Abstract
The immunogenicity of SARS-CoV nucleocapsid DNA vaccine and the immunoregulatory activity of interleukin-2 (IL-2) were investigated. DNA vaccine plasmids, pcDNA-N and pcDNA-IL2, were constructed and inoculated into BALB/c mice with or without pcDNA-IL2 by intramuscular injection. Cellular and humoral immune responses were assessed by indirect ELISA, lymphocyte proliferation assays, ELISPOT and FACS. The nucleocapsid DNA vaccine had good immunogenicity and can induce specific humoral and cellular immunity in BALB/c mice, while IL-2 plays an immunoadjuvant role and enhances specific immune responses. This study provides a frame of reference for the design of DNA vaccines against SARS-CoV.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
An international outbreak of severe acute respiratory syndrome (SARS), which has a strong infectivity and pathogenicity, spread through more than 30 countries and infected more than 8000 people and killed more than 700 from its emergence in mid-November 2002 until 7 August 2003 (Fouchier et al. 2003). The causative agent for SARS has subsequently been identified as a novel coronavirus, namely the SARS-coronavirus (SARS-CoV; Drosten et al. 2003; Peiris et al. 2003) SARS-CoV is phylogenetically different from other members of the coronaviridae family and is the only coronavirus currently known to cause severe morbidity and mortality in humans (Lee et al. 2003). SARS-CoV encodes four main structural proteins: spike (S) glycoprotein, membrane (M) protein, envelope (E) protein and nucleocapsid (N) protein (Rota et al. 2003; Stadler et al. 2003).
N protein in many coronaviruses is highly immunogenic and abundantly expressed during infection (Liu et al. 2001; Narayanan et al. 2003). The N protein of the feline infectious peritonitis virus (FIPV) has been used as subunit vaccines to induce protective immunity and to prevent the progression of diseases in a cat model (Hohdatsu et al. 2003). In porcine coronavirus, transmissible gastroenteritis virus (TGEV), the N protein is a representative antigen for the T-cell response and may induce both cellular and humoral immune response (Anton et al. 1996). These results indicate that a good N protein vaccine candidate should elicit strong immune responses. Moreover, SARS-CoV N protein is highly conserved (99%) within different isolates, and shares 20–30% amino acid homology with the N proteins of other coronaviruses (Rota et al. 2003). Thus, the N protein is an important target for SARS-CoV vaccine development.
The immunogenicity of SARS-CoV N protein has been under investigation for a long time. Antibodies against the N protein are lasting longer and occur in greater abundance in SARS patients than antibodies against other viral components such as the S, M and E proteins (Chen et al. 2004; Kim et al. 2004; Tan et al. 2004; Timani et al. 2004; Zhu et al. 2004). This phenomenon might be due to the higher expression of the N protein as compared with other viral proteins after SARS-CoV infection (Rota et al. 2003). N protein may therefore play a crucial role in antibody response during SARS-CoV infection, and might be useful in early diagnosis of the infection and for vaccine development. Moreover, vaccination with a plasmid expressing nucleocapsid protein is capable of generating strong N-specific humoral and cellular immune responses in vaccinated mice (Kim et al. 2004; Zhao et al. 2005; Zhu et al. 2004). Therefore, based on these observations, we initially focused on the SARS-CoV N protein as the target antigen for our DNA vaccine development.
DNA vaccines induce both cellular and humoral immune responses to produce long-lasting immunity against infectious diseases. However, the low immunogenicity of DNA-based vaccines could compromise its application. In recent years, many efforts have been directed towards enhancing the immune responses elicited by DNA vaccines, including the co-expression of cytokines, the use of heterologous prime-boost regimes, and the conventional route of delivery the DNA vaccine. Several cytokines act as immunological adjuvants for inactivated and subunit vaccines as well as DNA vaccines (Barouch et al. 2002; Chow et al. 1998; Kim et al. 2000, 2001; Nobiron et al. 2001). The immune responses to DNA vaccines can be enhanced dramatically by co-administration of plasmids encoding the IL-2 gene; for example in DNA vaccination against bovine herpesvirus 1 virus (Kuhnle et al. 1996), hepatitis C virus (Geissler et al. 1997), hepatitis B virus (Chow et al. 1998), bovine viral diarrhea virus (Nobiron et al. 2001), HIV (Moore et al. 2002), foot-and-mouth disease (Wong et al. 2002), and measles virus (Premenko-Lanier et al. 2003). In our current study, we have demonstrated that IL-2 gene as a cytokine adjuvant could increase the immune responses of the SARS-CoV nucleocapsid DNA vaccine.
