Abstract
Understanding the distribution of invasive species and their reproductive area is crucial for their managements after invasion. While catch and observation surveys are still embraced, environmental DNA (eDNA) has been increasingly utilized as an efficient tool for identifying these species in the wild. In this study, we developed a Bufo-specific eDNA assay for detecting an invasive, toxic, and terrestrial toad species Bufo japonicus formosus in Hokkaido, Japan, and applied it to their reproductive area at watershed scale. The eDNA assay was field-validated in ponds where B. japonicus were observed, as well as in rivers downstream of the reproductive ponds. Thus, the assay provided us an opportunity to screen watersheds that include their reproductive area by collecting downstream water samples. Applying it to the Ishikari river basin, the largest river basin in Hokkaido (c.a., 14,330 km2), we detected toad eDNA at 32 out of 73 sampling sites. They are composed of eleven sites with species observation records nearby (all the sites with observation records within a 500 m radius) and 21 sites without such records. And those eDNA detections were from twelve out of 31 river systems in the entire river basin. A Bayesian, multiscale occupancy model supported high eDNA detectability among those sites. These results suggest that the eDNA assay can efficiently estimate the presence of reproductive area of the terrestrial toad even from a distant downstream of the watershed, and that it provides a powerful means of detecting new reproductive area and monitoring further spread of invasive species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Difficulties in monitoring of current distribution of invasive species, especially at the front-line of their invasions, are one of the biggest issues in biodiversity management. To address this issue, detections of the presence of the invasive species through visual and acoustic assessments have been employed for a variety of taxa including amphibians (e.g., Boback et al. 2020; Diamond and Ross 2020). However, they are often ineffective to grasp distribution and reproductive area of the invasive species because of many restrictions (large potential distributions, high labour cost, difficulty in species identification, especially at their early developmental stages, etc.) (Lepczyk 2005; Ficetola et al. 2008). Therefore, developing effective means of detecting them in the wild is urgently required for better understanding of the current distribution and reproductive area of invasive species and for the conservation of native fauna (Martinez et al. 2020).
Environmental DNA (eDNA) techniques have been developed rapidly since they were first applied to American bullfrog (Lithobates catesbeianus) in Europe in 2008 (Ficetola et al. 2008). In particular, eDNA techniques based on river water samples are useful for screening the presence of focal species at large spatial scales because eDNA can be carried from their actual habitat to downstream by the river current (Deiner et al. 2016). In addition, large watershed systems are commonly composed of lotic and lentic waterbodies such as rivers and ponds, respectively, that are inter-connected spatiotemporally. Therefore, eDNA from pond-dwelling species can be presumably detected by collecting river water at distant downstream, although empirical evidence to support this idea is scarce at best (Deiner et al. 2016; Bista et al. 2017).
Eastern-Japanese common toad (Bufo japonicus formosus) is one of such pond-dwelling species at its egg and larval stages. It is a subspecies of Japanese common toad (B. japonicus japonicus), and both subspecies are non-native to Hokkaido, the second largest island of Japan. Note that Bufo j. formosus is the only Bufo species known to inhabit in Hokkaido at the moment. Bufo j. formosus was introduced from mainland Japan into Hokkaido as pets several decades ago (Sarashina and Yoshida 2015; Naito and Shiga 2016). After the introduction, B. j. formosus were repeatedly released into Hokkaido especially in Ishikari river basin (c.a. 14,330 km2 basin, length of mainstream: c.a. 268 km) (Ministry of Land, Infrastructure, Transport and Tourism of Japan 2008). This species has a neurotoxin (bufotoxin) in their body through their life cycle, and it has been reported that their embryos and hatchlings have lethal toxic effects on larvae of native amphibian species in Hokkaido (i.e., Hokkaido brown frog Rana pirica, Hokkaido salamander Hynobius retardatus) (Kazila and Kishida 2019; Oyake et al. 2020; Okamiya et al. 2021). The toxic effects (Kazila and Kishida 2019) suggest the potential threats of B. j. formosus as a substantial cause of declines of the native predators in invaded regions.
Bufo j. formosus spend most of their life in terrestrial habitats (e.g., forests and mountains) after metamorphosis. The adult individuals get close to waterbodies only in their reproductive season in spring (April to May in Hokkaido), and their egg and larval stages in May to June are their primary aquatic life stages. For controlling their further spread, identification of their reproductive area is crucial. However, identification of the actual reproductive ponds is difficult because of the following reasons. First, there is a large number of potential reproductive ponds. The adult toads are known to utilize various types and sizes of waterbodies such as natural and artificial ponds (Kusano et al. 1995), and they are located widely along rivers and their branches. Second, B. j. formosus breed within a few days, and they leave reproductive ponds immediately after breeding (Ishii et al. 1989; Kusano et al. 1995). Third, they are cryptic in the reproductive ponds. Adult’s mating call is weak, so that one can hear it only at the peak of their reproductive season with many adults. Moreover, the adult toads deposit their egg clutches on the bottom of reproductive ponds, making it difficult to find them from the surface (Kishida O, unpublished data).
In this study, we aimed to develop a Bufo-specific eDNA assay that works not only with the water samples collected directly from their reproductive ponds, but also with river water samples collected from a distant downstream of their reproductive ponds. It allows us to estimate the presence of B. japonicus reproductive area in the watershed without spotting each reproductive pond (hereafter, B. japonicus represents both species B. j. formosus and B. j. japonicus). We also aimed to estimate the current reproductive areas of the terrestrial toad in a wide-range of the river basin at watershed scale. To achieve these objectives, we set three goals: 1) establishing a Bufo-specific eDNA detection assay, 2) verifying its detectability at their reproductive ponds and the downstream of these ponds during their larval period, and 3) applying it to river water samples from 31 river systems within the Ishikari river basin. Given the eDNA detections in the river water samples, a Bayesian, multiscale occupancy modelling approach (EDNAOCCUPANCY, Dorazio and Erickson 2018) was employed for evaluating eDNA capture/detection probabilities and their covariates including the distance from observation record, water temperature, and conductivity.
