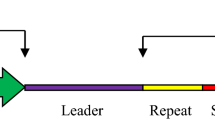
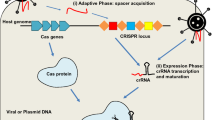
Abstract
Clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system allows biologists to edit genomic DNA of any cell in precise and specific way, entailing great potential for crop improvement, drug development and gene therapy. The system involves a nuclease (Cas9) and a designed guide RNA that are involved in wide range of applications such as genome modification, transcriptional modulation, genomic loci marking and RNA tracking. The limitation of the technique, in view of resistance of thymidine-rich genome to Cas9 cleavage, has now been overcome by the use of Cpf1 nuclease. In this review, we present an overview of CRISPR nucleases (Cas9 or Cpf1) with particular emphasis on human genome modification and compare their advantages and limitations. Furthermore, we summarize some of the pros and cons of CRISPR technology particularly in human therapeutics.


Similar content being viewed by others
Change history
07 October 2021
A Correction to this paper has been published: https://doi.org/10.1007/s10529-021-03189-9
References
Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, Zhang F (2016) C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353:6299
Ain QU, Chung JY, Kim YH (2015) Current and future delivery systems for engineered nucleases: ZFN, TALEN and REGN. J Control Release 205:120–127
Anantharaman V, Makarova KS, Burroughs AM, Koonin EV, Aravind L (2013) Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol Direct 8:15
Anders C, Niewoehner O, Duerst A, Jinek M (2014) Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513:569–573
Baltes NJ, Gil-Humanes J, Cermark T, Atkins PA, Voytas DF (2014) DNA replicons for plant genome engineering. Plant Cell 26:151–163
Baltimore D, Berg P, Botchan M, Carroll D, Charo RA, Church G, Corn JA, Daley JQ, Doudna JA, Fenner M, Greely ST, Jinek M, Martin GS, Penhoet E, Puck J, Sternberg SH, Weissman JS, Yamamoto KR (2015) A prudent path forward for genomic engineering and germline gene modification. Science 348:36–38
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineanu S, Romero DA, Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712
Bassett AR, Tibbit C, Ponting CP, Liu JL (2013) Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep 4:220–228
Bolotin A, Quinquis B, Sorokin A, Ehrlich SD (2005) Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151:2551–2561
Bondey-Denomy J, Garcia B, Strum S, Du M, Rollins MF, Reyes YH, Wiedenheft B, Maxwell KL, Davidson AR (2015) Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins. Nature 526:136–139
Bosely KS, Botchan M, Bredenoord AL, Carroll D, Charo RA, Charpentier E, Cohen R, Corn J, Doudna J, Feng G, Greely HT, Isasi R, Ji R, Kim JS, KnoppersELJ Badge RL, Martin GS, Moreno J, Naldini L, Pera M, Perry ACF, Venter JC, Zhang F, Zhou Q (2015) CRISPR germline engineering-the community speaks. Nat Biotechnol 33:478–486
Bronus SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964
Brooks C, Nekrasov V, Lippman ZB, Van Eck J (2014) Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol 166:1292–1297
Burgess DJ (2016) Genome editing for lineage tracing. Science 17:435
Callaway E (2016) Embryo editing gets green lights. Nature 530:18
Caplan AL, Parent B, Shen M, Plunkett C (2015) No time to waste-the ethical challenges created by CRISPR. EMBO Rep 16:1421–1426
Carroll D (2011) Genome engineering with zinc-finger nucleases. Genetics 188:773–782
Carroll D, Charo RA (2015) The social opportunities and challenges of genome editing. Geno Biol 16:242
Carroll D, Van Eenennaam AL, Taylor JF, Seger J, Voytas DF (2016) Regulate genome-edited products, not genome editing itself. Nat Biotechnol 34:477–479
Chaudhary K, Pratap D, Sharma PK (2016) Transcription activator-like effector nucleases (TALENs): an efficient tool for plant genome editing. Eng Life Sci 16:330–337
Chaudhary K, Chattopadhyay A, Pratap D (2018) Anti-CRISPR proteins: counterattack of phages on bacterial defense (CRISPR/Cas) system. J Cell Physiol 233:57–59
Chen X, Gonsalves MAFV (2016) Engineered viruses as genome editing devices. Mol Ther 24:447–457
Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, Huang B (2013) Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155:1479–1491
