Abstract
The zebra mussel, Dreissena polymorpha is an aquatic nuisance invasive species originally native to the Ponto-Caspian region where it is found in lakes and delta areas of large rivers draining into the Black and Caspian seas. The dispersal of D. polymorpha began at the end of the 18th century, at a time when shipping trade become increasingly important and many canals were built for linking different navigable river systems in Europe. Over the past 200 years, zebra mussels spread to most of the lakes, rivers and waterways in Europe by a combination of natural and anthropogenic dispersal mechanisms. D. polymorpha invaded Spain around 2001, being found for the first time in the Riba-roja reservoir at the lower part of the Ebro River, North-East Spain. The relatively late invasion of Spain was most likely caused by the presence of the Pyrenees, which isolated the Iberian Peninsula from the rest of the European continent, and acted as a barrier to the dispersal of D. polymorpha. In recent studies, molecular genetic methods have successfully been used to determine phylo-geographic relationships, which may reflect invasion corridors and can help retrace source populations. Zebra mussels from populations in Great Britain, The Netherlands, Belgium, France, Germany, Spain, Italy, Romania and North America were analyzed using PCR based amplified fragment length polymorphism (AFLP)-fingerprinting to determine the source population of D. polymorpha in Spain. The phylogenetic analyses and pair-wise genetic distances revealed that the recent invasion of zebra mussels in Spain is most likely from France.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The zebra mussel (Dreissena polymorpha) is a successful invasive bivalve that originates from the Ponto-Caspian region. Its rapid spread throughout Europe and North America (Kinzelbach 1992; Bij de Vaate et al. 2002) has been facilitated by a favourable combination of life-history characteristics (free-swimming larval stages, formation of byssus threads, etc.) and human-mediated dispersal mechanisms (Carlton 1993; Johnson and Carlton 1996). Once they are introduced into a new area, zebra mussels often form dense aggregations (reaching densities of up to 700,000 individuals m−2), a common feature of the zebra mussel that has led to many adverse socio-economic and ecological consequences (Pimentel et al. 2000). In North America, industrial facilities using surface water for production processes and drinking water companies had to take costly and time-consuming measures to prevent clogging of their intake pipes, with cost estimates ranging up to several billions of dollars by the turn of the century (Roberts 1990; Ludyanskiy et al. 1993; Van der Velde et al. 1994; Hushak 1996; Pimentel et al. 2000). The epizoic colonisation of snails, crayfish and unionid bivalves by this exotic species has led to a decline, and in many cases to a total eradication, of native species (Mackie 1991; Haag et al. 1993; Schloesser et al. 1996). Seston removal by filter-feeding of this organism has resulted in the transformation of turbid to clear waters (Reeders et al. 1989; Reeders and Bij de Vaate 1990, 1992; Reeders et al. 1993), not only resulting in severe ecological impacts due to complete changeovers of local communities, but also in negative socio-economic impacts on commercial- and sport fisheries due to the decline of commercially important fish species such as the Walleye (Sander vitreus) (Mackie 1991; Griffiths 1993; Van der Velde et al. 1994; Strayer et al. 1999).
D. polymorpha colonised Spain around 2001, where it was found for the first time in the Riba-roja reservoir in the lower part of the Ebro River, North-East Spain (Ruiz-Altaba et al. 2001). The relatively late invasion of Spain was most likely caused by the presence of the Pyrenees, which isolate the Iberian Peninsula from the rest of the European continent, and act as a dispersal barrier for D. polymorpha. Peribañez (2005) has tried to infer the origin of invasion by using the presence of D. polymorpha’s parasites in the Ebro River as origin tracers (Palau, unpubl.). They recorded the presence of the helminth Phyllodistomum folium in the branchiodes of D. polymorpha. This helminth species is widely known in Europe but, to date, not known in North America, suggesting a European rather than North American origin of the D. polymorpha population in the Ebro. However, the exact source region in Europe remained uncertain (refer to Burlakova et al. 2006 for more information on the use of parasites to infer origin of invasion by adult zebra mussels).
Recent studies have revealed the usefulness of genetic markers (e.g., RAPD, AFLP and microsatellites) as a tool to study the genetic structure of populations and infer source regions of invasion of Dreissena species (Wilson et al. 1999; Stepien et al. 2002; Pollux et al. 2003; Elderkin et al. 2004; Astanei et al. 2005; Therriault et al. 2005; May et al. 2006). Phylogeographic analysis, expressing the hierarchical descent of populations, allows the identification of source regions by comparing the genetic similarity between the newly established population and potential surrounding source populations (Pollux et al. 2003). In this study, the PCR-based AFLP-fingerprinting method (Vos et al. 1995) was applied to determine the most likely origin of the D. polymorpha population in the Ebro River in Spain.
