Abstract
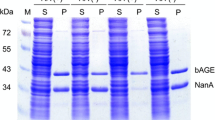
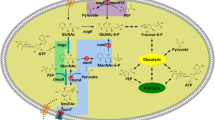
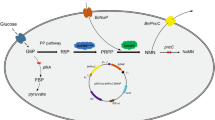
N‑Acetyl‑d‑neuraminic acid (Neu5Ac) is the crucial compound for the chemical synthesis of antiflu medicine Zanamivir. Chemoenzymatic synthesis of Neu5Ac involves N-acetyl-d-glucosamine 2-epimerase (AGE)-catalyzed epimerization of N-acetyl-d-glucosamine (GlcNAc) to N-acetyl-d-mannosamine (ManNAc), and aldolase-catalyzed condensation between ManNAc and pyruvate. Host optimization plays an important role in the whole-cell biotransformation of value-added compounds. In this study, via single-plasmid biotransformation system, we showed that the AGE gene BT0453, cloned from human gut microorganism Bacteroides thetaiotaomicron VPI-5482, showed the highest biotransformation yield among the AGE genes tested; and there is no clear Neu5Ac yield difference between the BT0453 coupled with one aldolase coding nanA gene and two nanA genes. Next, Escherichia coli chromosomal genes involved in substrate degradation, product exportation and pH change were deleted via recombineering and CRISPR/Cas9. With the final E. coli BL21(DE3) ΔnanA Δnag ΔpoxB as host, a significant 16.5% yield improvement was obtained. Furthermore, precursor (pyruvate) feeding resulted in 3.2% yield improvement, reaching 66.8% molar biotransformation. The result highlights the importance of host optimization, and set the stage for further metabolic engineering of whole-cell biotransformation of Neu5Ac.




Similar content being viewed by others
References
Campeotto I, Bolt AH, Harman TA et al (2010) Structural insights into substrate specificity in variants of N-acetylneuraminic acid lyase produced by directed evolution. J Mol Biol 404:56–69
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645
Deng MD, Severson DK, Grund AD et al (2005) Metabolic engineering of Escherichia coli for industrial production of glucosamine and N-acetylglucosamine. Metab Eng 7:201–214
Deng MD, Grund AD, Wassink SL et al (2006) Directed evolution and characterization of Escherichia coli glucosamine synthase. Biochimie 88:419–429
Gao X, Zhang F, Wu M, Shang G (2019) Production of N-acetyl-d-neuraminic acid by whole cells expressing Bacteroides thetaiotaomicron N-acetyl-d-glucosamine 2-epimerase and Escherichia coli N-acetyl-d-neuraminic acid aldolase. J Agric Food Chem 67:6285–6291
Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA (2013) RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31:233–239
Kao C, Chen Y, Wang L, Lee YC (2018) Production of N-acetyl-d-neuraminic acid by recombinant single whole cells co-expressing N-acetyl-d-glucosamine-2-epimerase and N-acetyl-d-neuraminic acid aldolase. Mol Biotechnol 60:427–434
Klermund L, Groher A, Castiglione K (2013) New N-acyl-d-glucosamine 2-epimerases from cyanobacteria with high activity in the absence of ATP and low inhibition by pyruvate. J Biotechnol 168:256–263
Kragl U, Gygax D, Ghisalba O, Wandrey C (1991) Enzymatic two-step synthesis of N-acetyl-neuraminic acid in the enzyme membrane reactor. Angew Chem Int Ed Engl 30:827–828
Kuhlman TE, Cox EC (2010) Site-specific chromosomal integration of large synthetic constructs. Nucl Acids Res 38:e92
Lee YC, Chien HC, Hsu WH (2007) Production of N-acetyl-d-neuraminic acid by recombinant whole cells expressing Anabaena sp. CH1 N-acetyl-d-glucosamine 2-epimerase and Escherichia coli N-acetyl-d-neuraminic acid lyase. J Biotechnol 129:453–460
Li J, Sun J, Gao X, Wu Z, Shang G (2019) Coupling ssDNA recombineering with CRISPR-Cas9 for Escherichia coli DnaG mutations. Appl Microbiol Biotechnol 103:559–3570
Lin B, Zhang Z, Liu W, Dong Z, Tao Y (2013) Enhanced production of N-acetyl-d-neuraminic acid by multi-approach whole-cell biocatalyst. Appl Microbiol Biotechnol 97:4775–4784