Materials and methods
Development of the eDNA assay for detecting Bufo-specific DNA
There are three Bufo species in Japan (B. japonicus [B. j. formosus and B. j. japonicus], B. gargarizans, and B. torrenticola), although none of them are native to Hokkaido (Matsui 1984; Sarashina and Yoshida 2015). Because of their genetic similarities, complexities in taxonomic classification, and hybridization among species (including the three Bufo species in Japan and B. bankorensis in Taiwan, Kawamura et al. 1980; Igawa et al. 2006), we developed genus-specific eDNA primers and probe, instead of species- or subspecies-specific ones. We first collected mitochondrial DNA (mtDNA) sequence information for the 12S ribosomal RNA region in the NCBI nucleotide database. They included 174 mtDNA sequences from six Bufo species (B. japonicus, B. gargarizans, B. torrenticola, B. bankorensis, B. bufo, and B. stejnegeri) and 14 other amphibian genera (Table S1). These sequences were aligned using BIOEDIT (https://bioedit.software.informer.com/), and primers were designed using PrimerQuest (https://sg.idtdna.com/Primerquest/Home/Index) and OrigoAnalyzer (https://sg.idtdna.com/calc/analyzer). We set the following criteria for primer selection, obtaining 5'–CCTTGTGGTCAGTTATTACTTACTA–3' as a candidate of forward primer and 5'–TAGGTGTCTTGGGCGGAT–3' as a candidate of reverse primer: (1) no mismatch with Bufo DNA sequences in the database, (2) at least one mismatch within five base pairs at the 3'–end of each primer for the other species’ DNA sequences, (3) GC content being 35–65% for the primer binding sequences, (4) the size of primers being 17–25 bp, (5) the size of PCR amplicon being less than 150 bp (actual amplicon size: 124 bp) including the primer binding sequences, and (6) an annealing temperature being ≧ 58 °C. The probe (5'–FAM–ATTAGGGACAAGGAGCCGGTATCAGG–TAMRA–3') was then designed on the expected B. japonicus amplicon sequences by these primers so that the probe binding sequences should have one or more mismatches with the aligned sequences of non-Bufo species at the same time.
After the designing of the primers and probe, we tested their performance using DNA extracted from tissue samples using DNeasy Blood and Tissue kits (Qiagen, Hilden, Germany) and following its extraction protocol. After DNA extraction, we conducted quantitative real-time PCR (qPCR) amplifications (See DNA extraction and detection for more details) with the tissue-oriented DNA from eight amphibian species that were known to be distributed in Hokkaido, B. j. formosus, R. pirica, H. retardatus, Dryophytes japonica, Glandirana rugosa, Lithobates catesbeianus, Pelophylax nigromaculatus, and P. porosus (Natural Environmental Division of Hokkaido 2015). There is one more amphibian species in Hokkaido (Siberian salamander Salamandrella keyserlingii), but we could not obtain the tissue sample from this species, and their DNA sequence on the database (Accession No. AY593113) had three and four mismatches in the forward and the reverse primers, respectively. Before the performance test, we measured the DNA concentrations of the tissue-oriented DNA (template DNA for qPCR) from one individual per species using Qubit® 3.0 Fluorometer (Thermo Fisher Scientific, Inc., Massachusetts, USA). The MIQE checklist (Bustin et al. 2009) for quantitative real-time PCR assays quality is provided in Table S2.
Sampling sites
Field sampling for eDNA analysis was conducted in the Ishikari river basin from 2018 to 2019. To verify the applicability of our eDNA assay to environmental water, we first collected water samples from eight ponds around Asahikawa on June 14th, 2018 (Naidaibu pond, Naidaibu Masuda pond, Ino experimental pond, Ino Okunoike pond, Ubun gate mountain side pond, Ubun gate river side pond, Ubun up weir pond and Ubun down weir pond). In those ponds, we observed larvae of B. japonicus at the time of water samplings (Table S3). In addition, we collected river water samples from two branches of the Ishikari river (Ino river and Kitanosawa river), of which watersheds included reproductive ponds of B. japonicus. The water collection sites on the two rivers were located 2000 m and 2500 m apart from the B. japonicus reproductive ponds (i.e., ponds with B. japonicus eggs in the same year of water collection) in Ino river and Kitanosawa river, respectively, according to the observations reported by Takai K. and Kishida O. (unpublished data) and Natural Environmental Division of Hokkaido (2020). These sites were within 500 m radius from public observation records of B. japonicus as well (Table S3), indicating high frequencies of their reproductive activities in the ponds along the rivers. At the water collection sites, we conducted three temporal samplings per site from the beginning to the end of June 2019 (Ino Riv. DS and Kitanosawa Riv.) in order to cover their reproductive season and egg/larval stages of their offspring. For the application on the wide-range screening of their reproductive area, we selected additional 71 sites from 29 river systems in the Ishikari river basin and collected river water samples from June 14th to 15th in 2018 and from June 5th to 26th in 2019 (Table S3). Including the two river sites for verification above, the number of sampling sites for the wide-range screening were 25 in 2018 and 48 in 2019 (73 from 31 river systems in total). In most of the sampling sites, we measured water temperature and conductivity using Pro30 Conductivity Meter (YSI Incorporated, Ohio, USA) for further analyses below (Table S3).