Materials and methods
Sampling and preservation
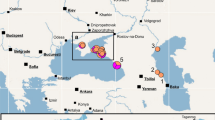
A total of 237 zebra mussels were sampled from eight locations throughout Europe and one location in North America (Table 1), covering a large part of their current range of geographic distribution (Fig. 1). Live zebra mussels were collected from submerged substrata and were immediately placed in 50 ml conical polypropylene tubes (Greiner Bio-One, Alphen aan den Rijn, Netherlands), filled to the rim with a saturated aqueous solution of Cetyltrimethylammonium-bromide (CTAB; Merck, Amsterdam, Netherlands) containing 20% Dimethylsulfoxide (DMSO; Merck) to fixate the DNA and stored at −20°C for further analysis.
Map showing the sampling locations for the present study. The numbers correlate with the locations mentioned in Table 1
DNA isolation
DNA was isolated, using the DNeasy Tissue Kit (Qiagen) following the DNeasy protocol for animal tissues, with some modifications. About 20 mg of fish muscle tissue is completely lysed by adding 30 μl of Proteinase K. Complete lysis was achieved after overnight incubation at 55°C. DNA was isolated following the procedures outlined by Qiagen DNeasy protocol. Final DNA dilutions were performed with two times 75 μl buffer instead of two times of 100 μl as mentioned in the protocol. The quality of the genomic DNA was checked by electrophoresis in 1.2% agarose gels.
The quality of the DNA is most important for the AFLP analysis because the process is dependent upon the complete enzymatic digestion of the DNA via endonucleases. AFLP techniques require 100–500 ng of relatively pure DNA (Saunders et al. 2002). These fragments are subsequently used in PCR amplification, and therefore, it is necessary to have complete digestion of the DNA to get reproducible patterns between replicate samples.
AFLP procedures
AFLP analyses were applied as described by Vos et al. (1995) with modifications as described in De Roos (2003). Restriction—Ligation reactions were performed in a single reaction at 37°C for 2 h, in a total volume of 10 μl containing 100 ng DNA, 1 × T4 Ligase Buffer (Life Technology, Invitrogen), 0.05 M NaCl, 5 U EcoRI—Enzyme (New England BioLabs), 5U MseI—Enzyme (New England BioLabs), 0.045 M bovine serum albumine (BSA, New England BioLabs), 0.2 μM EcoRI—adapter (5′-CTCGTAGACTGCGTACC, CATCTGACGCATGGTTAA-′5), 2.0 μM MseI—adapter (5′-GACGATGAGTCCTGAG, TACTCAGGACTCAT-′5) and 1 U T4—Ligase (Life Technology, Invitrogen). PCR amplifications were performed on a T-gradient thermocycler (Biometra) in two separate amplification steps. Pre-amplification PCR reactions were conducted in a 20 μl volume (containing 4 μl 20× diluted Restriction–Ligation mix, 0.5 μM EcoRI/+ A primer (Applied Biosystems), 0.5 μM MseI/+ C primer (Applied Biosystems) and 15 μl AFLP Amplification Core Mix (Applied Biosystems), with the following temperature profile: an initial denaturation step of 2 min 94°C; 20 cycles with 20 s 94°C, 30 s 56°C, 2 min 72°C; followed by 2 min 72°C and 30 min 60°C. Selective amplifications were performed in a 10 μl volume (containing 1.5 μl 10× diluted Pre-amplification product, 0.05 μM Dye labeled EcoRI/+ ACC primer (Applied Biosystems), 0.25 μM MseI/+ CAT primer (Applied Biosystems) and 7.5 μl AFLP Amplification Core Mix (Applied Biosystems), with the following temperature profile: an initial denaturation step of 2 min 94°C; 10 cycles with 20 s 94°C, 30 s 66°C decreasing with 1°C per cycle, 2 min 72°C; 25 cycles with 20 94°C, 30 s 56°C, 2 min 72°C; followed by 30 min 60°C. Amplified fragments were analyzed on a CEQ™ 8000 Genetic Analysis System (Beckman Coulter Inc. 2002). Fragments between 100 and 350 bp were scored as present (1) or absent (0) using the Fragment Analysis Software Module of the CEQ™ 8000 (AFLP settings: bin width = 1.0 nt; Ythreshold = 400 RFU). Prior to the analyses the AFLP data were transformed to a binary 1/0 character matrix, indicating the presence and absence of bands, respectively. Loci with a band frequency below 5% were considered as potential artifacts and omitted from further analysis. Altogether, 12 different primer combinations are tested for the AFLP studies of D. polymorpha on the reduced set of samples with a goal to pick good pattern. The primer combination of AAC/CAT is selected based on the average number of reliable fragment length peaks in basepairs.