Materials and methods
Construction of DNA vaccine expression plasmids
pcDNA-N and pcDNA-IL2 plasmids were used in this study. A DNA fragment encoding the full-length N gene was amplified from the cDNA of SARS-CoV WH-20 strain (GenBank accession No. AY772062). This DNA fragment was subcloned into a mammalian expression vector, pcDNA3.1 (+) (Invitrogen, Carlsbad, CA), to construct the recombinant plasmid pcDNA-N. The IL-2 gene was kindly provided by Prof. Cui Bo-an (Henan Agricultural University, Zhengzhou, People’s Republic of China) and subcloned into pcDNA3.1 (+) to construct the recombinant plasmid pcDNA-IL2. The accuracy of the constructs were confirmed by restriction digestion and sequencing. Large-scale plasmid preparation and purification were conducted using an EndoFree Plasmid Maxi Kit (Qiagen Inc., Chatsworth, CA) according to the manufactures’ protocol. The DNA was dissolved in endotoxin-free phosphate-buffered saline (PBS, Sigma) to a final concentration of 2 μg/μl, and stored at −20°C.
Animals and immunization
The pcDNA-N and pcDNA-IL2 constructs were transiently transfected to 293 cells and the protein expression efficiency was detected before the animal immunization was conducted. For the animal experiment, our employment of laboratory animals was approved by Hubei Province Laboratory Animal Management Committee (SCXK (Ea) 2003–2004) in China. Briefly, eight-week-old BALB/c mice were purchased from the Experimental Animal Center of Hubei Medical College, and randomly divided into five groups (13 mice each). Animals received pathogen-free water and food for maintenance. Mice were immunized three times with pcDNA-N (100 μg) alone or pcDNA-N (100 μg) plus pcDNA-IL2 (50 μg) on days 0, 14 and 28. All immunizations were delivered into the quadriceps muscles in a total volume of 100 μl PBS. DNA dosages used in mice and the immune scheme were optimized through a series of preliminary experiments.
Analysis of humoral immune response
Antibody to SARS-CoV in serum was tested by enzyme-linked immunosorbent assay (ELISA) as described previously (Hu et al. 2007). Chemically killed SARS-CoV was kindly provided by Prof. Chen Z. (Wuhan Institute of Virology, Chinese Academy of Science, People’s Republic of China) and used as the detection antigen. An optimized concentration (5 μg/ml) of antigen was coated onto 96-well plates (Costar) overnight at 4°C. Plates were washed, and blocked with 1% bovine serum albumin (BSA) buffered solution for 1 h at 37°C prior to 2 h incubation with 1:100 diluted mouse sera at 37°C. Bound antibodies were detected with alkaline phosphatase-conjugated, goat-anti-mouse IgG (Sigma). Color was developed by adding p-nitrophenyl phosphate (pNPP) as substrate and reading at 410 nm on a plate reader. The value of mouse serum from experimental groups was considered positive when over or equal to 2.1-times of that obtained from the control group. Values lower than 0.05 were not included.
A similar ELISA protocol was followed to assess N-specific immunoglobulin IgG and its subclasses (IgG1 and IgG2a). In this case, recombinant N protein expressed in E. Coli was purified and used as the detection antigen. Horseradish peroxidase (HRP)-conjugated goat-anti-mouse IgG, IgG1 and IgG2a (Sigma) were used as secondary antibodies and the A 490 was then determined.