Environmental DNA sample collection
Water filtration was carried out as described in Yatsuyanagi et al. (2020) with minor modifications. In brief, surface water samples were collected using a new plastic bag or a bleached plastic bucket, and filtered using a Sterivex™–HV filter cartridge with a pore size of 0.45 μm (Merck Millipore, Inc.) and a sterile 50 mL syringe (TERUMO, Inc., Tokyo, Japan) or a Masterflex® (Yamato Scientific Co., Ltd., Tokyo, Japan). We filtered 250 mL or 500 mL environmental water per filter, depending on the water turbidity (Table S3). After filtration, we added 2 mL RNAlater (Thermo Fisher Scientific, Inc., Massachusetts, USA) into the Sterivex cartridge for DNA preservation. At each sampling site we collected two replicated filter samples. After finishing the day’s sampling, we prepared negative control samples by filtering 500 mL pure water with the same manner to the other samples. All sampling equipment was bleached by soaking in a 10% sodium hypochlorite solution and rinsed with sterilized distilled water after each sampling. All filtered samples were stored around -20 °C at field stations and at -80 °C in the lab until further analyses.
Environmental DNA extraction and amplification
Environmental DNA extractions were conducted using DNeasy Blood and Tissue kits (Qiagen, Hilden, Germany) following Yatsuyanagi et al. (2020) and eluted in 100 uL AE buffer. The final volume of the extracted DNA was 100 µL/sample. In the DNA extraction step, an extraction blank consisting of a new Sterivex™–HV filter processed using the same extraction protocol as field samples was included. Quantitative real-time PCR were carried out in 20 µL volumes with 200 nM of Bufo specific primer and probe in a Brilliant III Ultra-Fast qPCR Master Mix with Low ROX (Agilent Technologies, Inc.) and 2 µL of the extracted DNA as a template. Triplicates PCR reactions × two filter samples per site were carried out on Mx3000P v4.10 (Agilent Technologies, Inc.). Thermal-cycling regime was 95 °C for 3 min, followed by 50 cycles of 95 °C for 10 s and 60 °C for 20 s. For the quantification of environmental samples, we applied synthetic linear DNA of the target region containing 2.0 × 101, × 102, × 103, × 104 and × 105 copies per tube as standards in all qPCR assays (n = 10, average R2 = 0.96, SD = 0.02, and average PCR efficiency = 1.10, SD = 0.21). In addition, sterilized distilled water samples were applied in all qPCR assays as PCR negative controls.
In a spike-in test for evaluating a potential effect of PCR inhibitors, we selected seven sampling sites (14 samples) without eDNA detection, but with observation record within a 1000 m radius (Chubetsu Riv. MS1, Chubetsu Riv. MS2, Chitose Riv. DS, Toyohira Riv. US, Subetsu Riv., Biei Riv. DS, Biei Riv. MS, see Results and Table S3). The synthetic linear DNA of 1.0 × 104 copies per tube was added into the samples after confirming no DNA detection from the original samples.
Positive detection was defined as at least one detection in six PCR replicates per site (two filters/site and three qPCR replicates/filter), and lowest detection limit was set at 0.01 copy per 2 µL, following Mizumoto et al. (2020). We had three temporal samples in Ino Riv. DS and Kitanosawa Riv., so we assumed as the positive detection sites when we had at least one positive detection samples out of three temporal samples in each site. Estimated DNA concentrations were represented as per 1000 mL for adjusting the variation in filtered water volume per sample.
Analyses on the wide-range application
For analyzing the eDNA detection results in river water samples, we referred public observation records of B. japonicus reported by Natural Environmental Division of Hokkaido (2020). In this study, we defined a sampling site with observation record nearby when the site had at least one observation record within a 500 m radius. By this definition, we found seven sites with observation record nearby. In addition, we included four additional sampling sites (Ino Riv. US, Naidaibu Riv. MS-3, Naidaibu Riv. DS and Ubun Riv.) with observation record nearby because those sites were located in a 500 m radius from the ponds where we ourselves observed larvae of B. japonicus on the same days as the water samplings. In total, we had eleven sites with observation record nearby and 62 sites without (Table S3). We then tested a null hypothesis that there is no relationship between eDNA detections and observation records within a 500 m radius using Fisher’s exact test. Statistical analyses were performed using R3.6.1 (R core team. 2020).
Based on a Bayesian multiscale occupancy model, we further investigated our eDNA detection probabilities. Using EDNAOCCUPANCY (Dorazio and Erickson 2018) with two filter samples per site and three qPCR replicates/sample, three probabilities were estimated: a site occupancy probability (ψ), a conditional capture probability of a filter sample given the occupancy at the site (θ), and a conditional detection probability of a qPCR replicate given the capture in the sample (ρ). In the analysis, three potential covariates, distance between sampling site and the closest observation record of toad (Dist), water temperature at the sampling (Temp), conductivity of river water at the sampling (Cond) were included. In total, we constructed eight models hypothesizing effects of Dist and Temp on the capture probability and an effect of Cond on detection probability (See Sengupta et al. (2019) for more details). All models were fitted by running a Markov chain Monte Carlo (MCMC) algorithm in EDNAOCCUPANCY for 110,000 iterations after a burn-in of 10,000 iterations. The models were then ranked according to the posterior-predictive loss criterion (PPLC) under squared-error loss (Gelfand and Ghosh 1998) and the widely applicable information criterion (WAIC, Watanabe 2010, 2013). Note that two sampling sites (Uryu Riv. MS & DS) were excluded from the analyses because of no Temp and Cond records at these sites, resulting in 71 sampling sites in the analysis.
Results
Establishment of the Bufo-specific eDNA assay
In silico evaluation of the selected primers suggested that Bufo was the only genera from which DNA sequences had no mismatch within five base pairs at the 3’–end of both primers. Note, however, that DNA sequences of European toad (B. bufo) and Korean water toad (B. stejnegeri) had one mismatch within five base pairs at the 3’–end of either one of the primers as exceptions (Table S4). And the probe DNA sequence was unique to Bufo species according to the in silico evaluation, although one mismatch was accepted within the 26 bp-long probe DNA sequence among five accessions from B. japonicus (Accession No. AB159468, AB159469, AB159470, AB159471 and AB303363) out of 27 B. japonicus Accessions in the database (Tables S4).
