Abstract
Arion subfuscus sensu lato (s. l.) refers to a cryptic slug species complex that is native to Europe and which, from there, has been introduced into the northeastern U.S. The species complex was detected in California for the first time in 2005 and recently 12 specimens were collected during statewide surveys. The genital morphology of these specimens suggested that only A. subfuscus sensu stricto (s. s.) was present and partial sequences of mitochondrial 16S rDNA (443 bp) showed that they all belonged to a single haplotype of the mitochondrial lineage, S1 sensu Pinceel et al. (Genetica 125: 155–171, 2005). This result was corroborated by a parallel analysis of a 655 bp COI DNA barcode. The 16S rDNA S1 haplotype (S1-03) of the Californian specimens is hitherto known only from the northeastern U.S. Hence, it is likely that populations may have been introduced to California from the northeastern states, rather than directly from the native area of the S1 haplogroup in Europe.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The slug, Arion subfuscus, was originally described from the Montagne Noire in southern France by Draparnaud (1805: as “Limax”). Since then, the species has traditionally been regarded as highly polymorphic, both in external morphology (e.g., Taylor 1907; Chichester and Getz 1973; Wiktor 1983), and in allozyme variation (McCracken and Selander 1980; Foltz et al. 1982), resulting in a long list of synonyms (Hesse 1926). Hence, several authors have suggested that the name A. subfuscus actually refers to a complex of cryptic species (e.g., Waldén 1976; Foltz et al. 1982; Backeljau et al. 1996; Falkner et al. 2002). In this context, Garrido et al. (1995) redescribed a series of species from the Iberian Peninsula including A. subfuscus and selected a neotype to definitively fix the name of the latter. Conversely, Pinceel et al. (2004) disentangled the species complex in northwest Europe by combining anatomical, allozyme and DNA sequence data, showing that in this region A. subfuscus comprises at least two distinct species, Arion subfuscus s. s. (based on the DNA analysis of the neotype) and Arion fuscus (Müller 1774). The gonad of the latter is small, deep dark and completely embedded in the digestive gland, while in the former the gonad is large, pale violet and located on the periphery of the gland. In addition, Pinceel et al. (2004, 2005) reported that A. subfuscus s. s. actually consists of, at least, five strongly divergent mitochondrial DNA lineages (=haplogroups; provisionally referred to as S1 to S5).
In the U.S., A. subfuscus s. l. was first reported from Boston in 1842 (Pilsbry 1948) and has since gradually colonized most of northeastern North America. It is now found in anthropogenic and natural habitats (Chichester and Getz 1969, 1973), where it is sometimes even more abundant than native slug and snail species (Ferguson 2004). Combining morphology, DNA (mitochondrial and nuclear) and allozyme variation, Pinceel et al. (2005) found that only one of their five European haplogroups was represented in specimens collected from northeastern states, the S1 haplogroup. In addition, a large loss in genetic variation was observed which was tentatively attributed to founder effects. The authors also found that the likely source area of the invasions comprised the British Isles and adjacent regions on the European mainland (e.g., Belgium, The Netherlands and northwest France), as based on the known distribution of the S1 lineage. Finally, Pinceel et al. (2005) suggested that of both species, A. subfuscus may be the more obvious invader, but A. fuscus may also occur in the U.S. because allozyme data indicated that certain North American strains of A. subfuscus s. l. were even more divergent than European A. subfuscus s. s. and A. fuscus (McCracken and Selander 1980; Foltz et al. 1982). This was confirmed by Barr et al. (2009) who relied on genital anatomy and mitochondrial 16S rDNA sequences to demonstrate the presence of A. fuscus in the northeast of the U.S. i.e., Vermont and Massachusetts (Norman Barr, Personal Communication).
In 2005, Arion subfuscus s. l. was collected in California (Half Moon Bay, San Mateo Co.) for the first time (Heike Reise and Janet Leonard, Personal Communication). The current investigation aims to (1) determine which of the cryptic taxa of the A. subfuscus complex are present in the state and (2) provide preliminary evidence about the possible source areas of this introduction. To this end we conducted a DNA sequence analysis of Californian specimens of A. subfuscus s. l. and used these data in a DNA barcoding approach by comparing them with existing sequence data from Pinceel et al. (2005) and Barr et al. (2009).
Materials and methods
Specimen collection and morphology-based identifications
During statewide invasive slug surveys of California in 2006, 12 specimens of A. subfuscus s. l. were discovered in San Mateo Co., Sonoma Co. and Humboldt Co. (Table 1). Specimens were collected by direct searching and kept alive in small plastic containers lined with damp paper towels and fed on carrots. On return to the laboratory, a small piece of tissue was snipped from the tails of the living slugs and preserved in 100% ethanol for DNA analysis. Next, the slugs were killed by placing them overnight in a sealed container filled with boiled water that had been allowed to cool. The following morning, the stretched slugs were dissected and preserved in 70% ethanol. Specimens were identified using the diagnostic genital characteristics given by Pinceel et al. (2004).