Data analysis
Genetic variation within populations was assessed by calculating Shannon’s index of diversity (I; Shannon and Weaver 1949), the number of polymorphic loci (N PL) and the proportion of polymorphic loci (P PL), using the software program POPGENE version 1.32 (Yeh et al. 1997).
Genetic diversity among populations and regional population structure was assessed by means of several methods: Firstly, Principal Component Analysis (PCA), a non-hierarchical grouping technique without prior knowledge of the source location of the sampled individuals, was performed using the GeneMaths XT software package, version 1.6.1 (Applied Maths BVBA, Sint-Martens-Latem, Belgium), to visualise the grouping of individuals into clusters. Secondly, phylogenetic analyses were performed using two different approaches: a neighbour-joining (NJ) distance analysis (Saitou and Nei 1987) and a Bayesian analysis (Huelsenbeck and Ronquist 2001). Preliminary analyses showed that the MR group clearly showed the earliest divergence when a closely related species Dreissena rostriformis bugensis was included as an outgroup. The North American (MR) population was therefore used as an out-group to compare the European populations. For the NJ analysis, the genetic distances were estimated using the algorithm of Nei and Li (1979), with the PHYLIP package (Felsenstein 2002). The Bayesian inference that evaluates posterior probabilities of clades was performed using the program MRBAYES version 3.1.2 (Ronquist and Huelsenbeck 2003). The character state transition model used is the one implemented in the program, the F81-like model, where the rate of the state transitions is calculated by the stationary state frequencies. Markov chain Monte Carlo from a random starting tree was initiated and run for 20 million generations. Trees were sampled every 1000th generation. The first 25% of the samples were discarded as ‘burn-in’, and the rest of the samples were used for inferring a Bayesian tree. The convergence diagnostic PSRF (potential scale reduction factor) value over the runs sampled approached 1. Examination of the log-likelihood and the observed consistency with the similar likelihood values between the two independent runs suggest that the run reached stationarity and that these burn-in periods were sufficiently long. Thirdly, an Analysis of Molecular Variance (AMOVA) was performed to assess the degree of molecular variation within and among populations, using the program ARLEQUIN v2.000 (Schneider et al. 2000), which performs a nested ANOVA using the matrix of Euclidean genetic distances as input (Excoffier et al. 1992). The level of population subdivision was estimated by calculating pairwise genetic distances between populations using Φ statistics (that are directly analogous to Wright’s F statistics; Weir and Cockerham 1984; Excoffier et al. 1992). The pair-wise genetic distances were calculated according to Lynch and Milligan (1994), using AFLP-SURV v1.0 under the assumption of a Hardy–Weinberg equilibrium (Vekemans et al. 2002). Exact tests of population differentiation (Raymond and Rousset 1995) were calculated with TFPGA (Tools for Populations Genetic Analysis version 1.3; Miller 1999). Analyses were performed with pairwise combinations of populations (using 20 batches and 2000 permutations), based on observed marker frequencies and assuming linkage equilibrium among loci (Miller 1999).
Results
Descriptive analyses
A total of 122 putative gene loci (i.e. clear and reproducible AFLP fragments) were resolved from the 237 D. polymorpha samples. The genetic diversity within populations was lowest in the Thames river, Great Britain (N pl = 32.79%; I = 0.0847 ± 0.1893) and highest in the Mississippi river, USA (N pl = 62.30%; I = 0.1800 ± 0.2409; Table 1).
Phylogeography and PCA analysis
In the Principal Component Analysis (PCA), more than 89% of the variation was explained by the first three components. The first component of PCA already accounted for 79% of the total variation. The first and fifth components clearly separate the nine Dreissena polymorpha populations into five geographically defined groups: G1 (Mississippi river, USA), G2 (Thames river, Great Britain), G3 (Danube river, Romania), G4 (the Waal river, the Meuse river and the Untersee in the neighbouring countries of the Netherlands, Belgium and Germany, respectively) and G5 (the Ebro river, the Petit Rhone river and the Po river in the neighbouring countries of Spain, France and Italy, respectively; Fig. 2).