Lymphocyte proliferation assay (LPA)
Antigen-specific T-cell proliferation (LPA) was performed as described previously (Froebel et al. 1999). In brief, 2 weeks following the final injection, mice were sacrificed and single-cell suspension was prepared from spleens for each group. 2 × 105 splenocytes per well in RPMI-1640 medium (Sigma) supplemented with 10% FBS were seeded in a 96-well plate in triplicate. Cultures were stimulated under various conditions for 60 h at 37°C and 5% CO2: 5 μg/ml Concanavalin A (positive control), 5 μg/ml purified N antigen (specific antigen), 5 μg/ml BSA (irrelevant antigen), or medium alone (negative control). 20 μl of CellTiter 96 Aqueous One Solution Reagent (Promega, USA) was added into each well according to the manufacturer’s protocol. Following 4 h incubation at 37°C, absorbance was read at 490 nm. Proliferative activity was estimated using the stimulation indexes (SI) defined as the mean OD 490 of antigen-containing wells divided by mean OD 490 of wells without antigen.
SARS-CoV N-specific ELISPOT assay
Cellular immune responses to SARS-CoV were assessed by IFN-γ and IL-4 enzyme-linked immunospot (ELISPOT) assay using mouse splenocytes. Assays were performed according to the instruction manual (U-CyTech, Netherlands). Ninety-six-well plates were coated with 100 μl/well of 5 μg/ml rat anti-mouse IFN-γ or IL-4 in PBS overnight. Plates were then washed three times with PBS containing 0.25% Tween-20 and blocked with PBS containing 5% FBS for 2 h at 37°C. After three washes with PBS containing 0.25% Tween-20, 105 splenocytes in a 100 μl of reaction buffer containing 2 μg purified N protein/ml were added into each well. Plates were incubated 16 h at 37°C in 5% CO2 and then washed 10 times with PBS. Biotinylated anti-mouse IFN-γ or IL-4 mAb at 1:500 dilutions was subsequently added and plates were incubated for 2 h at room temperature. After washing, avidin-horseradish peroxidase was then added for another 1 h incubation at room temperature. Following five washes with PBS, individual IFN-γ or IL-4 plates were developed as dark spots after a 10 min reaction with the peroxidase substrate-AEC. Reactions were stopped by rinsing plates with demineralised water. Plates were air-dried at room temperature and read using an ELISPOT reader (Hitech Instruments). Spot forming cells (SFC) per 106 splenocytes were calculated. Medium backgrounds were consistently <10 SFC per 106 splenocytes.
Determination of CD4+ and CD8+ cells in PBMC
Flow cytometry was used to monitor the expression of T cell surface markers. Direct immunofluorescence was performed according to the standard techniques as described elsewhere (Caruso et al. 1997). Two weeks following the final injection, blood was collected from the mouse eye pit with each vaccine groups. Peripheral blood mononuclear cells (PBMCs) were isolated by Isopaque-Ficoll (Lymphoprep; Nycomed, Oslo, Norway) following the manufacture’s instructions. Lymphocytes isolated from PBMCs were stained with the following mAbs: FITC-labeled anti-mouse CD3 antibody, PECy-5-labeled anti-mouse CD4 antibody, PE-labeled anti-mouse CD8 antibody (BD PharMingen), or the corresponding isotype controls. The stained lymphocytes were then washed three times with PBS containing 4% fetal bovine serum. 105 cells were acquired on a FACSCalibur flow cytometer (Beckton Dickinson, Lincoln Park, NJ) and data were analyzed using the WinMDI software.
Statistical analysis
All data are presented as the mean ± SD of immunized mice per group. Difference in humoral and cellular immune responses between groups was assayed by using single factor analysis of variances. The LSD t-test was used for between-group comparison. Values of P < 0.05 were considered statistically significant.
