Abstract
Microorganisms produce siderophores, which are secondary metabolites with a high affinity for iron. Siderophores have received significant attention due to their diverse applications in ecological and clinical research. In this study, siderophores production by Escherichia coli OQ866153 was optimized using two-stage statistical approach involving Plackett–Burman design (PBD) and response surface methodology (RSM) using central composite design (CCD). Out of 23 variables, succinate, tryptophan, Na2HPO4, CaCl2, agitation, and KH2PO4 were found to have the most significant effect on siderophores production in the first optimization stage with the highest SU% of 43.67%. In the second stage, RSM using CCD was utilized, and the optimal conditions were determined to be 0.3 g/l succinate, 0 g/l tryptophan, 6 g/l Na2HPO4, 0.1 g/l CaCl2, 150 RPM agitation, and 0.6 g/l KH2PO4, resulting in a maximum siderophore units (SU%) of 89.13%. The model was significant, as indicated by the model f-value of 314.14 (p-value = 0.0004) and coefficient of determination R2 of 0.9950. During validation experiments, the obtained maximum SU% was increased up to 87.1472%, which was two times as the value obtained under ordinary conditions (46.62%). The produced siderophores were purified and characterized using 1H, 13C NMR, IR spectroscopy. The obtained results indicated that the compound was enterobactin and entABCDEF genes were further detected in Escherichia coli OQ866153 extracted DNA. To our knowledge, this is the first report of statistical optimization for enterobactin synthesis by an E. coli strain isolated from a clinical source in Egypt.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Iron (Fe), a key component of several fundamental cellular processes, is the fourth most ample element in Earth’s crust (Soares 2022). However, due to its low bioavailability, it may be a rare element. In order to alleviate Fe depletion, microorganisms and plants biosynthesis secrete and reuptake certain organic secondary metabolites with low molecular masses (500–1500 Da) called siderophores that specifically chelate Fe (III) ions (Soares 2022). More than 500 siderophores have been reported, of which several hundreds have been purified and structurally characterized (using HPLC, NMR, IR, and Mass-Spectroscopy), and continually new structures are discovered (Gama et al. 2021). Siderophores are typically categorized by the functional groups with which they bind iron. Catecholate, hydroxamate, and α-hydroxycarboxylate are the three main bidentate ligands. Mixed siderophores (pyoverdines) have multiple ligand types. On siderophores, ligands may have a linear or cyclic arrangement. Because cyclization improves complex stability, cyclic siderophores are more stable than linear complexes (Hofmann et al. 2020).
Siderophores and their substituted derivatives have a large number of applications in medical and environmental sciences. In agriculture, various types of siderophores have been found to act as a biological control agent against harmful phytopathogens and enhance the growth of several plant species (Molnár et al. 2023). They can also be used for detoxifying heavy-metal-contaminated samples, making them useful in bioremediation (Gupta et al. 2023). Furthermore, siderophores have the ability to detect iron content in various environments, serving as a biosensor (Khasheii et al. 2021). In the medical field, siderophores use the "Trojan horse strategy" to form complexes with antibiotics, enabling selective delivery of antibiotics to antibiotic-resistant bacteria (Rayner et al. 2023). Siderophores can also be utilized in the treatment of certain iron overload diseases, such as sickle cell anemia (Ribeiro et al. 2022). Siderophores can also be used to treat malaria, get rid of transuranic elements from the body (Handore et al. 2022), and have anticancer activity (Pita-Grisanti et al. 2022).
To achieve the given wide range of applications, it is essential to optimize siderophores production by studying the factors that influence their production. Concentration of siderophores may extensively vary in the culture medium, depending on the bacteria, culture medium, cultural conditions, iron-phosphorous concentrations, carbon–nitrogen sources, and the presence of metals (Soares 2022). Various experimental designs can be employed to optimize cell media for growth and production. Statistical experimental designs (such as full or partial factorial designs, Latin rectangles, central composite, and Plackett–Burman) offer a methodical and effective approach to conduct experiments to achieve specific objectives while simultaneously studying multiple factors in a short period of time and comprehending the interactions between various variables (Sahu and Prakash 2021).
The production of most siderophores has a significant challenge due to the presence of enantiomorphs, limiting steps of the chromophore synthesis, non-proteinogenic amino acids, and their molecular complexity. Therefore, this study aimed to optimize and characterize the siderophore production by E. coli strain OQ866153 through two-stage statistical approach based on Plackett–Burman design (PBD) and response surface methodology (RSM) via central composite design (CCD). The genes responsible for biosynthesis of the produced siderophore was also investigated.