In the phylogenetic analyses, the North American lineage was clearly separated from the European populations. With respect to the phylogenetic affinities of the European populations, the Romanian (DR) population appeared to be an early diverging lineage in both of the analyses (Fig. 3). The population from Great Britain (TR) forms a distinct lineage within the rest of the European populations, and their phylogenetic affinity was not quite clear. The NJ tree suggests that the German (US), Dutch (WR) and Belgium (MN) populations group together, including several Spanish (ER) and Italian (PR) samples, which was not supported in the Bayesian analysis. In the Bayesian inference, a weak support was found in the monophyly of the cluster including Spanish (ER), French (RR), and Italian (PR) groups. Although the detailed phylogenetic relationships are not clearly resolved, both of the phylogenetic analyses suggest that the Spanish populations are more closely related to the French populations (Fig. 3a, b).
Phylogenetic relationship of the nine Dreissena polymorpha populations. The Spanish (EB) populations and the French (RR) populations are enclosed in the boxes. a Neighbor-joining (NJ) analysis. The numbers at nodes represent the bootstrap percentages (values < 50 not shown) from 100 replicates. b Bayesian analysis. The numbers at nodes represent the posterior probabilities (values < 0.5 not shown.). The scale bars indicate the estimated evolutionary distance. Populations are encoded following Table 1. Numbers refer to original sample numbers
Population subdivision
The Analysis of Molecular Variance (AMOVA) showed that the overall population differentiation was high (ΦST = 0.69479, P < 0.0001), suggesting that the populations did not form a single panmictic unit. Of the total genetic variation partitioned in the nine D. polymorpha populations, 69.5% was attributed to the differences among populations, whereas 30.5% was attributed to the differences among individuals within populations (Table 2). The pairwise genetic distances (ΦST) between populations varied widely, ranging from 0.0683 (between the Ebro river in Spain and the Petit Rhone river in France) to 0.7847 (between the Thames river in Great Britain and the Mississippi river in the USA; Table 3).
The clustering inferred from the phylogeographic and PCA analyses is strongly supported by the pairwise genetic distances (ΦST) between the zebra mussel populations, that are much lower among populations within clusters (ranging 0.0683–0.1991 and 0.0930–0.2065 in G4 and G5, respectively), than among populations from different clusters (ranging 0.2675–0.7847; Table 3). The exact tests of pairwise population differentiation (implemented in TFPGA), furthermore, showed that population pairs within clusters G4 and G5 were not significantly differentiated (P > 0.05; Table 3), while population pairs from different clusters were highly significantly differentiated (P < 0.01; Table 3). In cluster G5, moreover, the more than two-fold higher ΦST values between the Spanish-Italian populations (ΦST = 0.1622) compared to the Spanish-French populations (ΦST = 0.0683), suggests that the Spanish population is more closely related to the French population than to the Italian population (Table 3).
Discussion
The invasion of zebra mussel D. polymorpha in Spanish waters has resulted in considerable ecological and economical consequences (Palau et al. 2009). The introduction of this nuisance species throughout Europe and North America has mainly been caused by unintentional, anthropogenic spread (Carlton 1993). Retracing invasion pathways and identifying potential mechanisms of unintentional introductions is a necessary first step towards the implementation of management strategies aimed at preventing future invasions. In this study, AFLP fingerprinting was used to examine the genetic relationships between zebra mussel populations throughout Europe and North America with the purpose of identifying the source of the invasion to Spain.
The Principle Component Analysis (PCA) carried out in this study clearly shows the existence of five clusters: G1 (USA), G2 (Great Britain), G3 (Romania), G4 (the Netherlands, Belgium and Germany) and G5 (Spain, France and Italy). This clustering is consistent with the genetic differentiation among populations, as genetic distances (ΦST) are low and non-significant among populations within clusters while being high and significant between populations from different clusters. The close genetic similarity among the populations within cluster G4 (the Netherlands, Belgium and Germany) most likely reflects their linear connectivity along two large interconnected river systems in North-West Europe: the Rhine and Meuse rivers. As opposed to the other populations, which inhabit separate, unconnected catchments (USA, Romania, Spain, France, Italy and Great Britain), the populations from the Netherlands, Belgium and Germany inhabit the Rhine-Meuse catchments: Population US (Germany) is situated in the upper reaches of the Rhine River, population MN (Belgium) in the middle reach of the Meuse River (near Namur, Belgium) and population WR lies at the confluence of the Rhine and Meuse Rivers (near Vuren, the Netherlands) (Fig. 1). The relatively low and non-significant ΦST values suggest that the populations from the Netherlands, Belgium and Germany are readily connected by gene flow (Table 3), with a weak indication that the rate of gene flow among these three populations may be related to the distance between them: the Netherlands—Belgium (river distance ± 200 km; ΦST = 0.0930), the Netherlands—Germany (±1,200 km; ΦST = 0.1862), and Belgium—Germany (±1,400 km; ΦST = 0.2065).