Results
Antibody responses to SARS-CoV
After the expression of the DNA constructs were analyzed in vitro (data not shown), BALB/c mice were inoculated intramuscularly (i.m.) three times in each quadricep muscle separately with recombinant DNA plasmids at 2-week intervals. Antibody to SARS-CoV was analyzed by indirect ELISA on sera samples collected 0, 14, 28, 42 and 56 days after the first immunization. Blood sera from mice immunized either with pcDNA-N or pcDNA-N plus pcDNA-IL2 could be strongly detected by SARS-CoV (Fig. 1). Blood sera collected from control groups injected with pcDNA 3.1, pcDNA-IL2, and PBS, respectively, did not show any significant level of immune response. On day 14, the pcDNA-N plus pcDNA-IL2 immunized group showed a detectable level of immune response, whereas the pcDNA-N group showed limited immune response. In all the immune process, the antibody levels of co-administered pcDNA-IL2 groups were significantly higher than those in the pcDNA-N alone groups after the second immune boost (P < 0.05).
Antibody response to SARS-CoV induced by vaccinations with or without pcDNA-IL2 in mice. Mice were immunized three times with pcDNA-N (100 μg) alone or pcDNA-N (100 μg) plus pcDNA-IL2 (50 μg) on days 0, 14 and 28. Serum was collected 2-week intervals after vaccination from each group (n = 13). Antibody was detected by indirect ELISA assay. Data are presented as mean ± SD. A value ≥2.1 was considered as positive by calculating the absolute ratio of Post/naïve serum
N-specific antibodies and antibody subclasses
Two weeks after the final immunization, the N-specific antibodies were measured by ELISA (Fig. 2). All vaccinated mice developed substantial N-specific antibody responses, whereas animals in the control groups did not show any detectable N-specific antibody response. The expression of N-specific IgG antibodies was consistent with the expression pattern of antibodies to the SARS-CoV in the immunized groups (Fig. 1).
IgG1 and IgG2a were measured in order to detect humoral and cytotoxic immune responses, respectively. As shown in Fig. 2, anti-N-specific IgG subtype profile revealed that both IgG1 and IgG2a were induced in the pcDNA-N and pcDNA-N plus pcDNA-IL2 immunization regimens. Antibody levels in the combined group showed higher degrees of increase compared to that in pcDNA-N vaccinated group, although the antibody subclass profiles were similar in both groups. Co-administration of pcDNA-IL2 with pcDNA-N mainly enhanced the antibody response, but did not change antibody subclasses.
N-specific T-cell proliferation
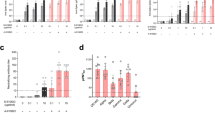
To determine whether T-cell proliferation response to the DNA vaccine encoding N gene may be boosted by IL-2 expressing vector, splenocytes from the vaccinated mice were examined for antigen-specific T-cell proliferation. As shown in Fig. 3, the DNA vaccine plus pcDNA-IL2 group, as well as the DNA vaccine group, exhibited higher levels of T cell proliferative response when re-stimulated with purified SARS-CoV N protein, while mice vaccinated with the pcDNA3.1, pcDNA-IL2 or PBS did not respond to the N protein (P < 0.05). Co-immunization with pcDNA-IL2 induced a substantial increase in T-cell proliferation response compared to the pcDNA-N vaccinated group alone (P < 0.05).
