Abstract
Purpose
We purified and characterized a novel ene-reductase (KaDBR1) from Kazachstania exigua HSC6 for the synthesis of dihydro-β-ionone from β-ionone.
Methods
KaDBR1 was purified to homogeneity by ammonium sulfate precipitation and phenyl-Sepharose Fast Flow and Q-Sepharose chromatography. The purified enzyme was characterized by measuring the amount of dihydro-β-ionone from β-ionone with LC–MS analysis method.
Results
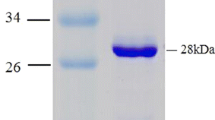
The molecular mass of KaDBR1 was estimated to be 45 kDa by SDS-PAGE. The purified KaDBR1 enzyme had optimal activity at 60 °C and pH 6.0. The addition of 5 mM Mg2+, Ca2+, Al3+, Na+, and dithiothreitol increased the activity of KaDBR1 by 25%, 18%, 34%, 20%, and 23%, respectively. KaDBR1 favored NADH over NADPH as a cofactor, and its catalytic efficiency (kcat/Km) toward β-ionone using NADH was 8.1-fold greater than when using NADPH.
Conclusion
Owing to its unique properties, KaDBR1 is a potential candidate for the enzymatic biotransformation of β-ionone to dihydro-β-ionone in biotechnology applications.





Similar content being viewed by others
References
Adalbjornsson BV, Toogood HS, Fryszkowska A, Pudney CR, Jowitt TA, Leys D, Scrutton NS (2010) Biocatalysis with thermostable enzymes: structure and properties of a thermophilic ‘ene’-reductase related to old yellow enzyme. ChemBioChem 11(2):197–207
Aloum L, Alefishat E, Adem A, Petroianu G (2020) Ionone is more than a violet’s fragrance: a review. Molecules 25(24):5822
Athéna PO, Bradford S, Jon DS (2014) Pichia stipitis OYE 2.6 variants with improved catalytic efficiencies from site-saturation mutagenesis libraries. Bioorg Med Chem 22(20):5628–5632
Beekwilder J, van Rossum HM, Koopman F, Sonntag F, Buchhaupt M, Schrader J, Hall RD, Bosch D, Pronk JT, van Maris AJA, Daran JM (2014) Polycistronic expression of a beta-carotene biosynthetic pathway in Saccharomyces cerevisiae coupled to beta-ionone production. J Biotechnol 192:383–392
Belsito D, Bickers D, Bruze M, Calow P, Greim H, Hanifin JM, Rogers AE, Saurat JH, Sipes IG, Tagami H (2007) A toxicologic and dermatologic assessment of ionones when used as fragrance ingredients. Food Chem Toxicol 45:S130–S167
Berger RG (2009) Biotechnology of flavours-the next generation. Biotechnol Lett 31(11):1651–1659
Black MJ, Biegasiewicz KF, Meichan AJ, Oblinsky DG, Kudisch B, Scholes GD, Hyster TK (2020) Asymmetric redox-neutral radical cyclization catalysed by flavin-dependent ’ene’-reductases. Nat Chem 12(1):71–75
Brige A, Van den Hemel D, Carpentier W, De Smet L, Van Beeumen JJ (2006) Comparative characterization and expression analysis of the four old yellow enzyme homologues from Shewanella oneidensis indicate differences in physiological function. Biochem J 394(Pt1):335–344
Cataldo VF, Lopez J, Carcamo M, Agosin E (2016) Chemical vs biotechnological synthesis of C-13-apocarotenoids: current methods, applications and perspectives. Appl Microbiol Biotechnol 100(13):5703–5718
Chaparro-Riggers JF, Rogers TA, Vazquez-Figueroa E, Polizzi KM, Bommarius AS (2007) Comparison of three enoate reductases and their potential use for biotransformations. Adv Synth Catal 349(8–9):1521–1531
Chapuis C, Jacoby D (2001) Catalysis in the preparation of fragrances and flavours. Appl Catal A 221(1):93–117
Chen BS, Médici R, van der Helm MP, van Zwet Y, Gjonaj L, van der Geest R, Otten LG, Hanefeld U (2018) Rhodococcus strains as source for ene-reductase activity. Appl Microbiol Biotechnol 02(13):5545–5556
Czajka JJ, Nathenson JA, Benites VT, Baidoo EEK, Cheng QS, Wang YC, Tang YJ (2018) Engineering the oleaginous yeast Yarrowia lipolytica to produce the aroma compound β-ionone. Microb Cell Fact 17(1):136
Heckenbichler K, Schweiger A, Brandner LA, Binter A, Toplak M, Macheroux P, Gruber K, Breinbauer R (2018) Asymmetric reductive carbocyclization using engineered ene reductases. Angew Chem Int Ed 57(24):7240–7244
Iorgu AI, Hedison TM, Hay S, Scrutton NS (2019) Selectivity through discriminatory induced fit enables switching of NAD(P)H coenzyme specificity in old yellow enzyme ene-reductases. FEBS J 286(16):3117–3128
Knowles WS (2002) Asymmetric hydrogenations (Nobel lecture). ChemInform 33(40):1999–2007
Lalko J, Lapczynski A, McGinty D, Bhatia S, Letizia CS, Api AM (2007) Fragrance material review on dihydro-β-ionone. Food Chem Toxicol 45(1):S225–S228
Lin F, Liu XL (2017) A preliminary study on preparation of two hydrogen β-ketone by hydrogenation. Biol Chem Eng 3(5):53–56
Noyori R (2002) Asymmetric catalysis: science and opportunities (Nobel lecture). Angew Chem 41(12):2008