The advective transportation of free-swimming planktonic larvae may partly account for the high levels of gene flow among populations in the Rhine-Meuse catchments. Zebra mussel larvae have only limited swimming capabilities and therefore rely on currents for their passive spread. The length of the planktonic phase, which may vary from 2 to 5 weeks, allows passive downstream dispersal of veliger larvae over considerable distances (Stoeckel et al. 1997; Jantz and Neumann 1998; Stoeckel et al. 2004), despite high mortality rates during transit (Horvath and Lamberti 1999; Schneider et al. 2003). In addition, the transportation of adult mussels attached to the hulls of commercial ships and recreational craft may also account for a both upstream and downstream spread of the zebra mussel along these rivers. Today, the Rhine and Meuse rivers are important water highways for shipping traffic that transport cargo to and from the Rotterdam harbour in the Netherlands (one of the main European trade ports) and cities located upstream along the Rhine and Meuse River. This concurs with the findings of Elderkin et al. (2004) who argued that boat mediated upstream transportation led to the bidirectional spread of zebra mussels along the Mississippi River (USA), thus accounting for an absence of a unidirectional model of gene flow.
The PCA clustering, the phylogenetic analyses and the pair-wise genetic distances show that the populations from France, Italy and Spain (cluster G5) are also genetically very similar, despite the fact that these populations inhabit unconnected catchments that are spatially separated by the presence of large mountain ranges: the Pyrenees (between France and Spain) and the Alps (between France and Italy) (Fig. 1). This close genetic similarity is most likely the result of recent introductions, in which one location served as the source for the introduction to the other locations. Pollux et al. (2003), for example, showed that the close genetic similarity inferred from AFLP loci among Irish and British zebra mussel populations most likely resulted from a recent introduction of zebra mussels from the Great Britain to Ireland, which was later confirmed in a study by Astanei et al. (2005) using microsatellite markers. The zebra mussel reached Spain around 2001, being found for the first time in the Riba-roja reservoir in the lower part of the Ebro River, North-East Spain (Ruiz-Altaba et al. 2001). The close genetic similarity among the Spanish, French and Italian populations (cluster G5) suggests that either France or Italy may have served as the origin of invasion to Spain around 2001. The genetic population differentiation is considerably lower between the Spanish-French populations (ΦST = 0.0683) compared to the Spanish-Italian populations (ΦST = 0.1622) and suggest that France most likely acted as the source region from which the zebra mussel was introduced to Spain.
During the 19th century the zebra mussel colonised large parts of North and Central Europe, including many of France’s major river systems e.g. Seine River 1855, Loire River 1863, Rhône River 1865 and Garonne River 1866 (Kinzelbach 1992). Its dispersal further southward was initially impeded by large mountain ranges like the Alps and the Pyrenees. However, the increasing popularity of recreational water sports after the Second World War led to the spread of zebra mussels, attached to recreational craft transported on trailers, to high alpine lakes (e.g. Lakes Geneva, Zurich and Constance in Switzerland) around 1960s (Kinzelbach 1992). In a similar way, zebra mussels from France are thought to have crossed the large mountain ranges (Alps and Pyrenees) resulting in the colonisation of more isolated South European countries such as Yugoslavia (1970s; Ludyanskiy et al. 1993), Italy (1969–1970; Giusti and Oppi 1972; Annoni et al. 1978) and most recently Spain (2001; Araujo and Álvarez 2001; Bij de Vaate et al. 2002). The zebra mussel has a high aerial exposure tolerance and is likely to survive a 3–6 day journey attached to the hulls of boats transported on trailers (Pollux et al. 2009). Not surprisingly, the transportation of recreational craft is deemed one of the most important vectors for the spread of zebra mussels among isolated catchments and water bodies (Padilla et al. 1996; Johnson et al. 2001; Minchin et al. 2002).