N-specific lymphocyte proliferation assay. Two weeks following the final injection, pooled splenocytes were obtained from mice (n = 5) immunized with DNA vaccine. Splenocytes were stimulated in vitro with purified N protein as test groups, with ConA as positive control and with BSA as the irrelevant antigen control. Splenocytes from the control groups (pcDNA-IL2, pcDNA3.1 or PBS) were stimulated with N protein served as the negative control and the sham control. The stimulation index (SI) was calculated by the following formula: SI = (mean OD of Con A- or antigen-stimulated proliferation)/(mean OD of non-stimulated proliferation). Each bar represents the mean SI ± SD of five mice
N-specific Th1 and Th2 type response
The effects of pcDNA-IL2 on the magnitude of IFN-γ and IL-4-producing T-cells induced by the pcDNA-N or pcDNA-N plus pcDNA-IL2 plasmid were investigated using ELISPOT assay. The protein dosage used for stimulation and the number of splenocytes were optimized to induce IFN-γ and IL-4 T-cell responses (data not shown). Splenocytes from vaccinated mice were harvested 2 weeks after the final vaccination, and N-specific IFN-γ and IL-4 ELISPOTs were enumerated. As shown in Fig. 4a, b, only a low number of non-specific IFN-γ and IL-4 ELISPOTs were detected in the control groups of PBS and pcDNA3.1 (Spots ⩽10/106 cells). Whereas for the control group of pcDNA-IL2, the number of INF-γ or IL-4-secreting cells were higher than that of the negative control groups of PBS and pcDNA3.1 (P < 0.05), but much lower than that of pcDNA-N or pcDNA-N plus pcDNA-IL2 groups (Fig. 4). Compared with the negative control groups, significant numbers of N-specific IFN-γ and IL-4 ELISPOTs were detected in the pcDNA-N and the pcDNA-N plus pcDNA-IL2 immunized groups (P < 0.01). Immunizations with pcDNA-N induced 453 ± 84 SFC/106 N-specific IFN-γ cells. By comparison, with the adjuvant pcDNA-IL2, N-specific IFN-γ T-cell frequency increased to 1230 ± 216 SFC/106 cells which is a 2.7-fold increase over the number of T-cells primed with immunizations of pcDNA-N (P < 0.05). However, the IL-4-secreting cell number in the vaccine groups was not as high as the IFN-γ-secreting cell number. The co-administrated pcDNA-IL2 also appreciably enhanced the IL-4-secreting cell number with a 2.3-fold increase compared to that for pcDNA-N immunization alone.
SARS-CoV N protein-specific IFN-γ (a) and IL-4 (b) ELISPOT. Graph shown the number of INF-γ-secreting cells in spleen of mice harvested two weeks following the final injection and stimulated in vitro with N protein. The results represent the average of triplicate wells of five mice and expressed as mean ± SD
CD8+ and CD4+ lymphocyte responses
As activated CD4+ and CD8+ T lymphocytes are among the most crucial components of antiviral effectors, those cell subsets in the PBMC of the vaccinated mice were assessed (Fig. 5). Flow cytometry analysis of unstimulated cells was used to standardize the background responses and there was little variation in non-immunized mice. pcDNA-N and pcDNA-N plus pcDNA-IL2 vaccinations significantly increased the percentages of activated CD4+ and CD8+ cells compared to the control groups (P < 0.01). The number of CD4+ and CD8+ activated cells increased in all the immunized groups. CD8+/CD4+ ratio in pcDNA-N plus pcDNA-IL2 groups was higher than that in pcDNA-N group.
Discussion
SARS-CoV N protein is an important target for cellular and humoral immune responses against SARS-CoV. Previous studies have shown that both the N protein and the DNA encoded N gene elicit antibody and cytotoxic T-cell responses in mice (Barouch et al. 2002; Liu et al. 2006; Zhu et al. 2004). Kim et al. (2004) also demonstrated that vaccination with the N DNA vaccine could successfully induce a SARS-CoV antigen-specific CD8+ T-cell response and distinctly reduce the titer of recombinant vaccinia virus expressing SARS-CoV N protein after the challenge and that the co-expression of calreticulin (CRT) with N gene could enhance its ability to protect against viral challenges (Kim et al. 2004). These results support the basic role for the N protein as an important component of any SARS-CoV vaccine. Therefore, in our present experiment, we focused on the SARS-CoV N protein as the target antigen for DNA vaccine development.
In our previous study, SARS-CoV N DNA vaccination approaches were investigated using different immunization routes in mouse model. We clearly showed that the administration of the N DNA vaccine candidate via different immunization routes could induce a qualitatively different immune response profile (Hu et al. 2009). Here, we have shown that intramuscularly vaccination with SARS-CoV N DNA can elicit SARS-CoV N-specific humoral and cellular immune responses, and these responses could be significantly enhanced by co-administrating an IL-2-expressing vector. Thus, the ability of the DNA immune strategy to enhance cellular and humoral immune responses has been confirmed in two distinct systems.