Chiruvella KK, Liang Z, Wilson TE (2013) Repair of double-strand breaks by end joining. Cold Spring Harb Prespect Biol 5:a012757
Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim J (2014) Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 24:132–141
Chylinski K, Makarova KS, Charpentier E, Koonin EV (2014) Classification and evolution of type II CRISPR-Cas system. Nucleic Acids Res 42:6091–6105
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, LA Jiangw Maraffind, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Cyranoski D (2015) Embryo editing divides scientists. Nature 519:272
Davis AJ, Chen DJ (2013) DNA double strand break repair via non-homologous end-joining. Transl Cancer Res 2:130–143
Daya S, Berns KI (2008) Gene therapy using adeno-associated virus vectors. Clin Microbial Rev 21:583–593
Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E (2011) CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471:602–607
Dominguez AA, Lim WA, Qi LS (2016) Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol 5:5–15
Dong D, Ren K, Qiu X, Zheng J, Guo M, Guan X, Liu H, Li N, Zhang B, Yang D, Ma I, Wang S, Dan W, Ma Y, Fan S, Wang J, Gao N, Huang Z (2016) The crystal structure of Cpf1 in complex with CRISPR RNA. Nature 532:522–526
Doudna JA, Charpentier E (2014) Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096
East-Seletsky A, O’Connell MR, Knight SC (2016) Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538:270–273
Endo M, Mikami M, Toki S (2015) Multigene knockout utilizing off-target mutations of the CRISPR/Cas9 system in rice. Plant Cell Physiol 56:41–47
Friedland AE, Tzur YB, Esvelt KM, Colaiacovo MP, Church GM, Culorco JA (2013) Heritable genome editing in C.elegans via a CRISPR-Cas9 system. Nat Methods 10:741–743
Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31:822–826
Fu Y, Sander JD, Reyon D, Cascio VM, Joung J (2014) Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32:279–284
Gaj T, Gersbach CA, Barbas CF III (2013) ZFN, TALEN and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31:397–405
Gardner MJ, Shallom SJ, Carlton JM, Salzberg Steven L, Nene V, Shoaibi A, Ciecko A, Lynn J, Rizzo M, Weaver B, Jarrahi B, Brenner M, Parvizi B, Tallon L, Moazzez A, Granger D, Fujii C, Hansen C, Pederson J, Feldblyum T, Peterson J, Suh B, Angiuoli S, Pertea M, Allen A, Selengut J, White O, Cummings LM, Smith HO, Adams MD, Venter JC, Carucci DJ, Hoffman SL, Fraser CM (2002) Sequence of Plasmodium falciparum chromosomes 2, 10, 11 and 14. Nature 419:531–534
Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Ginossar SN, Brandman O, Whitehed EH, Doudna JA, Lim WA, Scott SDA, Mikkelsen TS, Hecl D, Edert BL, Root DE, Waissman JS, Qi LS (2013) CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154:442–451
Gori JL, Hsu PD, Maeder ML, Maeder ML, Shen S, Welstead GG, Bumcrot D (2015) Delivery and specificity of CRISPR/Cas9 genome editing technologies for human gene therapy. Hum Gene Ther 26:443–451
Gratz SJ, Cumming AM, Nguyen JN, Hamm DC, Donhuc LK, Harrison MM, Wildonager J, O’connor-Giles KM (2013) Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194:1029–1035
Hashimoto M, Takemoto T (2015) Electroporation enables the efficient mRNA delivery into the mouse zygotes and facilitates CRISPR/Cas9-based genome editing. Sci Rep 5:11315
Hayden EC (2016) Tomorrow’s children. Nature 530:402–405
Hilton IB, D’lppolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA (2015) Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33:510–517
Hirano H, Gootenberg JS, Horii T, Abudayyeh OO, Kimura M, Hsu PD, Nakane T, Ishitani R, Hatada I, Zhang F, Nishimasu H, Nureki O (2016) Structure and engineering of Francisella novicida Cas9. Cell 164:950–961
Hsu PD, Scot DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F (2013) DNA-targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31:827–832
Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278
Huang S, Weigel D, Beachy RN, Li J (2016) A proposed regulatory framework for genome-editing crops. Nat Genet 48:109–111
Hur JK, Kim K, Been KW, Baek G, Ye S, Hur JW, Ryu SM, Lee YS, Kim JS (2016) Targeted mutagenesis in mice by electroporation of Cpf1 ribonucleoproteins. Nat Biotechnol 34:807–808