Mahmoudian M, Noble D, Drake CS et al (1997) An efficient process for production of N-acetylneuraminic acid using N-acetylneuraminic acid aldolase. Enzyme Microb Technol 20:393–400
Maru I, Ohta Y, Murata K, Tsukada Y (1996) Molecular cloning and identification of N-acyl-d-glucosamine 2-epimerase from porcine kidney as a renin-binding protein. J Biol Chem 271:16294–16299
Maru I, Ohnishi J, Ohta Y, Tsukada Y (1998) Simple and large-scale production of N-acetylneuraminic acid from N-acetyl-d-glucosamine and pyruvate using N-acyl-d-glucosamine 2-epimerase and N-acetylneuraminate lyase. Carbohydr Res 306:575–578
Moxley WC, Eiteman MA (2021) Pyruvate production by Escherichia coli by use of pyruvate dehydrogenase variants. Appl Environ Microbiol 87:e0048721
Murphy KC (2016) λ recombination and recombineering. EcoSal plus. https://doi.org/10.1128/ecosalplus.ESP-0011-2015
Posfai G, Plunkett G III, Feher T et al (2006) Emergent properties of reduced-genome Escherichia coli. Science 312:1044–1046
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York
Sola-Carvajal A, Sanchez-Carron G, Garcia-Garcia MI, Garcia-Carmona F, Sanchez-Ferrer A (2012) Properties of BoAGE2, a second N-acetyl-d-glucosamine 2-epimerase from Bacteroides ovatus ATCC 8483. Biochimie 94:222–230
Tabata K, Koizumi S, Endo T (2002) Production of N-acetyl-d-neuraminic acid by coupling bacteria expressing N-acetyl-d-glucosamine 2-epimerase and N-acetyl-d-neuraminic acid synthetase. Enzyme Microb Technol 30:327–333
Takahashi S, Takahashi K, Kaneko T, Ogasawara H, Shindo S, Kobayashi M (1999) Human renin-binding protein is the enzyme N-acetyl-d-glucosamine 2-epimerase. J Biochem 125:348–353
van Oh JH, Pijkeren JP (2014) CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucl Acids Res 42:e131
von Itzstein M (2007) The war against influenza: discovery and development of sialidase inhibitors. Nat Rev Drug Discov 6:967–974
Wu S, Snajdrova R, Moore JC, Baldenius K, Bornscheuer UT (2021) Biocatalysis: enzymatic synthesis for industrial applications. Angew Chem Int Ed Engl 60:88–119
Yan Q, Fong SS (2018) Design and modularized optimization of one-step production of N-acetylneuraminic acid from chitin in Serratia marcescens. Biotechnol Bioeng 115:2255–2267
Zhu D, Wu J, Zhan X, Zhu L, Zheng Z, Gao M (2017) Phosphoenolpyruvate-supply module in Escherichia coli improves N-acetyl-d-neuraminic acid biocatalysis. Biotechnol Lett 39:227–234
Acknowledgements
We thank Dr. Thomas Kuhlman for providing pTKRed used in this research.
Supplementary Information
Supplementary file 1. Table S1: Bacterial strains and plasmids used in this research. Table S2: PCR primers and oligonucleotides used in the study.
Figure S1: Schematic presentation of the recombineering and I-SceI-mediated gene deletion. Figure S2: Confirmation of Δnan mutant. Figure S3: Confirmation of Δman mutant. Figure S4: Confirmation of Δnag mutant. Figure S5: Confirmation of ΔpoxB mutant. Figure S6: Outline of CRISPR/Cas9-assisted ssDNA recombineering for gene deletion. Figure S7: Confirmation of ΔldhA mutant. Figure S8: Confirmation of ΔackA mutant. Figure S9: HPLC analysis of Neu5Ac biotransformation.
Funding
This research was financially supported by the National Natural Science Foundation of China (No. 82073742).
Author information
Authors and Affiliations
Contributions
GS was responsible for the experimental design and drafted the manuscript. QZ, JZ and YS performed the experiments. All authors participated in data analysis, read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors report no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, Q., Zhang, J., Shao, Y. et al. Escherichia coli BL21(DE3) optimized deletion mutant as the host for whole-cell biotransformation of N‑acetyl‑d‑neuraminic acid. Biotechnol Lett 45, 1521–1528 (2023). https://doi.org/10.1007/s10529-023-03426-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-023-03426-3