Molecular analysis
DNA was extracted from the tail tissue pieces of 12 specimens, using the QIAamp Tissue Kit (Qiagen Inc., Valencia, CA). Cell lysis was achieved overnight in a 56°C constant temperature shaker with proteinase K and Qiagen lysis buffer. The QIAamp Tissue Kit then adsorbed the DNA onto a silica membrane in a spin column that was cleaned with two wash buffers. Finally, purified template DNA was eluted in AE storage buffer. Polymerase chain reaction (PCR) was used to amplify a 443 bp fragment of the mitochondrial 16S rDNA gene and a 655 bp fragment of cytochrome c oxidase (COI; Table 2). Amplified DNA was cleaned using the Wizard® PCR Preps DNA Purification System (Promega, Madison, WI). The PCR product was first purified using a purification buffer and resin, and then washed using 80% isopropanol. Clean DNA was eluted in double distilled water and then directly sequenced in both directions at the University of California, Riverside Genomics Institute Core Instrumentation Facility using an Applied BioSystems 3730 DNA Analyser with a Big-Dye® V3.1 kit (Applied Biosystems, Foster City, CA). Raw sequences were trimmed by removing the primers and aligned manually in Bioedit v 7.0.5.3 (Hall 1999). The aligned 16S and COI sequences were then compared with those for the different mitochondrial haplogroups of A. subfuscus s. s. published by Pinceel et al. (2005) and Barr et al. (2009). Neighbor-Joining (NJ) trees based on Kimura-2-parameter distances, 1000 bootstrap replicates and using Arion lusitanicus and Arion rufus as outgroups were used to confirm the association of the sequences with haplogroups. The NJ tree reconstruction and analysis were carried out with MEGA v. 4 software (Kumar et al. 2008). The COI and 16S sequences (Table 1) for the A. subfuscus specimens were deposited in GenBank (Benson et al. 2008) and DNA extracts of the 12 specimens have been deposited in the collections of the Royal Belgian Institute of Natural Sciences under the general inventory number I.G. 31556.
Results and discussion
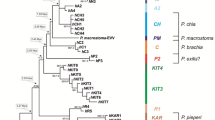
The gonads of all of the specimens collected during this study were large, pale violet and located on the periphery of the digestive gland which indicated that only A. subfuscus s. s. was collected in California. This was confirmed by the DNA data, which further specified that the Californian specimens belonged to haplogroup S1, since for the 16S rDNA they had one and the same haplotype that was identical to S1-03 as defined by Pinceel et al. (2005) (cf. GenBank accession number AY861459), while for COI, they shared a single haplotype that differed in only one nucleotide from the COI haplotype of A. subfuscus with GenBank accession number AY987905 (a specimen from Kortrijk Belgium). In the NJ tree of the COI sequences, both the sequence AY987905 and that of the Californian specimen fell in the S1 haplogroup with a 100% bootstrap support (Fig. 1).
Assignment of the partial COI gene sequences (655 bp) of Californian specimens of Arion subfuscus s. s. to the S1 haplogroup (100% bootstrap support) using a neighbor-joining tree based on Kimura-2-parameter distances, 1000 bootstrap replicates and both Arion lusitanicus and Arion rufus as outgroups
A. subfuscus s. l. is native to Europe and the S1 lineage has been collected only in Ireland, Great Britain, Belgium and France, while it was introduced into northeastern North America (Pinceel et al. 2004). It is, therefore, logical to assume that the Californian specimens of A. subfuscus s. l. should originate from one of these two areas. However, since the S1-03 16S rDNA haplotype is hitherto known only from the northeastern U.S. (Pinceel et al. 2005), it seems most likely that the Californian specimens were introduced from that area and not directly from Europe. Given that (1) this introduction appears be very recent i.e., the first specimens of the species were collected in the state during 2005 and (2) transport within the U.S. has been responsible for the spread of several other alien terrestrial gastropods (e.g., Chichester and Getz 1969; Dundee 1970; Mead 1971), it is possible that other taxa of A. subfuscus s. l. may show up in California. Indeed Barr et al. (2009) recently demonstrated that A. fuscus and A. subfuscus S2 are also present in the U.S. Nevertheless, the current study illustrates how molecular techniques (i.e., DNA barcoding) can be easily and successfully applied to identify cryptic taxa, even if these are not yet fully defined. Moreover, the study emphasizes the need for continuous monitoring to determine how and where exotic gastropod species might spread within new regions should they evade detection at ports of entry and establish viable populations in the wider environment.
Finally, the 12 Californian specimens of A. subfuscus in this study were all identical both in their 16S rDNA and their COI, thus representing only one single composite haplotype. This may be the result of a founder effect acting upon the already impoverished gene pool of A. subfuscus s. l. in the northeastern U.S., where Pinceel et al. (2005) observed a significant reduction in genetic diversity among 219 American specimens of this species compared to European populations. In fact Pinceel et al. (2004) reported in Europe no less than 53 different 16S rDNA haplotypes distributed over five haplogroups, while in the U.S. they found only two haplotypes, both of haplogroup S1. Even with the addition of the recent A. subfuscus s. s. data of Barr et al. (2009), the reduction in genetic diversity in the American populations compared to the European ones, remains overwhelming. The further reduction of genetic diversity in the Californian material is, hence, no surprise considering their probable recent introduction and the likely very small number of founders.
