844-0