Transport via ballast tanks of big commercial vessels was probably a further mechanism by which the D. polymorpha were successfully introduced in many areas (Minchin et al. 2002). Carlton (1993) has suggested that transport through ballast water from a European port is considered to be the route by which zebra mussels (most likely as veliger larvae) were introduced into the Great Lakes of North America (Hebert et al. 1989). Similarly, Bij de Vaate et al. (2002) have discussed the role of ballast-water transport in the spread of a number of Ponto-Caspian species through the mainland of Europe, including the zebra mussel. However, this is relatively unlikely because of the absence of big vessels coming from the ocean into the Ebro river system.
The third possibility of introduction is through transport of exotic fish species to the Ebro river. Historically, there have been introductions of fishes from Central Europe to Spain (Palau et al. 2009) examples being the bleak (Alburnus alburnus) and the zander (Sander lucioperca). According to Palau et al. (2006), the introduction of the zebra mussel in Spain coincides with introduction of such exotic fish species first reported in the lower reaches of the river Ebro. Fish tanks could have been filled with water from D. polymorpha infested rivers and mussel larvae could have easily survived the short journey without any problems (Minchin et al. 2003; Bidwell 2009).
Conclusions
Phylogenetic analyses and pair-wise genetic distances determined from the PCR based AFLP fingerprinting have revealed that the recent invasion of zebra mussels of Spain has most likely been from France. The introduction of D. polymorpha to Spain has been possibly attributed to the transport of recreational boats with mussel-fouled hulls or by fish transport across the Pyrenees (Bij de Vaate et al. 2002; Minchin et al. 2003; Pollux et al. 2003).
References
Annoni D, Bianchi I, Girod A et al (1978) Introduction of Dreissena polymorpha (Pallas) (Mollusca, Bivalvia) in the coastal mollusc beds of Lake Garda (North Italy). Ouaderni Civica Stazione Idrobiologica Milano 6:77–84
Araujo R, Álvarez RM (2001) El mejillón cebra en el Ebro: un grave caso de riesgo ambiental en Aragón. Naturaleza Aragonesa 8:39–46
Astanei I, Gosling E, Wilson J et al (2005) Genetic variability and phylogeography of the invasive zebra mussel, Dreissena polymorpha (Pallas). Mol Ecol 14:1655–1666
Beckman Coulter Inc (2002) CEQ™ 8000 Genetic analysis system
Bidwell JR (2009) Range expansion of the zebra mussel, Dreissena polymorpha: a review of major dispersal vectors in Europe and North America. In: Van der Velde G, Rajagopal S, Bij de Vaate A (eds) The Zebra Mussel in Europe. Backhuys Publishers, Leiden, pp 73–83
Bij de Vaate A, Jazdzewski K, Ketelaars HAM et al (2002) Geographical patterns in range extension of Ponto-Caspian macro-invertebrate species in Europe. Can J Fish Aquat Sci 59:1159–1174
Burlakova LE, Padilla DK, Karatayev AY et al (2006) Endosymbionts of Dreissena polymorpha in Ireland: evidence for the introduction of adult mussels. J Mollusc Stud 72:207–210
Carlton JT (1993) Dispersal mechanisms of the zebra mussel (Dreissena polymorpha). In: Nalepa TF, Schloesser DW (eds) Zebra mussels: biology, impacts, and control. Lewis Publishers, CRC Press, Boca Raton, USA, pp 677–698
De Roos K (2003) CEQ™ 8000 AFLP Protocol. Beckman Coulter Netherlands BV, Mijdrecht, The Netherlands
Elderkin CL, Perkins EJ, Leberg PL et al (2004) Amplified fragment length polymorphism (AFLP) analysis of the genetic structure of the zebra mussel, Dreissena polymorpha, in the Mississippi River. Freshw Biol 49:1487–1494
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Felsenstein J (2002) PHYLIP (Phylogeny Inference Package) version 3.6a3. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle
Giusti F, Oppi E (1972) Dreissena polymorpha (Pallas) nuovamente in Italia. Memorie del Museo Civico di Storia Naturale di Verona 10:45–49
Griffiths RW (1993) Effects of zebra mussel (Dreissena polymorpha) on the benthic fauna of Lake St. Clair. In: Nalepa TF, Schloesser DW (eds) Zebra mussels: biology, impacts and control. Lewis Publishers, Boca Raton, Fla, pp 415–437