Although DNA sequence from Anaxyrus punctatus, formally called Bufo punctatus (Accession No. AY680237), had no mismatch within five base pairs at the 3’–end of forward primer, the reverse primer had two mismatches within five base pairs at the 3’–end of the A. punctatus DNA sequence (The C at positions 3 and 4 from the 3'–end of the reverse primer was replaced with T.). This species is mainly distributed to southwestern United States to northwestern Mexico, and there is no report of establishment of the species in Japan (www.nies.go.jp/biodiversity/invasive/DB/detail/40180e.html).
In the performance test of the eDNA assay using DNA from tissue samples, qPCR DNA amplification was observed only from the target species (B. j. formosus) among the eight co-existing amphibian species in Hokkaido (B. j. formosus, D. japonica, G. rugosa, H. retardatus, L. catesbeianus, P. nigromaculatus, P. porosus and R. pirica, Online Resource 1), despite the high concentrations of the tissue-oriented DNA from the eight species (1.8–56.8 ng/µL). It is also noteworthy that none of the Bufo species is native to Hokkaido, and B. j. formosus is the only subspecies that has been identified in Hokkaido after introduction (Natural Environmental Division of Hokkaido 2015).
Field validation
In the validation of the Bufo-specific eDNA assay based on pond water samples, we detected eDNA from seven out of eight pond water samples that were collected from where larvae of B. j. formosus were observed (Table 1). The average eDNA concentrations were in the range of 6.65 × 103 and 8.28 × 106 copies per 1000 mL of pond water sample (Table 1). One exception (site without any detection of eDNA) was Ubun gate river side pond, where many B. japonicus larvae were observed at the time of water collection (See Discussion for more detail).
At the two river sites with observation records of B. japonicus within a 500 m radius (Table S3), we detected eDNA at both sites (Fig. 1). eDNA concentrations estimated in Ino Riv. DS and Kitanosawa Riv. samples were the highest at the beginning of June (1.02 × 105 and 3.04 × 104 copies/1000 mL × site, respectively), and decreased toward the end of June (exact Wilcoxon-Mann–Whitney test, Ino Riv. DS: 7th June vs 16th June and 16th June vs 23rd June, p < 0.01; Kitanosawa Riv.: 5th June vs 18th June, p < 0.01, 18th June vs 26th June, p > 0.05) (Fig. 1). On June 23rd in Ino Riv. DS, the estimated eDNA concentration became 5.23% of that on June 7th. And in Kitanosawa Riv., it become zero on June 26th. The estimated eDNA concentrations in Ino Riv. DS samples were significantly higher than that of Kitanosawa Riv. throughout June, presumably reflecting different biomass of reproductive populations between them (exact Wilcoxon-Mann–Whitney test, p < 0.01) (Fig. 1).
Estimated eDNA concentrations in Ino Riv. DS and Kitanosawa Riv. in June 2019. X-axis represents sampling date, and Y-axis represents the log-transformed eDNA concentrations per 1000 mL environmental water. Meshed boxes and white boxes represent results from Ino Riv. DS and Kitanosawa Riv., respectively. Error bars represent standard errors. Different alphabets upon each box represent significant differences (p < 0.01) and the same alphabets represent no significant differences (p > 0.05) in the exact Wilcoxon-Mann–Whitney test
Application to the Ishikari river basin
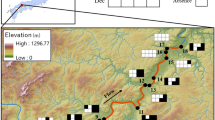
Overall, we had no positive detection from all negative control samples taken on the field, DNA extraction, and qPCR. Out of the 73 river water samples collected in the Ishikari river basin in 2018 and 2019, we had 32 positive eDNA detections (eleven black circles, 19 Gy circles and two gray stars: Fig. 2). Comparing with the observation records, we detected eDNA from all the eleven sites where the observation records existed within a 500 m radius (eleven black circles in Fig. 2, Table 2). In addition, we detected the eDNA from 21 out of 62 sites without any observation records nearby (19 Gy circles and two gray stars). They belonged to twelve river systems including Ishikari river mainstem (Aibetsu, Bibai, Ino, Kumaana, Naidaibu, Pankehoronai, Penkeutashinai, Sorachi, Toyohira, Ubun, Uryu, and Ishikari river), and two of them had no observation record throughout the river systems (Aibetsu and Bibai river). We found a significant association between eDNA detections and observation records nearby (p < 0.001 by Fisher’s exact test).
Map of the Ishikari river basin including the water sampling sites. A left upper map represents the whole Hokkaido. Samplings were conducted from June 14th to 15th in 2018 and from June 5th to 26th in 2019 within the river basin. Thick gray line represents mainstream of the Ishikari river and thin gray lines represent its branches from which we collected water samples. Eleven black circles represent positive eDNA detection sites with observation records of Natural Environmental Division of Hokkaido (2020), 19 Gy circles represent these without observation record within a 500 m radius, and 41 open circles represent negative eDNA detection sites (and no observation record). Two gray stars represent Aibetsu Riv. MS-1 and Bibai Riv. US where there were no previous observation records throughout the branch river systems. A double circle represents city office of Asahikawa. Twelve river lines with the italic alphabet in the open squares represent twelve river systems that did not have observation records nearby, but had positive eDNA detections (A: Aibetsu Riv., B: Bibai Riv., C: Ino Riv., D: Ishikari Riv., E: Kumaana Riv., F: Naidaibu Riv., G: Pankehoronai Riv., H: Penkeutashinai Riv., I: Sorachi Riv., J: Toyohira Riv., K: Uryu Riv. and L: Ubun Riv.). Data of the rivers, coastline and administrative divisions were provided by the National Land Information Division, Ministry of Land, Infrastructure, Transport and Tourism of Japan, under the CC BY 4.0 license
When sampling sites with observation records within a 1000 m radius were included, on the other hand, we found seven sites without the eDNA detection (Table 2). In order to evaluate a potential influence of PCR inhibitors in these samples, we applied them to the spike-in test with a 1.0 × 104 copies of the synthetic linear DNA as a standard (Online Resource 2). The qPCR analysis resulted in estimations of 0.83–1.33 × 104 copies/qPCR reaction (1.15 × 104 ± 0.16 × 104 on average), suggesting no extensive influence of PCR inhibitors among the river water samples.