From our results, both humoral- and cellular-mediated N-specific responses were induced in mice by N DNA plasmid. N-specific IgG, IgG1 and IgG2a antibodies were elicited in all the immunized animals. Compared to the level of IgG1 antibody, a higher level of IgG2a antibody was induced. In addition, the T cell proliferation, the ratio of CD4+ and CD8+ activated cells were also successfully evoked after N DNA vaccination. We also found that antigen-specific T cells were capable of secreting a high level of Th1 cytokine IFN-γ and a moderate level of Th2 cytokine IL-4 upon in vitro stimulation with SARS-CoV N protein. These findings suggest that the N DNA vaccine is effective in the activation of both B- and T-cells to generate anti-N antibodies and cellular immune (mainly Th1) responses in mouse model. And our results are consistent with the former studies of N protein DNA vaccine (Liu et al. 2006; Zhao et al. 2005).
IL-2 is critical for the proliferation and clonal expansion of antigen-specific T cells. Several lines of evidence suggest that co-administration of plasmids encoding the IL-2 gene and a given antigen results in enhancement of both humoral and cell-mediated immune responses to that antigen, and mostly favors Th1 cell differentiation (Geissler et al. 1997; Kuhnle et al. 1996; Moore et al. 2002; Nobiron et al. 2001; Sin et al. 1999; Wong et al. 2002). In current study, on the basis of our examination of the immune response induced by a N DNA vaccine, we have tested the ability of IL-2 to improve the immunogenicity of a SARS-CoV N DNA vaccine. Our results clearly demonstrate that the immunogenicity of the N DNA vaccine was significantly enhanced by co-administration of an IL-2 expressing plasmid. In the studies on the antigen-specific humoral immune response of the co-immunized group, the antibody response induced by co-administration of pcDNA-N plus pcDNA-IL2 was significantly higher than that induced by vaccination with pcDNA-N alone (P < 0.05). The production of antibody induced by co-administration of the plasmids began 1 week earlier than that induced by pcDNA-N administration alone (Fig. 1). From this observation, it suggests that the antibody silent period becomes shorter when IL-2 is used as an adjuvant, this could be very helpful in the control of infection soon after SARS-CoV infection.
Moreover, IL-2 is known to promote a Th1-type response and may be most useful as a vaccine adjuvant in diseases where cell-mediated immunity plays an important role in protection against pathogens (Chow et al. 1998). Published data show that the N gene plays an important role in cellular immune response of SARS-CoV infection (Kim et al. 2004; Zhao et al. 2005; Zhu et al. 2004). In this study, the antigen-specific cellular immune response of T lymphocytes, and the ELISPOT assay have been employed to assess the frequency of cytokine producing cells and the kind of T cell produced in cultures of lymphocytes isolated from co-immunized mice. Our results show that immunization with pcDNA-N plus pcDNA-IL2 elicited significant levels of T-cell responses, and the response was higher than that in the pcDNA-N alone group. ELISPOT and the FACS data further confirmed this result. Using the ELISPOT assay, we measured the number of antigen-specific T-cells per million splenocytes and found that pcDNA-IL2 significantly increased the stimulated function of Th1 (as reflected by an increased release of IFN-γ) and of Th2 cells (as reflected by a increased release of IL-4). There was higher level of IFN-γ production compared to IL-4 (Fig. 4), suggesting that vaccine combination induced a Th1-dominated immune response. This finding is consistent with the ability of combined vaccines to activate CD4+ and CD+ 8 T cells, with a strong CD8+ and a mild CD4+ cell increase. Thus, the present results indicate that synergistic action of IL-2 results in a strong activation of SARS-CoV N-specific Th1 lymphocytes, witnessed by a high IFN-γ production. This might be sufficient for viral clearance in animals and humans with SARS-CoV infection.