Opperman DJ, Sewell BT, Litthauer D, Isupov MN, Littlechild JA (2010) Crystal structure of a thermostable old yellow enzyme from Thermus scotoductus SA-01. Biochem Biophys Res Commun 393(3):426–431
Parmeggiani F, Brenna E, Colombo D, Gatti FG, Tentori F, Tessaro D (2022) “A Study in Yellow”: investigations in the Stereoselectivity of Ene-Reductases. ChemBioChem 23(1):e202100445
Pei XQ, Xu MY, Wu ZL (2015) Two “classical” old yellow enzymes from Chryseobacterium sp. CA49: broad substrate specificity of Chr-OYE1 and limited activity of Chr-OYE2. J Mol Catal B 123(1):91–99
Qi ZP, Tong XY, Zhang XM, Lin HF, Bu S, Zhao LG (2022) One-pot synthesis of dihydro-β-ionone from carotenoids using carotenoid cleavage dioxygenase and enoate reductase. Bioprocess Biosyst Eng 45(5):891–900
Romano D, Contente ML, Molinari F, Eberini I, Ruvutuso E, Sensi C, Amaretti A, Rossi M, Raimondi S (2014) Recombinant S. cerevisiae expressing old yellow enzymes from non-conventional yeasts: an easy system for selective reduction of activated alkenes. Microb Cell Fact 13:60
Santosh KP, Despina JB, Jon DS (2009) Site-saturation mutagenesis of tryptophan 116 of Saccharomyces pastorianus old yellow enzyme uncovers stereo complementary variants. J Am Chem Soc 131(9):3271–3280
Schittmayer M, Glieder A, Uhl MK, Winkler A, Zach S, Schrittwieser JH, Kroutil W, Macheroux P, Gruber K, Kambourakis S (2011) Old yellow enzyme-catalyzed dehydrogenation of saturated ketones. Adv Synth Catal 353:268–274
Simona RM, Niero M, Loprete G, Cendron L, Bergantino E (2021) A new thermophilic ene-reductase from the filamentous anoxygenic phototrophic bacterium Chloroflexus aggregans. Microorganisms 9(5):953
Stuermer R, Hauer B, Hall M, Faber K (2007) Asymmetric bioreduction of activated C=C bonds using enoatereductasesfromthe old yellow enzyme family. Curr Opin Chem Biol 11(2):203–213
Szczepańska E, Colombo D, Tentori F, Olejniczak T, Brenna E, Monti D, Boratyński F (2021) Ene-reductase transformation of massoia lactone to δ-decalactone in a continuous-flow reactor. Sci Rep 11(1):18794
Toogood HS, Scrutton NS (2014) New developments in ‘ene’-reductase catalysed biological hydrogenations. Curr Opin Chem Biol 19:107–115
Toogood HS, Scrutton NS (2019) Discovery, characterisation, engineering and applications of ene reductases for industrial biocatalysis. ACS Catal 8(4):3532–3549
Toogood HS, Gardiner JM, Scrutton NS (2010) Biocatalytic reductions and chemical versatility of the old yellow enzyme family of flavoprotein oxidoreductases. Chem Cat Chem 2(8):892–914
Tsuji N, Honda K, Wada M, Okano K, Ohtake H (2014) Isolation and characterization of a thermo tolerant ene-reductase from Geobacillus sp. 30 and its heterologous expression in Rhodococcus opacus. Appl Microbiol Biotechnol 98(13):5925–5935
Yang G, Hong S, Yang P, Sun Y, Wang Y, Zhang P, Jiang W, Gu Y (2021) Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria. Nat Commun 12(1):790
Zhang CQ, Chen XX, Lindley ND, Too HP (2018a) A “plug-n-play” modular metabolic system for the production of apocarotenoids. Biotechnol Bioeng 115(1):174–183
Zhang XS, Liao SY, Cao FL, Zhao LG, Pei JJ, Tang F (2018b) Cloning and characterization of enoate reductase with high β-ionone to dihydro-β-ionone bioconversion productivity. BMC Biotechnol 18(1):26
Supporting information
Supplementary Fig. 1—Mass spectrometry analysis of the digested KaDBR1. The amino acid sequences of the ene-reductase from Kazachstania exigua (Accession No. AEP22543.1) and those of the eight fragments of purified KaDBR1 determined by MALDI-TOF-MS are shown.
Supplementary Fig. 2—HPLC analysis of the catalytic product of purified KaDBR1. (A) authentic β-ionone, (B) authentic dihydro-β-ionone, (C) HPLC analysis of the reaction products.
Supplementary Fig. 3—In vitro kinetic analysis of purified KaDBR1 with β-ionone as substrate. Reaction velocity (mmol/min/L) as a function of substrate concentration is plotted for 0–55 μM NADH (A), 0–80 μM NADPH (B) and 0–10 mM β-ionone (C).
Funding
This work was supported by grants from the Zhongyuan Science and Technology Innovation Leading Talent Project (224200510017), and the Science and Technology Project of China Guangxi Tobacco Industry Co. Ltd. (2020450000340027).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Human and animal rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Long, Z., Li, K., Xue, Y. et al. Purification and biochemical characterization of a novel ene- reductase from Kazachstania exigua HSC6 for dihydro-β-ionone from β-ionone. Biotechnol Lett 45, 499–508 (2023). https://doi.org/10.1007/s10529-023-03355-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-023-03355-1