Materials and Methods
The used Bacterial Strain
E. coli 49 was kindly provided by Professor Mona Mabrouk, National Organization for Drug Control and Research, Giza, Egypt. In a previous study, this strain was found to be antibiotic resistant, and siderophores producer and its identity was confirmed as an E. coli strain by using 16S rRNA sequence analysis (Khazaal et al. 2022). Recently, the sequence was deposited at the National Center for Biotechnology Information (NCBI) database with an accession numbers OQ866153.
Quantitative Determination of Siderophores
Siderophores production by E. coli OQ866153 was previously determined (Khazaal et al. 2022) using Chrome Azurol Sulfonate (CAS) broth assay of Alexander and Zuberer (1991) after growth in M9 medium at 37 °C for 24 h. Bacterial cells were then removed by centrifugation at 6000 rpm at room temperature for 10 min. The supernatant was filter sterilized through 0.45 µm membrane filter and then added to CAS assay solution which consisted of [Solution 1; 21.9 mg of hexadecyltrimethylammonium bromide (HDTMA) in 25 ml of double-distilled water, 1.5 ml of 1 mM FeCl3.6H2O in 10 mM HCl, 7.5 ml of 2 mM CAS, Solution 2; 9.76 g of 2-[N-morpholino] ethanesulfonic acid (MES buffer) was dissolved in 50 ml and the pH adjusted to 5.6 using 50% KOH)] in (1:1) and mixed with shuttle (87.3 mg of 5-sulfosalicylic acid) solution (100:1). The obtained blue mixture was incubated in the dark for 15 min at room temperature, after which the degree of color change was measured using T60 UV–Vis Spectrophotometer (PG Instruments LTD) at 630 nm. Siderophores concentration in the culture supernatant was calculated and expressed as siderophores unit percent (SU%) using the following formula: [SU% = (Ar−As/Ar) × 100], where Ar is the reference solution absorbance and As is the sample solution absorbance, at 630 nm.
Optimization of Siderophores Production
Plackett–Burman Design (PBD) for Selecting Significant Variables
Twenty-three independent variables were used in PBD; four of them were temperature, incubation time, pH, and agitation as physical variables, eighteen chemical variables were used as glycerol, glucose, glutamate, succinate, NH4Cl, (NH4)2SO4, urea, asparagine, phenylalanine, tyrosine, tryptophane, KH2PO4, K2HPO4, MgSO4, FeSO4, NaCl, Na2HPO4, and CaCl2, and one dummy variable. Two levels (low (− 1) and high (+ 1)) of each variable, as indicated in Table 1, were submitted to the Design Expert version 13 program, and twenty-five trials were obtained (Table 2). The PB runs were carried out in duplicate, and the produced siderophores were determined quantitatively as siderophores unit percent (SU%) by CAS shuttle assay according to Alexander and Zuberer (1991). PBD depends on the first-order model Y = β0 + ΣβiXi where Y represents the siderophores unit percent (SU%), while β0 denotes the model intercept, βi represents the linear coefficient, and Xi indicates the level of the independent variable.
Optimization of Significant Variables Using Response Surface Methodology (RSM)
One of the RSM is the central composite design in which the interactions between the significant factors obtained from PBD results were studied (Korany et al. 2021). In this investigation, six variables (succinate, tryptophane, Na2HPO4, CaCl2, agitation, and KH2PO4) at five levels (-alpha (-α), low (−1), middle (0), high (+1) and + alpha (+α)) were used (Table 3). Fifty-five trials were obtained, the experiments done in duplicate with fixing the remained variables at their low levels and siderophores unit percent (SU%) was obtained as a response (Table 4). The following polynomial second-order equation was used to fit the experimental results: Y = β0 + ∑ βi Xi + ∑ βii Xi2 + ∑ βij Xi Xj where Y is the predicted response, β0 is the regression coefficients, βi is the linear coefficient, βii is the quadratic coefficients, βij is the interaction coefficients, and Xi is the coded levels of independent variable.
Validation of Central Composite Design
Once the data were analyzed, the model was tested for its adequacy and precision. The software solution shown in Fig. 1c was applied in duplicate.