Hwang WY, Fu Y, Reyon D, Marder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JRJ, Joung JK (2013) Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31:227–229
Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A (1987) Nucleotide sequence of the iap gene, responsible for alkaline phosphatise isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 169:5429–5433
Jacobs TB, LaFayette PR, Schmitz RJ, Parrott WA (2015) Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol 15:16
Janson R, Embden JD, Gaastra W, Schouls LM (2002) Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol 43:1565–1575
Jao LE, Wente SR, Chen W (2013) Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci 110:13904–13909
Jiang W, Zhou H, Bi H, Fromm M, Yong B, Weeks DP (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modifications in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res 41:e188
Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, Kaplan M, Lavarone AT, Charpentier Nagales E, Doudna JA (2014) Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343:1247997
Joung JK, Sander JD (2013) TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol 14:49–55
Kabadi AM, Gersbach CA (2014) Engineering synthetic TALE and CRISPR/Cas9 transcription factors for gene expression. Methods 69:188–197
Kang X, He W, Huang Y, Yu Q, Chen Y, Gao X, Sun X, Fan Y (2016) Introducing precise genetic modifications into human 3PN embryos by CRISPR/Cas-mediated genome editing. J Assist Reprod Genet 33:581–588
Kim H, Kim JS (2014) A guide to genome engineering with programmable nucleases. Nat Rev Genet 15:321–334
Kim D, Kim J, Hur JK (2016) Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol 34:863–868
Kleinstiver BP, Tsai SQ, Prew SM, Nguyen NT, Welch MM, Lopez JM, McCaw ZR, Aryee MJ, Joung JK (2016) Genome-wide specificites of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotechnol 34:869–874
Kotterman MA, Schaffer DV (2014) Engineered adeno-associated viruses for clinical gene therapy. Nat Rev Genet 15:445–451
Lamphire E, Urnov F, Haecker SE, Werner M, Smolenski J (2015) Don’t edit the human germ line. Nature 519:410–411
Laufer BI, Singh SM (2015) Strategies for precision modulation of gene expression by epigenome editing: an overview. Epigenet Chromat 8:34
Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Na G, Wang L, Lu X, Zha Y, Liu M (2013) Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol 31:681–683
Liang P, Xu Y, Zhang X, Ding C, Huang R, Zhang Z, Lv J, Xie X, Chen Y, Li Y, Sun Y, Bai Y, Songyang Z, Ma W, Zhou C, Huang J (2015) CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Prot Cell 6:363–372
Lieber MR (2010) The mechanism of double-strand break repairs by the non-homlogous DNA end-joining pathway. Annu Rev Biochem 79:181–211
Liu F, Song Y, Liu D (1999) Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther 6:1258–1266
Liu L, Li X, Wang J, Liu Liang, Wang Xueyan LiJiuyu, Wang Min, Chen Peng, Wang Maolu Yin Jiazhi LiGang Sheng Yanli (2017) Two distinct catalytic sites are responsible for C2c2 RNase activities. Cell 168:121–134. https://doi.org/10.1016/j.cell.2016.12.031
Lozano-Duran R (2016) Geminivirus for biotechnology: the art of parasite taming. New Phytol 210:58–64
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang ZF, Li H, Lin Y, Xie Y, Shen R, Wang SZ, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu YG (2015) A robust CRISPR/Cas9 system for convinent, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8:1274–1284
Ma H, Tu LC, Naseri A, Huisman M, Zhang S, Grunwald D, Pederson T (2016) Multiplexed labelling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat Biotechnol 34:528–530
Mach J (2014) Geminivirus vectors deliver reagents for plant genome engineering. Plant Cell 26:2
Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK (2013) CRISPR RNA-guided activation of endogenous human genes. Nat Methods 10:977–981
Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJS, Barrangou R, Brouns JJ, Charpentier E, Haft DH, Horvarth P, Moineau S, Mojica FJM, Terns R, Terns MP, White MF, Yakunin AF, Garrett RA, vander Oost J, Backrfen R, Koonin EV (2015) An updated evolutionary classification of CRISPR-Cas system. Nat Rev Microbiol 13:722–736