The Bayesian approach for multiscale occupancy models supported that the three covariates (Dist, Temp, and Cond) had significant effects either on eDNA capture or detection probability (Fig. 3, Table S5): As expected, the posterior medians of the eDNA capture probability of a filter sample (\(\stackrel{\sim }{\theta }\)) were higher when the distance between sampling site and the closest observation record (Dist) was shorter. Although \(\stackrel{\sim }{\theta }\) were generally high (\(\stackrel{\sim }{\theta }\)= 0.58–0.96) in the samples with the Dist shorter than 1000 m, it clearly decreased in the samples with Dist > 2000 m (Fig. 3). Temp and Cond also had negative effects on \(\stackrel{\sim }{\theta }\) and the posterior medians of the eDNA detection probability \(\stackrel{\sim }{\rho }\), respectively, but low water temperature did not necessarily result in high \(\stackrel{\sim }{\theta }\) according to Fig. 3. In the best-fitting model, the overall posterior medians of the probability of occurrence, capture, and detection were 59.4%, 60.9%, and 79.8%, respectively (Table S5).
Posterior medians of the eDNA capture/detection probability and their relationship with covariates estimated in the best-fitting occupancy model. a Relationship between the capture probability and distance between the sampling site and the closest observation record, b that between the capture probability and water temperature, and c that between detection probability and conductivity. The graphs were created using EDNAOCCUPANCY (Dorazio and Erickson 2018)
Discussion
In our study, we aimed to develop an eDNA assay for detecting Bufo-specific DNA and to estimate the distribution of reproductive area of the terrestrial invasive terrestrial toad at watershed scale. Our results suggest that the development of the eDNA assay was successful, and eDNA was detected from seven out of eight ponds (Table 1). In addition, we found eDNA from 32 sites from river systems within the Ishikari river basin without sighting the reproductive ponds of B. japonicus.
Environmental DNA techniques have been applied for understanding distributions of invasive species in the world, mainly for aquatic organisms (Ficetola et al. 2008; Adrian-Kalchhauser and Burkhardt-Holm 2016; Larson et al. 2017; Crookes et al. 2020; LeBlanc et al. 2020). For terrestrial organisms, eDNA was detected from ponds and lakes that were sporadically distributed in the forests or mountains (Bista et al. 2017; Ushio et al. 2017; Tingley et al. 2019). In addition, some previous studies aimed to detect eDNA of terrestrial species from stream water samples (Cannon et al. 2016; Deiner et al. 2016). In the present study, distributions of the invasive terrestrial toad and their reproductive area at the watershed scale was efficiently estimated from a distant downstream of the riverine system. It provides positive evidence for the hypothesis that a riverine system, through the transport of eDNA, acts as conveyer belts of species presence information over broad scales (Deiner et al. 2016). In addition, our study suggests that eDNA collected downstream is potentially useful to screen the presence of reproductive ponds of terrestrial amphibians and the other pond-dwelling species at large spatial scale. For preventing further spread of invasive species, one can take advantage of eDNA detections from a distance, for example by concentrating eradication efforts on invasion frontlines and source populations. However, since many factors influence the detectability of eDNA from terrestrial and pond-dwelling species, such as dilution of pond water when it joined into rivers (Jane et al. 2015), future studies are required to investigate how hydrological features of the riverine system affect the aggregation and transport of eDNA in combination with the other environmental factors affecting the detectability (i.e., biotic and abiotic conditions) (Pilliod et al. 2014; Curtis et al. 2020).
Despite the high detectability in ponds (Table 1), we failed to detect the Bufo eDNA in one pond. A potential cause of the detection error is PCR inhibitors (e.g., humic acid, tannic acid). This specific pond was surrounded by silt and turbid environment, and it is known that pond water sometimes includes much PCR inhibitors (Lakay et al. 2007; Fujii et al. 2019). In fact, we could recover the detection of eDNA from this pond when the sample was diluted 100 times in our additional analysis (Table S6), although we do not recommend the 100-time dilution in the wide-scale analyses due to generally low eDNA concentration in river water samples (e.g., average eDNA concentration in the positive detections of eDNA from the river samples was only 6.18 copies/mL).
To our surprise, we found eDNA detections at all the sites with the previous observation records nearby in the river samples (11/11 sites with observation records within a 500 m radius, Table 2). It suggests that eDNA assays work efficiently to estimate reproductive area of terrestrial amphibians from a distance if their reproductive seasons and larval periods are well understood. The average distance between the sampling sites and the nearest observation points was 191.8 m in the eleven samples (23.6–393.6 m, one standard deviation 130.1 m, Table S3). Accepting observation records within a 1000 m radius, on the other hand, we found seven sites with observation records without detecting eDNA (eDNA detections at 18 out of 25 sites with observation records nearby). Nevertheless, it was also true that within a 1000 m radius, there still were 14 cases in which eDNA was detected without observation record (Table 2).