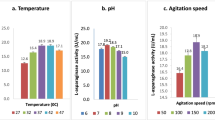
Optimization of siderophores production by E. coli OQ866153. a Pareto chart showing the most significant independent variables on siderophores production. A Temperature, B incubation time, C pH, D agitation, E glycerol, F glucose, G glutamate, H succinate, J NH4Cl, K (NH4)2SO4, L urea, M asparagine, N phenylalanine, O tyrosine, P tryptophane, Q KH2PO4, R K2HPO4, S MgSO4, T FeSO4, U NaCl, V Na2HPO4, W CaCl2, X dummy factor. b Three-dimensional graph (3D) for the most significant interaction between succinate (a) and tryptophan (b) showed the highest predicted SU% ~ 88.13%. c The model solution that actually validated with the highest SU% = 87.15%
Statistical Analysis
Design-expert 13 software (Stat-Ease Inc. Minneapolis, USA) was used for the experimental design and data analysis.
Siderophores Biomass Production
According to Fiedler et al. (2001), siderophores from E. coli OQ866153 strain were extracted as follows: in an Erlenmeyer flask, 100 ml of M9 growth medium was inoculated with OQ866153 strain and incubated overnight at 37 °C and 150 rpm. Then, 10 ml of previously prepared culture was added to one liter of the same medium and incubated for 24 h at 37 °C and 150 rpm with shaking. After incubation, the bacterial suspension was centrifuged at 4 °C and 10,000 rpm for 15 min using a Sigma 3-16PK centrifuge (Germany) and the supernatant was collected and filter sterilized using 0.45 µl Millipore cellulose nitrate membrane filter (CHM, Spain). The filtrate pH was adjusted to 3 using concentrated 12 N hydrochloric acid, then one liter ethyl acetate was added for 1 h at 10 °C and 100 rpm in a shaking incubator. Finally, the ethyl acetate extract was evaporated using a rotary evaporator (IKA, Germany) at 40 °C to obtain a dry crude and stored at − 20 °C until further use.
Extraction and Purification of Siderophore
By using a silica gel thin-layer chromatography (TLC) plate (Merck-Germany) and a Dichloromethane (DCM): Methanol (9:1 v/v) as a solvent system, the siderophore compound was extracted as follows: in a chromatographic jar was initially equilibrated with the solvent system for 20 min, the TLC plate that loaded with ethyl acetate extract was subsequently immersed in the solvent system to facilitate complete elution and separation of compounds. Then, the TLC plate was carefully removed and visualized under a UV lamp (VL-215 LC, Marne La Vallee, France) at 254/365 nm. The desired band that showed a bright blue fluorescence, was carefully scratched, transferred to another vial, and eluted several times using ethyl acetate to obtain the siderophore compound. This process was repeated multiple times to obtain the desired quantity of the compound, which was then purified using a sephadex LH-20 column and a DCM: Methanol (6:4 v/v) solvent system.
Characterization of the Purified Siderophore Compound
Nuclear Magnetic Resonance (NMR) Spectroscopy
The purified compound was characterized by 1H and 13C NMR spectroscopy (Moreno et al. 2018) using a Bruker Corp. spectrophotometer with frequencies of 400 and 100 MHz at the NMR Lab in the Microanalytical Unit (MAU) of the Faculty of Pharmacy at Cairo University, Egypt.
Fourier Transform Infrared (FTIR) Spectroscopy
The IR spectrum of the purified compound (Moreno et al. 2018) was performed using PerkinElmer L1600400 FTIR spectrum (UK) within a wavelength range and a resolution of 400–4000 cm−1 and 4 cm−1, respectively, at the central lab, Faculty of Science, Helwan University, Cairo, Egypt.
Detection of Enterobactin Biosynthesis Genes
The primers of enterobactin biosynthesis genes (entA, entB, entC, entD, entE, and entF) were designed using OLIGO 7.57 software (Molecular Biology Insights, Inc) and the E. coli strain NZ_CP050203 sequence was used as a reference (Table 7). Bacterial DNA was extracted following the DNA QIAamp mini kit (QIAGEN) instructions. After DNA extraction, the designed primers were used for the PCR amplification of the enterobactin biosynthesis genes and the PCR conditions were denaturation at 95 °C/5 min, 30 denaturation cycles at 94 °C/45 s, annealing at 59 °C/30 s, extension at 72 °C/30 s, and a final 72 °C/5 min extension step. The amplification products were separated by 1% agarose gel electrophoresis and visualized under UV transilluminator. Finally, the PCR products of enterobactin biosynthesis genes were sequenced using their forward primers in an automatic 3730XL ABI prism sequencer (Applied Biosystems, USA) at Korea Macrogen Inc. and Geneious prime 2021 software was employed to evaluate the BLAST results and build phylogenetic tree with the corresponding genes in gene bank database.