Mali P, Yang L, Esvelt KM, Aach J, Gulell M, Dicarlo JE, Norville JE, Church GM, Shah SA, Saunders J, Barrangou R, Brouns JJS, Charpentier E (2013) RNA-guided human genome engineering via Cas9. Science 339:823–826
Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL (2015) Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol 33:538–542
Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X, Wan J, Gu H, Jiu LI (2013) Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res 23:1233–1236
Mojica FJ, Diez-Villasenor C, Soria E, Juez G (2000) Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol Microbiol 36:244–246
Mou H, Keneddy Z, Anderson DG, Hao Y, Wen X (2015) Precision cancer mouse models through genome editing with CRISPR-Cas9. Genome Med 7:53
Nelles DA, Fang MY, O’Connell MR, Xu JL, Markmiller SJ, Doudna JA, Yeo GW (2016) Programmable RNA tracking in live cells with CRISPR/Cas9. Cell 165:488–496
Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohamae N, Ishitani R, Zhang F, Nureki O (2014) Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156:935–949
Nunez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna JA, Richards M, Boyaval P, Romero DAM, Harath P (2014) Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat Struct Mol Biol 21:528–534
O’Connell MR, Oakes BL, Strenberg SH, East-Seletsky A, Kaplan A, Doudna JA (2014) Progrmmable RNA recognition and cleavage by CRISPR/Cas9. Nature 516:263–266
Otieno MO (2015) CRISPR-Cas9 human genome editing: challenges, ethical concerns and implications. J Clin Res Bioeth 6:253
Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DA (2013) High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol 31:839–843
Pawluk A, Amrani N, Zhang Y, Garcia B, Reyes YH, Lee J, Edraki A, Shah M, Sontheimer EJ, Maxwell KL, Davidson AR (2016) Naturally occurring off-switches for CRISPR-Cas9. Cell 167:1829–1838
Perez-Pinera P, Kocak DD, Vockley CM, Polstein Thakore PI, Adler AF, Kabadi AM, Leorg LR, Guilak F, Crauford GE, Reddy TE, Gersbach CA (2013) RNA-guided gene activation by CRISPR-Cas9 based transcription factors. Nat Methods 10:973–976
Piatek A, Ali Z, Baazim H, Piatek A, Ali Z, Baazim H, Li L, Abulfaraj A, Al-Shareef S, Aouida M, Mahfouz MM (2014) RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol J 13:578–589
Pourcel C, Salvignol G, Vergnaud G (2005) CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151:653–663
Pulla P (2016) India nears putting GM mustard on the table. Science 352:1043
Qi LS, Larson MH, Gilbert LA, Dounda AJ, Weisiman JS, Arkia AP (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152:1173–1183
Qin W, Dion SL, Kutny PM, Zhang Y, Cheng AW, Jillette NL, Malhotra A, Geurts AM, Chen YG, Wang H (2015) Efficient CRISPR/Cas9-mediated genome editing in mice by zygote electroporation of nuclease. Genetics 200:423–430
Ramakrishna S, Dad ABK, Beloor J, Gopalappa R, Lee SK, Kim H (2014) Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Geno Res 24:1020–1027
Ramanan V, Shlomai A, Cox DBT, Schwartz RE, Michailidis E, Bhatta A, Scott DA, Zhang F, Rice CM, Bhatia SN (2015) CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci Rep 5:10833
Rauch BJ, Silvis MR, Hultquist JF, Waters CS, McGregor MJ, Krogan NJ, Bondy-Denomy J (2017) Inhibition of CRISPR-Cas9 with bacteriophage proteins. Cell 168:150–158
Ron M, Kajala K, Pauluzzi G, Wang D, Reynoso MA, Zumstein K, Garch J, Winter S, Masson H, Inagaki S, Federici F, Sinha N, Deal RB, Bailey-Scrres J (2014) Hairy root transformation using Agrobacterium rhizogenesis as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol 166:455–469
Sanchez-Rivera FJ, Jack T (2015) Applications of the CRISPR-Cas9 system in cancer biology. Nat Rev Cancer 15:387–395
Sander JD, Joung JK (2014) CRISPR-Cas system for editing, regulating and targeting genomes. Nat Biotechnol 32:347–355
Shalem O, Sanjana NE, Hartenian E, DA Hhix Scott, Mikkelsen TS, Heckl D, Edert BL, Root DE, Doench JG, Zhang F (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343:84–87
Shalem O, Sanjana NE, Zhang F (2015) High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet 16:299–311
Shan Q, Wang Y, Li J, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qin JL, Gao C (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31:686–688