In fact, the occupation modelling approach allowed us to estimate the eDNA capture and detection probabilities. The best-fitting model suggests not only the distance between the sampling site and the site with the closest observation record, but also water temperature negatively influenced the eDNA capture probability of a sample (Fig. 3). Furthermore, conductivity influenced the eDNA detection probability, suggesting that conductivity reflects dissolved solids including PCR inhibitors (although the effect size of the PCR inhibitors was seemingly small in most of the cases we studied. See Fig. 3c and Online Resource 2). It is obvious that eDNA detectability also depends on many other factors such as number of individuals in the reproductive ponds and their developmental stages, and that no eDNA detection does not necessarily mean no reproductive ponds in the focal watershed. Our analytical results based on the multiscale occupancy model suggest that with three qPCR replicates, we had 99.2% of average detection probability when the sample contained target DNA (= 1—[1—0.798]3). With two filter samples at a site, the average capture probability was 84.7% (= 1—[1—0.609]2). In theory, therefore, expected eDNA detection probability in one of the 2 × 3 replicates, given the occurrence of the target species, would be 84.0% (= 0.992 × 0.847). It means that we had a risk of false negative in 16% of the cases, which could be minimized mainly by increasing the number of filter samples (e.g., it drops to 3% if we had four filter samples). Thus, the detailed sampling design for an eDNA survey should be carefully evaluated for better understanding of the current distributions of invasive species, and for better management of them during the reproductive season in future studies.
For nailing the spread of invasive species, it is essential to understand hotspots of their reproductions. As the results of stationary monitoring in Ino Riv. DS and Kitanosawa Riv. aimed to verify the detectability of their reproductive seasons and/or larval stage, estimated eDNA concentrations were gradually decreased from the start to the end of our sampling period (Fig. 1). Reproductive season of B. j. formosus is quite limited in a few days and they leave their reproductive ponds immediately after their reproduction (Ishii et al. 1989; Kusano et al. 1995). According to a local researcher, the number of observed larvae of B. j. formosus in ponds decreased during our sampling period, and no larva was observed after June 18th, 2019, around the Kitanosawa (Tokuda T., personal communication). Furthermore, Villacorta-Rath et al. (2020) reported that cane toad eDNA drastically decreased within a few days after its last presence in aquaria. Thus, estimated eDNA seems to be mostly originated from larvae. It suggests that positive detections reflected the presence of their reproductive ponds. With regard to the estimated eDNA concentrations, Ino Riv. DS showed significantly higher concentrations of toad eDNA than Kitanosawa Riv. did. Two possible explanations are difference in biomass, and that in spawning timing between the two sites. Under certain conditions, concentrations of eDNA are known to reflect biomass of target species (Doi et al. 2017; Mizumoto et al. 2018; Tingley et al. 2019). Given the terrestrial characteristics and short spawning period of B. japonicus, however, it is equally likely that the microhabitat conditions influenced their phenology and the spawning timing at each area. To evaluate these possibilities, however, further studies on their phenology and microhabitat conditions.
Many ponds along the river are persistent by inflow water from spring, and the pond water overflow into the adjacent river in our survey area (Kishida O. and Takai K., unpublished data), which allowed us to estimate the presence of their reproductive ponds from a distance. The high consistency between the eDNA detections and previous observation records suggests that they provide similar quality of information regarding their distribution, and that the distribution does not change rapidly once they invaded in each watershed (The two kinds of information [eDNA and observation] had 0–8 years of time lag, Table S3). Furthermore, the eDNA detections at the sites without observation records nearby suggest relatively new reproductive ponds in the watersheds, which might include hubs of their further spread within the river basin. Given that B. j. formosus invaded round Asahikawa City several decade ago, the eDNA detection patterns in the Ishikari river basin (Fig. 2) represent a stable distribution of B. j. formosus reproductive ponds around the original invasion sites, as well as further spreads of them along the Ishikari river. It indicates the importance of wide-scale monitoring of their reproductive ponds along with the mainstem and branches of the Ishikari river with and without direct observation of B. j. formosus.
A potential pitfall of this study is the limited species-specificity of the developed eDNA assay. With many practical restrictions as shown above, we developed a genus-specific eDNA assay, instead of species-specific one. It might cause misidentifications especially in regions when more than one Bufo species exist. Nevertheless, it is noteworthy that to date, we have only a single Bufo species (B. j. formosus) in Hokkaido, the second largest island in Japan, and that the two subspecies of B. japonicus often distributes differently in the other part of Japan, according to the National Institute for Environmental Studies, (www.nies.go.jp/biodiversity/invasive/DB/etoc4_amphibians.html). Given that B. japonicus is endemic to Japan and that they could become a toxic alien species in any other regions in the world, our eDNA assay and its application to river water samples might provide a powerful means of screening their reproductive ponds at the early stage of their invasion, although further investigations are required to test sensitivity of the assay to other Bufo species that are endemic to such regions. With more technical advances and follow-up studies, including an update of the primers with higher species-specificity, eDNA assays can be a standard tool for estimating reproductive areas of amphibian species in general, with which biodiversity managements become much more efficient.
In conclusion, we detected B. japonicus eDNA not only from pond water samples but also from river water samples in the Ishikari river basin in this study. It suggests that our eDNA assay can be used for screening their presence in watershed, most likely reflecting their reproductive activities nearby. The watershed-scale screening of their reproductive area based on eDNA is a practical approach for grasping spread of front-line of the invasive species because it has a potential to save high labour cost for the pond-scale inspection by the direct observation methods. Although increasing number of eDNA studies have been conducted upon aquatic invasive and endemic species for their management (Adrian-Kalchhauser and Burkhardt-Holm 2016; Mizumoto et al. 2020; Mauvisseau et al. 2021), our study sheds light on an opportunity of their application to terrestrial species with limited period of time for reproduction in and around the watershed in a river basin.