Results
Optimization of Siderophores Production
PB- Design Results
Results of PB-design (25 runs) revealed that the percentage of siderophores unit (SU%) increased from 0 (run 24), 8.98% (Run 18), 13.73 (Run 22), 26.78% (Run 13), and 34.35% (Run 8), to the highest value 43.67% in run 14 as indicated in Table 2. Furthermore, the model p-value was 0.0094 (< 0.05), indicating its significance. The incubation temperature (A), incubation time (B), pH (C), agitation (D), glycerol (E), glutamate (G), succinate (H), NH4Cl (J), (NH4)2SO4 (K), urea (L), phenylalanine (N), tryptophane (P), KH2PO4 (Q), K2HPO4 (R), MgSO4 (S), NaCl (U), Na2HPO4 (V), and CaCl2 (W) variables were found to be with significant p-values of 0.0146, 0.0105, 0.0123, 0.0089, 0.0147, 0.0281, 0.0011, 0.0157, 0.015, 0.0183, 0.057, 0.0266, 0.0073, 0.0092, 0.0367, 0.0186, 0.0074, and 0.0087, respectively. The model Adeq precision ratio of 43.0822 indicated an adequate signal, where a ratio greater than 4 is desirable, and confirmed that the model can be used to navigate the design space (Table 5). Additionally, as in the Pareto chart (Fig. 1a), the variables succinate (H), tryptophan (P), Na2HPO4 (V), CaCl2 (W), agitation (D), and KH2PO4 (Q) were deemed the most significant on the production of siderophores, with f-values of 883.61, 135.63, 134.5, 114.06, 110.73, and 106.98, respectively.
CC- Design Results
Results of CC-design shown in Table 4 demonstrated that the interaction between the most significant variables of PB-design increased the SU% from 21.54% (Run 37), 37.33% (Run 35), 49.12 (Run 7), 56.77% (Run 55), and 64.12% (Run 27) to the highest value 89.13% (Run 22).CCD data analysis demonstrated the model p-value of 0.0004 and f-value of 314.14, indicating its significance. The coefficients for the linear terms were succinate (-14.34), tryptophan (-5.41), Na2HPO4 (5.69), CaCl2 (-5.13), Agitation (4.88), and KH2PO4 (-4.63); the interactive coefficients for the two-factor interactions were AB (1.21), AC (-0.6371), AD (0.5466), AE (-0.5865), AF (0.9310), BC (-0.3812), BD (0.3017), BE (-0.2227), BF (0.7244), CD (-0.2443), CE (0.0279), CF (-0.1671), DE (-0.2256), DF (0.2152), and EF (-0.3742), and the quadratic coefficients for A2, B2, C2, D2, E2, and F2 were 5.44, -6.50, -2.50, -0.52, 5.48, and -0.5769, respectively (Table 5). The model determination (R2) coefficient of 0.9950 indicated that it could explain approximately 99.50% of the variability in the response. The results of the model also determined the impact of each input parameter on siderophore production by applying the following actual second-order polynomial equation.
Interactions Between the Significant Variables
The interactions between the six significant variables for maximum siderophore production were studied through three dimensional plots. Different significant interactions were observed between the variables AB, AC, AD, AE, AF, and BF. The most significant one was between succinate and tryptophan (AB), where, at a constant level of Na2HPO4 (C = 6 g/l), CaCl2 (D = 0.1 g/l), agitation (E = 150 rpm), and KH2PO4 (F = 0.6 g/l), the presence of the lowest or highest tryptophan (B) levels with low level of succinate (A) resulted in a higher siderophore production (SU%) of 88.1 and 72.8%, respectively. On the other hand, siderophore production (SU%) lessened to 53.8 and 43.3% in the presence of the lowest or highest tryptophan (B) levels with high level of succinate (A), respectively (Fig. 1b).
The model was validated using one of the suggested solutions and the results showed SU% of 87.1472%, which deviated by − 1% from the predicted value. Nonetheless, this value was approximately doubled compared to the SU% obtained before optimization, which was 46.62%.