References
Backeljau T, Winnepenninckx B, Jordaens K, De Wolf H, Breugelmans K, Parejo C, Rodriguez T (1996) Protein electrophoresis in arionid taxonomy. BCPC Symp Proc 66:21–28
Barr N, Cook A, Elder P, Molongoski J, Prasher D, Robinson DG (2009) Application of a DNA barcode using the 16S rRNA gene to diagnose pest Arion species in the USA. J Moll Stud 75:187–191
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL (2008) Genbank. Nucleic Acids Res 36:25–30
Chichester LF, Getz LL (1969) The zoogeography and ecology of arionid and limacid slugs introduced into northeastern North America. Malacologia 7:313–346
Chichester LF, Getz LL (1973) The terrestrial slugs of northeastern North America. Sterkiana 51:11–42
Draparnaud JPR (1805) Histoire Naturelle des Mollusques Terrestres et Fluviatiles de la France. Atlas, Paris
Dundee DS (1970) Introduced Gulf Coast Mollusca. Tulane Stud Zool Bot 16:101–115
Falkner G, Ripken TEJ, Falkner M (2002) Mollusques continentaux de France—Liste de référence annotée et bibliographie. Patrimoines naturels 52:1–350
Ferguson SH (2004) Effects of poisoning nonindigenous slugs in a boreal forest. Can J Forest Res 34:449–455
Folmer O, Black M, Hoeh WR, Lutz R, Vrijenhoek RC (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotech 3:294–299
Foltz DW, Ochman H, Jones JS, Evangelist SM, Selander RK (1982) Genetic population structure and breeding systems in arionid slugs (Mollusca: Pulmonata). Biol J Linn Soc 17:225–241
Garrido C, Castillejo J, Iglesias J (1995) The Arion subfuscus complex in the eastern part of the Iberian peninsula, with redescription of Arion subfuscus (Draparnaud, 1805) (Gastropoda: Pulmonata: Arionidae). Arch Moll 124:103–118
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment [ed.], and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98. http://www.mbio.ncsu.edu/BioEdit/bioedit.html
Hesse P (1926) Die Nacktschnecken der palaearktischen Region. Abh Arch Moll 2:1–152
Kumar S, Nei M, Dudley N, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinformat 9:299–306
McCracken GF, Selander RK (1980) Self-fertilization and monogenic strains in natural populations of terrestrial slugs. Proc Natl Acad Sci USA 77:684–688
Mead AR (1971) Status of Achatina and Rumina in the United States. Biologist 53:112–117
Palumbi SR, Benzie J (1991) Large mitochondrial DNA differences between morphologically similar Penaeid shrimps. Mol Mar Biol Biotech 1:27–34
Pilsbry HA (1948) Land Mollusca of North America (north of Mexico). Academy of Natural Sciences of Philapelphia, Monograph 3 2:521–1113
Pinceel J, Jordaens K, Van Houtte N, De Winter AJ, Backeljau T (2004) Molecular and morphological data reveal cryptic taxonomic diversity in the terrestrial slug complex Arion subfuscus/fuscus. Biol J Linn Soc 83:23–38
Pinceel J, Jordaens K, Van Houtte N, Bernon G, Backeljau T (2005) Population genetics and identity of an introduced terrestrial slug: Arion subfuscus s. l. in the north-east USA (Gastropoda, Pulmonata, Arionidae). Genetica 125:155–171
Taylor JW (1907) Monograph of the land & freshwater Mollusca of the British Isles. Testacellidae, Limacidae & Arionidae. Taylor Brothers, Leeds
Waldén HW (1976) A nomenclatural list of the land Mollusca of the British Isles. J Conch 29:21–25
Wiktor A (1983) The slugs of Bulgaria (Arionidae, Milacidae, Limacidae, Agriolimacidae—Gastropoda, Stylommatophora). Ann Zool 37:71–206
Acknowledgments
This research was funded by the European Union under a Marie Curie Outgoing International Fellowship (MOIF-CT-2005-21592). Thierry Backeljau and Kurt Jordaens were supported by a Belgian Federal Science Policy Office grant (MO/36/017). We are grateful to A. Hansen for assistance with fieldwork and to G. Bernon and C. Williams for review comments.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Mc Donnell, R.J., Rugman-Jones, P., Backeljau, T. et al. Molecular identification of the exotic slug Arion subfuscus sensu stricto (Gastropoda: Pulmonata) in California, with comments on the source location of introduced populations. Biol Invasions 13, 61–66 (2011). https://doi.org/10.1007/s10530-010-9789-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-010-9789-5