Shimatani Z, Nishizawa-Yokoi A, Endo M, Tok S, Ternda R (2015) Postive-negative-selection-mediated gene targeting in rice. Front Plant Sci 5:748
Shmakov S, Abudayyeh OO, Makarova KS (2015) Discovery and functional characterization of diverse class 2 CRISPR-Cas system. Mol Cell 60:383–397
Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F (2016) Rationally engineered Cas9 nucleases with improved specificity. Science 351:84–88
Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA (2014) DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507:62–67
Suda T, Liu D (2007) Hydrodynamic gene delivery: its principles and applications. Mol Ther 15:2063–2069
Sun N, Zhao H (2013) Transcription activator-like effector nucleases (TALENs): a highly efficient and versatile tool for genome editing. Biotchnol Bioeng 110:1811–1821
Sun X, Hu Z, Chen R, Jiang Q, Song G, Znang H, Xi Y (2015) Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci Rep 5:10342
Sung P, Klein H (2006) Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol 7:739–750
Upadhyay SK, Kumar J, Alok A, Tuli R (2013) RNA-guided genome editing for target gene mutations in wheat. G3 3:2233–2238
Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PH (2010) Genome editing with engineered zinc finger nucleases. Nat Rev Genet 11:636–646
Waltz E (2016a) Gene-edited CRISPR mushroom escapes US regulation. Nature 532:293
Waltz E (2016b) GM salmon declared fit for dinner plates. Nat Biotechnol 34:7–9
Wang F, Wang C, Liu P, Lei C, Hao W, Gao Y, Liu YG, Zhao K (2016a) Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS ONE 11:e0154027
Wang Z, Pan Q, Gendron P, Zhu W, Guo F, Cen S, Wainberg MA, Liang C (2016b) CRISPR/Cas9-derived mutations both inhibit HIV-1 replications and accelerate viral escape. Cell Rep 15:481–489
Weinthal D, Tovkach A, Zeevi V, Tzfira T (2010) Genome editing in plant cells by zinc finger nucleases. Trend Plant Sci 15:306–321
Wiedenheft B, Zhou K, Jinek M, Coyle SM, Ma W, Doudna JA (2009) Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure 17:904–912
Wu Y, Liang D, Wang Y et al (2013) Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell 13:659–662
Wu X, Kriz AJ, Sharp PA (2014) Target specificity of the CRISPR-Cas9 system. Quant Biol 2:59–70
Xu R, Li H, Qin R, Wang L, LiL Wei P, Yang J (2014a) Gene targeting using the Agrobacterium tumefaciens-mediated CRISPR-Cas system in rice. Rice (N Y) 7:5
Xu T, Li Y, Van Nostrand JD, Van Nostrand Joy D, He Zhili, Zhou Jizhong (2014b) Cas9-based tools for targeted genome editing and transcriptional control. App Environ Microbiol 80:1544–1552
Yamano T, Nishimasu H, Zetsche B (2016) Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell 165:949–962
Yang H, Gao P, Rajashankar KR, Patel DJ (2016) PAM-dependent target DNA recognition and cleavage by C2c1 CRISPR-Cas endonuclease. Cell 167:1814–1828
Yin H, Wang H, Shivalila CS, Cheng AC, Shi L, Jaenish R (2013) One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154:1370–1379
Yin K, Han T, Liu G, Chen T, Wang Y, Yu AY, Liu Y (2015) A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci Rep 5:14926
Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymarker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, vander Oost J, Regev A, Koonin EV, Zhang F (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163:759–771
Zhang G, Budker V, Wolff JA (1999) High level of foreign gene expression in heptocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther 10:1735–1737
Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Zhang H, XuN ZhuJK (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J 12:797–807
Zhou H, Liu B, Weeks DP, Spalding MH, Yang B (2014) Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res 42:10903–10914
Acknowledgements
We would like to thank Elizabeth J. Sparke for excellent editing and helpful comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
The original version of the article was revised due to change in author name from “Kulbhushan Chaudhary” to “Kul Bhushan”.
Rights and permissions
About this article
Cite this article
Bhushan, K., Chattopadhyay, A. & Pratap, D. The evolution of CRISPR/Cas9 and their cousins: hope or hype?. Biotechnol Lett 40, 465–477 (2018). https://doi.org/10.1007/s10529-018-2506-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-018-2506-7