References
Adrian-Kalchhauser I, Burkhardt-Holm P (2016) An eDNA assay to monitor a globally invasive fish species from flowing freshwater. PLoS ONE 11:e0147558. https://doi.org/10.1371/journal.pone.0147558
Bista I, Carvalho GR, Walsh K et al (2017) Annual time-series analysis of aqueous eDNA reveals ecologically relevant dynamics of lake ecosystem biodiversity. Nat Commun 8:14087. https://doi.org/10.1038/ncomms14087
Boback SM, Nafus MG, Adams Y (2020) Use of visual surveys and radiotelemetry reveals sources of detection bias for a cryptic snake at low densities. Ecosphere 11(1):e03000. https://doi.org/10.1002/ecs2.3000
Bustin SA, Benes V, Garson JA et al (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. https://doi.org/10.1373/clinchem.2008.112797
Cannon MV, Hester J, Shalkhauser A et al (2016) In silico assessment of primers for eDNA studies using PrimerTree and application to characterize the biodiversity surrounding the Cuyahoga River. Sci Rep 6:22908. https://doi.org/10.1038/srep22908
Crookes S, Heer T, Castañeda RA et al (2020) Monitoring the silver carp invasion in Africa: a case study using environmental DNA (eDNA) in dangerous watersheds. NeoBiota 56:31–47. https://doi.org/10.3897/neobiota.56.47475
Curtis AN, Tiemann JS, Douglass SA et al (2020) High stream flows dilute environmental DNA (eDNA) concentrations and reduce detectability. Divers Distrib. https://doi.org/10.1111/ddi.13196
Deiner K, Fronhofer EA, Mächler E et al (2016) Environmental DNA reveals that rivers are conveyer belts of biodiversity information. Nat Commun 7:12544. https://doi.org/10.1038/ncomms12544
Diamond JM, Ross MS (2020) Overlap in reproductive phenology increases the likelihood of cavity nest usurpation by invasive species in a tropical city. The Condor 122(3), duaa013. https://doi.org/10.1093/condor/duaa013
Doi H, Inui R, Akamatsu Y et al (2017) Environmental DNA analysis for estimating the abundance and biomass of stream fish. Freshw Biol 62(1):30–39. https://doi.org/10.1111/fwb.12846
Dorazio RM, Erickson RA (2018) EDNAOCCUPANCY: An R package for multiscale occupancy modelling of environmental DNA data. Mol Ecol Resour 18:368–380. https://doi.org/10.1111/1755-0998.12735
Ficetola GF, Miaud C, Pompanon F et al (2008) Species detection using environmental DNA from water samples. Biol Lett 4(4):423–425. https://doi.org/10.1098/rsbl.2008.0118
Fujii K, Doi H, Matsuoka S et al (2019) Environmental DNA metabarcoding for fish community analysis in backwater lakes: A comparison of capture methods. PLoS ONE 14(1):e0210357. https://doi.org/10.1371/journal.pone.0210357
Gelfand AE, Ghosh SK (1998) Model choice: A minimum posterior predictive loss approach. Biometrika 85:1–11. https://doi.org/10.1093/biomet/85.1.1
Igawa T, Kurabayashi A, Nishioka M et al (2006) Molecular phylogenetic relationship of toads distributed in the Far East and Europe inferred from the nucleotide sequences of mitochondrial DNA genes. Mol Phylogenet Evol 38(1):250–260. https://doi.org/10.1016/j.ympev.2005.09.003
Ishii S, Yoneyama H, Inoue M et al (1989) Changes in plasma and pituitary levels of prolactin in the toad, Bufo japonicus, throughout the year with special reference to the breeding migration. Gen Comp Endocr 74(3):365–372. https://doi.org/10.1016/S0016-6480(89)80032-2
Jane SF, Wilcox TM, McKelvey KS et al (2015) Distance, flow and PCR inhibition: eDNA dynamics in two headwater streams. Mol Ecol Resour 15:216–227. https://doi.org/10.1111/1755-0998.12285
Kawamura T, Nishioka M, Ueda H (1980) Inter-and intraspecific hybrids among Japanese, European and American toads. Sci Rep Lab Amphib Biol Hiroshima Univ 4:1–125
Kazila E, Kishida O (2019) Foraging traits of native predators determine their vulnerability to a toxic alien prey. Freshw Biol 64(1):56–70. https://doi.org/10.1111/fwb.13194
Kusano T, Maruyama K, Kaneko S (1995) Post-Breeding Dispersal of the Japanese Toad Bufo Japonicus Formosus. J Herpetol 29(4):633–638. https://doi.org/10.2307/1564755
Lakay FM, Botha A, Prior BA (2007) Comparative analysis of environmental DNA extraction and purification methods from different humic acid-rich soils. J Appl Microbiol 102(1):265–273. https://doi.org/10.1111/j.1365-2672.2006.03052.x
Larson ER, Renshaw MA, Gantz CA et al (2017) Environmental DNA (eDNA) detects the invasive crayfishes Orconectes rusticus and Pacifastacus leniusculus in large lakes of North America. Hydrobiologia 800(1):173–185. https://doi.org/10.1007/s10750-017-3210-7
LeBlanc F, Belliveau V, Watson E et al (2020) Environmental DNA (eDNA) detection of marine aquatic invasive species (AIS) in Eastern Canada using a targeted species-specific qPCR approach. Manag Biol Invasions 11(2):201. https://doi.org/10.3391/mbi.2020.11.2.03
Lepczyk CA (2005) Integrating published data and citizen science to describe bird diversity across a landscape. J Appl Ecol 42(4):672–677. https://doi.org/10.1111/j.1365-2664.2005.01059.x
Martinez B, Reaser JK, Dehgan A et al (2020) Technology innovation: advancing capacities for the early detection of and rapid response to invasive species. Biol Invasions 22(1):75–100. https://doi.org/10.1007/s10530-019-02146-y
Matsui M (1984) Morphometric variation analyses and revision of the Japanese toads (Genus Bufo, Bufonidae). Contr Biol Lab Kyoto Univ 26(3–4), 209–428. http://hdl.handle.net/2433/156031
Mauvisseau Q, Kalogianni E, Zimmerman B et al (2021) eDNA-based monitoring: Advancement in management and conservation of critically endangered killifish species. Environmental DNA 2(4):601–613. https://doi.org/10.1002/edn3.92
Ministry of Land, Infrastructure, Transport and Tourism. (2008). Basic information of Ishikari River basin (in Japanese). Available: https://www.mlit.go.jp/river/toukei_chousa/kasen/jiten/nihon_kawa/0109_ishikari/0109_ishikari_00.html [Accessed July 6, 2021].