Extraction, Purification, and Characterization of the Synthesized Siderophores
E. coli OQ866153 strain was grown in M9 medium under the optimum growth condition. The synthesized siderophores were extracted and separated on TLC plates then examined under the UV lamp at 254/365 nm which showed a bright blue fluorescent band (Fig. 2a). This band was scratched and eluted, and the compound was characterized by using NMR (1H and 13C) and FTIR spectroscopy. The 1H NMR spectrum indicated the presence of proton chemical shift signals at δ 4.07, 4.71, 5.00, 6.77, 6.85, 7.35, and 8.31 ppm (Fig. 2b). Similarly, the 13C NMR spectrum recorded 10 different carbon signals at δ 52.41, 64.56, 115.95, 118.94, 119.46, 120.07, 145.43, 148.61, 169.44, and 170.40 ppm (Fig. 2c). The chemical shifts were compared with those reported in (Table 6). On the other hand, the results of FTIR revealed a broad band located at the wavenumber of 3000–3500 cm−1, which corresponded to the overlap between OH and NH (Fig. 2d). Additionally, the band observed at 1690 cm−1 could correspond to the amide-carbonyl group's characteristic. The bands at 1607 and 1532 cm−1 represented the C=C stretch and C3–H bend overlaps. The following IR region (1000 –1300 cm−1) was associated with C–OH, C–H, N–H, and OH. The results obtained indicated that the compound was enterobactin.
Detection of Enterobactin Biosynthesis Genes
Enterobactin designed primers were amplified using PCR for detection of enterobactin biosynthesis genes in E. coli OQ866153 extracted DNA and the amplicons were examined using agarose gel electrophoresis. Six bands corresponding to enterobactin biosynthesis (entA, entB, entC, entD, entE, and entF) genes with different sizes (Table 7) were detected in the gel (Fig. 3a). These bands were excised, eluted, purified, and sequenced by the sequencing services of Macrogen in Korea (Fig. S1). The obtained sequences were submitted separately to the gene bank and the accession numbers provided were entA: OR645470, entB: OR645471, entC: OR645472, entD: OR645473, entE: OR645474, and entF: OR645475. The sequenced DNA were then subjected to a Blast hit analysis against E. coli RH-045-MS chromosome (NZ_CP050203), as shown in Fig. 3b, where the sequenced genes were highlighted in black. The sequence alignment showed > 90% identity percentage to the corresponding genes in the gene bank (Table S1) and represented by phylogenetic tree (Fig. 4). These results indicated that the peptidic assembly of enterobactin could be attributed to the non-ribosomal peptide synthetases (entABCDEF) as demonstrated in Fig. 5.
a Agarose gel electrophoresis of amplified entABCDEF genes of E. coli OQ866153. Lanes M, 100 bp molecular weight DNA marker; 1 entA OR645470, 2 entB OR645471, 3 entC OR645472, 4 entD OR645473, 5 entE OR645474, 6 entF OR645475. b BLAST Hit of sequenced enterobactin production genes (black color) with E. coli NZ_CP050203
Discussion
Siderophores could be applied as biosensors, biocontrol, bioremediation, and chelation agents, in addition to weathering soil minerals and improving plant growth, (Ahmed and Holmström 2014; Khasheii et al. 2021).
For future applications of siderophores produced by E. coli OQ866153, statistical optimization of siderophores production by this strain was carried out. Different cultural conditions and physiological variables were used to optimize siderophores production. Two-step approaches were used in the optimization experiment: the first one was Plackett–Burman (PB) to recognize the most significant factors, and the second was central composite design (CCD) to determine the optimal concentration of selected variables and assess their interactions. The siderophores unit percent (SU%) was used as the response (Y) to maximize siderophore production in each trial. Gao and Bian (2022) suggested that the production of siderophores in microorganisms is influenced by the type of substrate used. As a result, a total of 23 independent variables were selected based on a component of M9 medium, succinate medium (Shaikh et al. 2016), and enterobactin biosynthesis pathway on Kegg pathway under accession number (Entry: map01053) in Plackett–Burman design experiment. The PB and CC models were both statistically significant, with p-values of 0.0094 and 0.0004, respectively. Pham et al. (2019) noted that smaller p -values and larger f -values represent the significance of the corresponding coefficients. PB results showed that Run 14 recorded the highest siderophores percentage (SU%) of 43.67% and revealed that the following variables succinate, tryptophan, Na2HPO4, CaCl2, KH2PO4, and agitation were the most significant. These results nearly agree with Kalyan et al. (2022) as they mentioned that the organic and inorganic nutrients present in the medium are significant predictors of siderophore production in a given organism. Different nitrogen sources, including NH4Cl, (NH4)2SO4, urea, phenylalanine, tryptophane, and asparagine, were tested and the results revealed that tryptophane was found to be the most effective with a p -value of 0.0073. These results agreed with Mushtaq et al. (2022) who found that L-tryptophane improves iron uptake in Acinetobacter calcoaceticus and Bacillus simplex, resulting