The above observations strongly suggest that IL-2 can enhance not only the humoral immune response but also the cell-mediated response triggered by DNA vaccination. The co-injection of the IL-2 gene during DNA immunization enhanced the development of Th1 and Th2 cells with the predominant Th1 cells. Our results are consistent with a previous report, which showed that the co-expression of the immune-stimulatory IL-2 could increase the immune response of coxsackievirus B3 principally towards Th1 (Henke et al. 2004). Conversely, Chow reported that the co-injection of the IL-2 gene during DNA immunization studies enhanced the development of Th1 cells, while the Th2 activation was not affected (Chow et al. 1998), which is in not agreement with our results. However, these different results could have been due to the different nature of the antigens used in the respective studies.
Our findings suggest that the reported strategy is useful for vaccination against SARS-CoV as well, and that IL-2 is a promising cytokine adjuvant for SARS-CoV DNA vaccines and perhaps other coronavirus DNA vaccines in the future. However, the effects in mice may be more dramatic than those in humans, and there are significant limitations with the current technologies that prevent the full effectiveness of DNA vaccines in larger animals. Whether this approach could be applied to other animal models is still unknown, and its immunogenicity in humans remains to be established. Therefore, it is very important to evaluate the efficacy of this SARS DNA vaccine strategy in some highly relevant translational models to demonstrate human immune responsiveness.
References
Anton IM, Gonzalez S, Bullido MJ et al (1996) Cooperation between transmissible gastroenteritis coronavirus (TGEV) structural proteins in the in vitro induction of virus-specific antibodies. Virus Res 46:111–124
Barouch DH, Santra S, Tenner-Racz K et al (2002) Potent CD4+ T cell responses elicited by a bicistronic HIV-1 DNA vaccine expressing gp120 and GM-CSF. J Immunol 168:562–568
Caruso A, Licenziati S, Corulli M et al (1997) Flow cytometric analysis of activation markers on stimulated T cells and their correlation with cell proliferation. Cytometry 27:71–76
Chen Z, Pei D, Jiang L et al (2004) Antigenicity analysis of different regions of the severe acute respiratory syndrome coronavirus nucleocapsid protein. Clin Chem 50:988–995
Chow YH, Chiang BL, Lee YL et al (1998) Development of Th1 and Th2 populations and the nature of immune responses to hepatitis B virus DNA vaccines can be modulated by codelivery of various cytokine genes. J Immunol 160:1320–1329
Fouchier RA, Kuiken T, Schutten M et al (2003) Aetiology: Koch’s postulates fulfilled for SARS virus. Nature 423:240
Froebel KS, Pakker NG, Aiuti F et al (1999) Standardisation and quality assurance of lymphocyte proliferation assays for use in the assessment of immune function. European concerted action on immunological and virological markers of HIV disease progression. J Immunol Methods 227:85–97
Geissler M, Gesien A, Tokushige K et al (1997) Enhancement of cellular and humoral immune responses to hepatitis C virus core protein using DNA-based vaccines augmented with cytokine-expressing plasmids. J Immunol 158:1231–1237
Henke A, Chiang CS, Zell R et al (2004) Co-expression of interleukin-2 to increase the efficacy of DNA vaccine-mediated protection in coxsackievirus B3-infected mice. Antiviral Res 64:131–136
Hohdatsu T, Yamato H, Ohkawa T et al (2003) Vaccine efficacy of a cell lysate with recombinant baculovirus-expressed feline infectious peritonitis (FIP) virus nucleocapsid protein against progression of FIP. Vet Microbiol 97:31–44
Hu H, Lu X, Tao L et al (2007) Induction of specific immune responses by severe acute respiratory syndrome coronavirus spike DNA vaccine with or without interleukin-2 immunization using different vaccination routes in mice. Clin Vaccine Immunol 14:894–901
Hu H, Huang X, Tao L et al (2009) Comparative analysis of the immunogenicity of SARS-CoV nucleocapsid DNA vaccine administrated with different routes in mouse model. Vaccine 27:1758–1763