Haag WR, Berg DJ, Garton DW et al (1993) Reduced survival and fitness in native bivalves in response to fouling by the introduced zebra mussel (Dreissena polymorpha) in Western Lake Erie. Can J Fish Aquat Sci 50:13–19
Hebert PDN, Muncaster BW, Mackie GL (1989) Ecological and genetic studies on Dreissena polymorpha (Pallas): a new mollusc in the Great Lakes. Can J Fish Aquat Sci 46:1587–1591
Horvath T, Lamberti GA (1999) Mortality of zebra mussel, Dreissena polymorpha, veligers during downstream transport. Freshw Biol 42:69–76
Huelsenbeck JP, Ronquist F (2001) MRBAYES. Bayesian inference phylogeny. Bioinformatics 17:754–755
Hushak L (1996) Zebra mussels costing $120 million in five years. Sea Grant Network, Zebra Mussel Update 27:4
Jantz B, Neumann D (1998) Growth and reproductive cycle of the zebra mussel in the River Rhine as studied in a river bypass. Oecologia 114:213–225
Johnson LE, Carlton JT (1996) Post-establishment spread in large-scale invasions: dispersal mechanisms of the zebra mussel Dreissena polymorpha. Ecology 77:1686–1690
Johnson LE, Ricciardi A, Carlton JT (2001) Overland dispersal of aquatic invasive species: a risk assessment of transient recreational boating. Ecol Appl 11:1789–1799
Kinzelbach R (1992) The main features of the phylogeny and dispersal of the Zebra mussel Dreissena polymorpha. In: Neumann D, Jenner HA (eds) The Zebra mussel Dreissena polymorpha. Gustav Fisher Verlag, Stuttgart, pp 5–17
Ludyanskiy ML, McDonald D, MacNeill D (1993) Impact of the zebra mussel, a bivalve invader. Bioscience 43:533–544
Lynch B, Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Mol Ecol 3:91–99
Mackie GL (1991) Biology of the exotic zebra mussel, Dreissena polymorpha, in relation to native bivalves and its potential impact in Lake St. Clair. Hydrobiologia 219:251–268
May GE, Gelembiuk GW, Panov VE et al (2006) Molecular ecology of zebra mussel invasions. Mol Ecol 15:1021–1031
Miller MP (1999) Tools for population genetic analyses (TFPGA): a windowa™ program for the analysis of allozyme and molecular population genetic data. Available at http://www.marksgeneticsoftware. net/_vti_bin/shtml.exe/tfpga.htm
Minchin D, Lucy F, Sullivan M (2002) Zebra mussel: impacts and spread. In: Leppäkoski E, Gollasch S, Olenin S (eds) Invasive aquatic species of Europe: distribution. impacts and management. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 135–146
Minchin D, Maguire C, Rosell R (2003) The zebra mussel (Dreissena polymorpha PALLAS) invades Ireland: human mediated vectors and the potential for rapid intranational spread. Biol Environ Proc R Ir Acad 103B:23–30
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Nat Acad Sci USA 76:5269–5273
Padilla DK, Chotkowski MA, Buchan LAJ (1996) Predicting the spread of zebra mussels (Dreissena polymorpha) to inland waters using boater movement patterns. Global Ecol Biogeogr Lett 5:353–359
Palau A, Casas R, Cia I et al (2006) The ENDESA control strategy of Zebra Mussels (Dreissena polymorpha) in the Riba-Roja reservoir. Transactions of the International Congress on Large Dams
Palau A, Cia I, Casas R et al (2009) Zebra mussel distribution and habitat preference in the lower Ebro river (North East Spain). In: Van der Velde G, Rajagopal S, Bij de Vaate A (eds) The Zebra Mussel in Europe. Backhuys Publishers, Leiden, pp 121–129
Peribañez MA (2005) Estudio del mejillón cebra, Dreissena polymorpha, en el río Ebro: Encuesta parasitológica y pruebas de control microbiológico en el laboratorio. Unpublished document. Universidad de Zaragoza—ENDESA. 3 pp
Pimentel D, Lach L, Zuniga R et al (2000) Environmental and economic costs of nonindigenous species in the United States. Bioscience 50:53–65
Pollux BJA, Minchin D, Van der Velde G et al (2003) Zebra mussels (Dreissena polymorpha) in Ireland, AFLP-fingerprinting and boat traffic both indicate an origin from Britain. Freshw Biol 48:1127–1139
Pollux BJA, Van der Velde G, Bij de Vaate A (2009) A perspective on global spread of the zebra mussel Dreissena polymorpha: a review on possibilities and limitations. In: Van der Velde G, Rajagopal S, Bij de Vaate A (eds) The Zebra Mussel in Europe. Backhuys Publishers, Leiden, pp 47–61