in better plant growth and not agreed with Sayyed et al. (2019) who reported that in the presence of urea, both Pseudomonas fluorescence and Pseudomonas putida exhibited maximum productivity of siderophores. Also, Srivastava et al. (2022) found that (NH4)2 SO4, NaNO3, and urea facilitate siderophore production with a SU value larger than 60%. The choice of carbon source is critical within the components of the culture medium (Vindeirinho et al. 2021). In this study, succinate as a carbon source was the most significant one (with f-value of 883.61) comparing with the other tested sources. Our findings were in consistent with those described that Pseudomonas fluorescens DSM 50090 significantly produced higher pyoverdine in minimal medium containing succinate than that with dextrose or glycerol (Vindeirinho et al. 2021). Furthermore, phosphorus is crucial in culture media because it is necessary for cellular metabolism and can act as a buffer. In the Pseudomonas genus, it has been reported that the concentration of phosphate can impact PVD production (Soares 2022). One of the most important aspects of our study was agitation, which had a significant, positive impact on siderophore production. Our findings corroborated those of Panda et al. (2017), who found that siderophore production was too low when conducted under static conditions but optimal at 150 rpm. According to RajeNimbalkar et al. (2022), the determination coefficient (R2) value which was closer to 1, indicating the strength of the model and its ability to predict the response. In this study, the R2 values were 0.9991 and 0.995 in both PB and CC, indicating that 99.91 and 99.5% of the variability in siderophore production could be explained by the model’s independent variables, respectively. From PB results, six significant factors were introduced to central composite design to determine their optimal range and study the interaction between them. Ezemagu et al. (2021) reported that a high coefficient of variation (CV) indicated low model experiment reliability. The precision of the experiments in this study was demonstrated by a CV of 2.91%. CC results showed increase in SU% from 21.5 in run 37 to 89.1 in run 22. The results nearly matched with RajeNimbalkar et al. (2022), who reported that the maximum siderophore production was 98% in succinate medium during CC results. A 3D graphs was plotted to perform the variables interactions and the results showed that the most significant one was between succinate and tryptophan and the highest siderophore production (SU%) of 88.1 was obtained with the presence of the lowest level of tryptophan and succinate at a constant level of Na2HPO4 (C = 6 g/l), CaCl2 (D = 0.1 g/l), agitation (E = 150 rpm), and KH2PO4 (F = 0.6 g/l), indicating that presence of the two factors together had a negative effect on the siderophores production. These findings were in close agreement with those of Vindeirinho et al. (2021), who found that succinate performed best among the carbon sources tested; however, increasing succinate concentration in the culture medium did not lead to an increase in PVD maximum production, despite leading to a rise in biomass. The statistical model was validated under the predicted conditions (0.3 g/l succinate, 0 mg/l tryptophan, 6 g/l Na2HPO4, 0.1 g/l CaCl2, 150 rpm agitation, and 0.6 g/l KH2PO4 and the siderophore production (SU%) was 87.1472% increased by two-fold compared to the value obtained from the ordinary conditions (46.62% before optimization). The results were almost in agreement with those reported by Srivastava et al. (2022), who found that after siderophores were optimized by Pseudomonas monteilii strain MN759447, the actual values were 89.9% siderophore units, which was 10% higher than the value obtained during CAS quantification (80.06%). Moreover, the maximum production of a siderophore compound produced by an endophytic fungus Talaromyces trachyspermus was 88.9% through the application of Plackett–Burman design and Response Surface Methodology via Central Composite Design.
To confirm the structure of the extracted and purified compound, 1H, 13C NMR, and IR spectroscopy were utilized. The use of spectroscopy, specifically 1H and 13C NMR, is a common method for characterizing siderophore compounds due to the distinct features they provide (Fraga-Corral et al. 2022). NMR spectroscopy aids in identifying structural properties, specific functional groups present in the sample, and determining conformation and saccharide ratio within the mixture. The obtained carbon and proton chemical shifts were compared with previously reported data (Moreno et al. 2018). The 1H NMR spectrum displayed three doublet of doublet signals representing the three protons of the aromatic ring, with peaks at δ 6.77, 6.85, and 7.35 ppm for protons 5, 4, and 6, respectively. The coupling constant indicated that proton 5 was in the ortho position to both 6 and 4, while protons 4 and 6 were in the meta position to each other. An amide proton signal appeared at δ 8.31 ppm and a multiplate signal was observed at δ 5.00 ppm. Additionally, two doublet of doublet signals were detected for methylene protons at carbon 10, indicating the presence of enterobactin compound.