Kim JJ, Yang JS, Montaner L et al (2000) Coimmunization with IFN-gamma or IL-2, but not IL-13 or IL-4 cDNA can enhance Th1-type DNA vaccine-induced immune responses in vivo. J Interferon Cytokine Res 20:311–319
Kim JJ, Yang JS, Dang K et al (2001) Engineering enhancement of immune responses to DNA-based vaccines in a prostate cancer model in rhesus macaques through the use of cytokine gene adjuvants. Clin Cancer Res 7:882s–889s
Kim TW, Lee JH, Hung CF et al (2004) Generation and characterization of DNA vaccines targeting the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J Virol 78:4638–4645
Kuhnle G, Collins RA, Scott JE et al (1996) Bovine interleukins 2 and 4 expressed in recombinant bovine herpesvirus 1 are biologically active secreted glycoproteins. J Gen Virol 77:2231–2240
Lee N, Hui D, Wu A et al (2003) A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med 348:1986–1994
Liu C, Kokuho T, Kubota T et al (2001) DNA mediated immunization with encoding the nucleoprotein gene of porcine transmissible gastroenteritis virus. Virus Res 80:75–82
Liu SJ, Leng CH, Lien SP et al (2006) Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine 24:3100–3108
Moore AC, Kong WP, Chakrabarti BK et al (2002) Effects of antigen and genetic adjuvants on immune responses to human immunodeficiency virus DNA vaccines in mice. J Virol 76:243–250
Narayanan K, Chen CJ, Maeda J et al (2003) Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J Virol 77:2922–2927
Nobiron I, Thompson I, Brownlie J (2001) Cytokine adjuvancy of BVDV DNA vaccine enhances both humoral and cellular immune responses in mice. Vaccine 19:4226–4235
Peiris JS, Lai ST, Poon LL et al (2003) Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319–1325
Premenko-Lanier M, Rota PA, Rhodes G (2003) DNA vaccination of infants in the presence of maternal antibody: a measles model in the primate. Virology 307:67–75
Rota PA, Oberste MS, Monroe SS et al (2003) Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394–1399
Sin JI, Kim JJ, Boyer JD et al (1999) In vivo modulation of vaccine-induced immune responses toward a Th1 phenotype increases potency and vaccine effectiveness in a herpes simplex virus type 2 mouse model. J Virol 73:501–509
Stadler K, Masignani V, Eickmann M et al (2003) SARS—beginning to understand a new virus. Nat Rev Microbiol 1:209–218
Tan YJ, Goh PY, Fielding BC et al (2004) Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin Diagn Lab Immunol 11:362–371
Timani KA, Ye L, Ye L et al (2004) Cloning, sequencing, expression, and purification of SARS-associated coronavirus nucleocapsid protein for serodiagnosis of SARS. J Clin Virol 30:309–312
Wong HT, Cheng SC, Sin FW et al (2002) A DNA vaccine against foot-and-mouth disease elicits an immune response in swine which is enhanced by co-administration with interleukin-2. Vaccine 20:2641–2647
Zhao P, Cao J, Zhao LJ et al (2005) Immune responses against SARS-coronavirus nucleocapsid protein induced by DNA vaccine. Virology 331:128–135
Zhu MS, Pan Y, Chen HQ et al (2004) Induction of SARS-nucleoprotein-specific immune response by use of DNA vaccine. Immunol Lett 92:237–243
Acknowledgments
This work was supported in part by a National “973 Project” grants (2005CB523000) and “863 Project” grants (2005AA219070) from the Ministry of Science and Technology, People’s Republic of China and Chinese Academy of Sciences (KSCX1-YW-R-07). We would like to thank Xuefang An (Wuhan Institute of Virology, Chinese Academy of science) for her help in the experiments and Dr. Zhanyong Wei (Henan Agricultural University, Zhengzhou, People’s Republic of China) for scientific editing of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hu, H., Tao, L., Wang, Y. et al. Enhancing immune responses against SARS-CoV nucleocapsid DNA vaccine by co-inoculating interleukin-2 expressing vector in mice. Biotechnol Lett 31, 1685–1693 (2009). https://doi.org/10.1007/s10529-009-0061-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-009-0061-y