Raymond M, Rousset F (1995) GENEPOP (V. 1.2): a population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Reeders HH, Bij de Vaate A (1990) Zebra mussels (Dreissena polymorpha): a new perspective for water quality management. Hydrobiologia 200/201:437–450
Reeders HH, Bij de Vaate A, Slim FJ (1989) The filtration rate of Dreissena polymorpha (Bivalvia) in three Dutch lakes with reference to biological water quality management. Freshw Biol 22:133–141
Reeders HH, Bij de Vaate A, Noordhuis R (1993) Potential of the Zebra nussel (Dreissena polymorpha) for water quality management. In: Nalepa TF, Schloesser DW (eds) Zebra mussel: biology, impacts, and control. Lewis Publishers, CRC Press, Boca Raton, USA, pp 439–451
Roberts L (1990) Zebra mussel invasion threatens US waters. Science 249:1370–1372
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Ruiz-Altaba C, Jiménez PJ, López MA (2001) El temido mejillón cebra empieza a invadir los ríos españoles desde el curso bajo del río Ebro. Quercus 188:50–51
Saitou N, Nei M (1987) The neighbour-joining method: a new method for reconstructing phylogenetic tree. Mol Biol Evol 4:406–425
Saunders JA, Mischke S, Hemeida AA (2002) The use of AFLP techniques for DNA fingerprinting in plants. Application information, Beckman Couter, California, pp 1–9
Schloesser DW, Nalepa TF, Mackie GL (1996) Zebra mussel infestation of unionid bivalves (Unionidae) in North America. Am Zool 36:300–310
Schneider S, Roessli D, Excoffier L (2000) Arlequin: a software for population genetics data analysis. Ver 2.000. Genetics and Biometry Lab, Department of Anthropology, University of Geneva
Schneider DW, Stoeckel JA, Rehmann CR et al (2003) A developmental bottleneck in dispersing larvae: implications for spatial population dynamics. Ecol Lett 6:352–360
Shannon CE, Weaver W (1949) The mathematical theory of communication. University of Illinois Press, Urbana
Stepien CA, Taylor CD, Dabrowska KA (2002) Genetic variability and phylogeographical patterns of a nonindigenous species invasion: a comparison of exotic vs. native zebra and quagga mussel populations. J Evol Biol 15:314–328
Stoeckel JA, Schneider DW, Soeken LA et al (1997) Larval dynamics of a riverine metapopulation: implications for zebra mussel recruitment, dispersal, and control in a large river system. J North Am Benthol Soc 16:586–601
Stoeckel JA, Rehmann CR, Schneider DW et al (2004) Retention and supply of zebra mussel larvae in a large river system: importance of an upstream lake. Freshw Biol 49:919–930
Strayer DL, Caraco NF, Cole JJ et al (1999) Transformation of freshwater ecosystems by bivalves. Bioscience 49:19–27
Therriault TW, Orlova MI, Docker MF et al (2005) Invasion genetics of a freshwater mussel (Dreissena rostriformis bugensis) in eastern Europe: high gene flow and multiple introductions. Heredity 95:16–23
Van der Velde G, Paffen BGP, Van den Brink FWB et al (1994) Decline of zebra mussel populations in the Rhine–Competition between two mass invaders (Dreissena polymorpha and Corophium curvispinum). Naturwissenschaften 81:2–34
Vekemans X, Beauwens T, Lemaire M et al (2002) Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Mol Ecol 11:139–151
Vos P, Hogers R, Bleeker M et al (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution Int J org Evolution 38:1358–1370
Wilson AB, Naish KA, Boulding EG (1999) Multiple dispersal strategies of the invasive quagga mussel (Dreissena bugensis) as revealed by microsatellite analysis. Can J Fish Aquat Sci 56:2248–2261
Yeh FC, Yang R, Boyle TBJ et al (1997) POPGENE, the user-friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Centre, University of Alberta, Canada
Acknowledgments
This study was funded by the European Commission in the Community’s Sixth Framework Programme (INCO project, Contract number: PL510658, Assessing impacts of TBT on multiple coastal uses). It is contribution number 470 of the Centre for Wetland Ecology (CWE).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Rajagopal, S., Pollux, B.J.A., Peters, J.L. et al. Origin of Spanish invasion by the zebra mussel, Dreissena polymorpha (Pallas, 1771) revealed by amplified fragment length polymorphism (AFLP) fingerprinting. Biol Invasions 11, 2147–2159 (2009). https://doi.org/10.1007/s10530-009-9495-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-009-9495-3