Ministry of Land, Infrastructure, Transport and Tourism. (2020). National regional planning bureau: digital national land information. Available: http://nlftp.mlit.go.jp/ksj-e/gml/gml_datalist.html [Accessed February 19, 2020].
Mizumoto H, Urabe H, Kanbe T et al (2018) Establishing an environmental DNA method to detect and estimate the biomass of Sakhalin taimen, a critically endangered Asian salmonid. Limnology 19:219–227. https://doi.org/10.1007/s10201-017-0535-x
Mizumoto H, Mitsuzuka T, Araki H (2020) An Environmental DNA Survey on Distribution of an Endangered Salmonid Species, Parahucho perryi, in Hokkaido. Japan Front Ecol Evol 8:569425. https://doi.org/10.3389/fevo.2020.569425
Naito H, Shiga K (2016) Immigrant of Bufo japonicus into the left bank region of Ishikari estuary, Hokkaido, Japan. Bulletin of Ishikari Sakyu No Kaze Museum 6:65–70 (in Japanese)
Natural Environmental Division of Hokkaido, Hokkaido Government (2015) Species and subspecies list of amphibians and reptiles in Hokkaido. http://www.pref.hokkaido.lg.jp/ks/skn/syuasyumokuroku_ryouseihatyuu2015.pdf. in Japanese
Natural Environmental Division of Hokkaido, Hokkaido Government (2020) Map of the observation information of Japanese toad. http://www.pref.hokkaido.lg.jp/ks/skn/alien/toad/azuma.html [Accessed February 19, 2020]. in Japanese
Okamiya H, Takai K, Kishida O (2021) Japanese common toad, Bufo japonicus formosus, contains venom in the egg stage. Curr Herpetol 40(1):103–106. https://doi.org/10.5358/hsj.40.103
Oyake N, Sasaki N, Yamaguchi A et al (2020) Comparison of susceptibility to a toxic alien toad (Bufo japonicus formosus) between predators in its native and invaded ranges. Freshw Biol 65(2):240–252. https://doi.org/10.1111/fwb.13417
Pilliod DS, Godberg CS, Arkle RS et al (2014) Factors influencing detection of eDNA from a stream-dwelling amphibian. Mol Ecol Resour 14:109–116. https://doi.org/10.1111/1755-0998.12159
R Core Team (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available via: http://www.R-project.org.
Sarashina M, Yoshida T (2015) Predation impacts of four invasive frogs in Hokkaido, Japan, assessed by an index of relative importance. Jpn J Conserv Ecol 20:15–26. (in Japanese)
Sengupta ME, Hellström M, Kariuki HC et al (2019) Environmental DNA for improved detection and environmental surveillance of schistosomiasis. Proc Nat Acad Sci 116(18):8931–8940. https://doi.org/10.1073/pnas.1815046116
Tingley R, Greenlees M, Oertel S et al (2019) Environmental DNA sampling as a surveillance tool for cane toad Rhinella marina introductions on offshore islands. Biol Invasions 21(1):1–6. https://doi.org/10.1007/s10530-018-1810-4
Ushio M, Fukuda H, Inoue T et al (2017) Environmental DNA enables detection of terrestrial mammals from forest pond water. Mol Ecol Resour 17(6):e63–e75. https://doi.org/10.1111/1755-0998.12690
Villacorta-Rath C, Adekunle AI, Edmunds RC et al (2020) Can environmental DNA be used to detect first arrivals of the cane toad, Rhinella marina, into novel locations? Environmental DNA 2:635–646. https://doi.org/10.1002/edn3.114
Watanabe S (2010) Asymptotic equivalence of Bayes cross validation and widely applicable information criterion in singular learning theory. J Machine Learning 11:3571–3594
Watanabe S (2013) A widely applicable Bayesian information criterion. J Machine Learning 14:867–897
Yatsuyanagi T, Ishida R, Sakata MK et al (2020) Environmental DNA monitoring for short-term reproductive migration of endemic anadromous species, Shishamo smelt (Spirinchus lanceolatus). Environmental DNA 2(2):130–139. https://doi.org/10.1002/edn3.50
Acknowledgements
We thank the members of staff and the members of Tomakomai Experimental Forest of the Field Science Center for Northern Biosphere Hokkaido University, and staff and the members of Animal Ecology Lab in Research Faculty of Agriculture, Hokkaido University. We also thank T. Aota and T. Tokuda for sharing local information and K. Fukaya and H. Doi for technical advises on the occupancy modelling. This study was funded by the Mitsui and Co. Environment Fund (R16‐0008), Asahi glass foundation to OK and the Environment Research and Technology Development Fund of the Environmental Restoration and Conservation Agency of Japan (JPMEERF20204004: HA). We declare that the experiments complied strictly with the current laws in Japan.
Funding
This study was funded by the Mitsui and Co. Environment Fund (R16–0008: OK) and the Environment Research and Technology Development Fund of the Environmental Restoration and Conservation Agency of Japan (JPMEERF20204004: HA).
Author information
Authors and Affiliations
Contributions
OK and HA conceived the original idea and acquired the funding. HM, KT, OK and HA collected samples and revised the manuscript, after HM, NM and HA analyzed data and HM and HA wrote the original draft. OK and HA administrated the project and gave final approval for publication.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declared that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mizumoto, H., Kishida, O., Takai, K. et al. Utilizing environmental DNA for wide-range distributions of reproductive area of an invasive terrestrial toad in Ishikari river basin in Japan. Biol Invasions 24, 1199–1211 (2022). https://doi.org/10.1007/s10530-021-02709-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-021-02709-y