Based on the 13C NMR data, it was observed that there are 10 carbon signals present in the compound. Two of these signals, namely C7 and C11, were identified as characteristic carbonyl signals with a chemical shift of δ 170 and 169, respectively. Additionally, two oxygenated carbons, C3 and C2, were identified at δ 145.4 and 148.6, respectively. Furthermore, three aromatic methines were identified at δ 118.9, 119.5, and 120.1 for C6, C5, and C4, respectively. A quaternary carbon signal was observed at δ 115.9 for C1, while a methylene carbon signal was identified at δ 64.6 for C10. Lastly, a signal at δ 52.4 was observed for C9. By comparing this data with reported literature Moreno et al. (2018) and analyzing the 1H and 13C NMR data, it was concluded that the compound was enterobactin.
FTIR is used to differentiate between organic and inorganic materials and to acquire information about the functional groups present in certain compounds (Leela and Anchana 2017). For instance, the wide band observed at 3000–3500 cm−1 is frequently linked to the overlap between OH and NH, as noted by Subi et al. (2022). Moreno et al. (2018) found that this broad band was made up of three overlapping OH stretching modes, which were NH stretching bands at higher frequencies and showed three separate medium-sharp bands that each belonged to a different catechol amide arm. According to Kaur et al. (2021), proton transfer to neighboring oxygen may cause some shifts in N–H IR bands. The 1740 cm−1 band in the IR spectrum was assigned to represent the crown ester and amide group on carboxylic acid derivatives (Wang et al. 2022). Workman and Weyer (2012) reported that a more electronegative substituent can increase the carbonyl carbon oxygen stretch frequency above the nominal frequency of 1715 cm−1 by up to 100 cm−1. Carbonyl stretching bands were also observed at 1770 and 1818 cm−1 in studies of carboxylic acid derivatives and alpha-Angelica lactones (Moreno et al. 2018). However, in this study, the band was observed at 1690 cm−1. Moreno et al. (2018) also reported that the frequency of ester IR bands varies depending on whether it is in a crown ester, as in enterobactin, or in an open chain. According to Moreno et al. (2018), the bands observed at 1607 and 1532 cm−1 are attributed to the C=C stretch and C3–H bend overlaps. The C–OH, C–H, N–H, and OH stretching from catechol are observed in the 1000–1300 cm−1 IR region, which is consistent with Moreno et al. (2018) findings. By comparing the obtained IR spectroscopy data with the literature reported by Moreno et al. (2018), the compound was identified as enterobactin.
Enterobactin is a cyclic trimer of 2,3-dihydroxybenzoyl-l-serine produced by bacteria such as Escherichia coli and Klebsiella pneumophila belonging to family Enterobacteriaceae to scavenge Fe (III) with high affinity (Soares 2022). In this study, enterobactin encoding genes were detected in the DNA extracted from E. coli OQ866153 using enterobactin designed primer. The obtained results revealed that six bands corresponding to enterobactin biosynthesis (entA OR645470, entB OR645471, entC OR645472, entD OR645473, entE OR645474, and entF OR645475) genes with the size ranging from 314 to 671 bp were detected. The sequence alignment of the purified amplicons showed identity percentage higher than 90% to the corresponding genes in the gene bank. Most siderophores are produced by non-ribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs) working along with NRPS modules, but a small number are produced by other mechanisms (Kramer et al. 2020). In this study, the peptidic assembly of enterobactin could be attributed to the non-ribosomal peptide synthetases (entABCDEF), where enterobactin synthetase, a two-module NRPS, is responsible for E. coli siderophores biosynthesis and under iron-deficient conditions, the NRPS (entB, entE, and entF) components are induced for enterobactin synthesis (Liu et al. 2023). Reitz et al. (2017) reported that the protein products of the entA, entB, entC, entD, entE, and entF genes are 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase, isochorismatase, isochorimate synthase, 4-phosphatetheinyl transferase, 2,3-dihydroxybenzoyl adenylate synthase, and enterobactin non-ribosomal peptide synthetase, respectively. Enterobactin is synthesized through a two-step process, starting with the conversion of chorismic acid into isochorismate, followed by its conversion into 2,3-dihydro-2,3-dihydroxybenzoate, and finally to 2,3-dihydroxybenzoic acid (DHB). These successive steps are catalyzed by entC, entB, and entA, respectively (Reitz et al. 2017). entD, entE, entF, and a C-terminal aryl carrier of entB are involved in the formation of the amide linkage between DHB and l-serine (Jaremko et al. 2020). Before serine binds onto entF peptidyl carrier protein domain, it is activated by adenylation (Reitz et al. 2017). Finally, after the intermolecular cyclization and hydrolysis of three molecules of DHB-Ser, the entF terminal thioesterase domain releases enterobactin (Jelowicki and Butler 2022). According to the research by Jelowicki and Butler 2022), the strain of Vibrio campbellii BAA-1116 harbors a cluster of genes (entA-F) that are similar to the enterobactin biosynthetic gene cluster (BGC). Also, they suggested that AebABCE is responsible for the production of 2,3-dihydroxybenzoic acid (2,3-DHBA). Furthermore, (Wellawa et al. 2022) noticed that the entABCDEF operon proteins, synthesize and converge three N-2,3-dihyroxybenzoyl-l-serines to accommodate Fe3+. The antibacterial protein enterobactin shields E. coli from the oxidative stress caused by agents like hydrogen peroxide and several agricultural pesticides (Peralta et al. 2022). Due to its potent iron chelating characteristics, iron-free enterobactin is able to selectively induce lethal effects on highly proliferative cells. As a result, enterobactin shows promise as a powerful anticancer agent (Saha et al. 2019).
Conclusion
In conclusion, using a two-stage statistical approach involving Plackett–Burman design (PBD) and response surface methodology (RSM) using central composite design (CCD) increased the production of siderophores by E. coli OQ866153. Out of 23 variables, succinate, tryptophan, Na2HPO4, CaCl2, agitation, and KH2PO4 were found to have the most significant effect on siderophores production in the first optimization stage with SU% response increased from 0 (Run 24) to the highest value of 43.67% (Run 14). In the second stage, CCD results showed increase in the SU% from 21.5 in run 37 to 89.1 in run 22., and the optimal conditions were determined to be 0.3 g/l succinate, 0 g/l tryptophan, 6 g/l Na2HPO4, 0.1 g/l CaCl2, 150 RPM agitation, and 0.6 g/l KH2PO4. Additionally, CCD results showed that the most significant variables interaction was between succinate and tryptophan with the highest siderophore production (SU%) of 88.1 in the presence of the lowest level of tryptophan and succinate at a constant level of Na2HPO4 (C = 6 g/l), CaCl2 (D = 0.1 g/l), agitation (E = 150 rpm), and KH2PO4 (F = 0.6 g/l), indicating that presence of the two factors together had a negative effect on the siderophores production. The statistical model was validated under the predicted conditions (0.3 g/l succinate, 0 mg/l tryptophan, 6 g/l Na2HPO4, 0.1 g/l CaCl2, 150 rpm agitation, and 0.6 g/l KH2PO4 and the siderophore production (SU%) was 87.1472% increased by two-fold compared to the value obtained from the ordinary conditions (46.62% before optimization). Based on 1H, 13C, and IR spectroscopy results, the purified siderophore was identified as enterobactin, a catecholate-type siderophore that is widely distributed among enteric bacteria. The presence of entABCDEF genes confirmed the biosynthetic pathway of enterobactin in E. coli OQ866153. To our knowledge, this is the first report of statistical optimization for enterobactin production by an E. coli strain isolated from clinical source in Egypt. This study provides valuable insights into the diversity and potential applications of siderophores in environmental and medical research.
Data Availability
All data generated or analyzed during this study are included in this manuscript and its supplementary information files.
References
Ahmed E, Holmström SJM (2014) Siderophores in environmental research: roles and applications. Microb Biotechnol 7:196–208. https://doi.org/10.1111/1751-7915.12117
Alexander DB, Zuberer DA (1991) Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol Fertil Soils 12:39–45. https://doi.org/10.1007/BF00369386
Ezemagu IG, Ejimofor MI, Menkiti MC, Nwobi-Okoye CC (2021) Modeling and optimization of turbidity removal from produced water using response surface methodology and artificial neural network. S Afr J Chem Eng 35:78–88. https://doi.org/10.1016/j.sajce.2020.11.007
Fiedler H-P, Krastel P, Müller J, et al (2001) Enterobactin: the characteristic catecholate siderophore of enterobacteriaceae is produced by Streptomyces species. FEMS Microbiol Lett 196:147–151. https://doi.org/10.1111/j.1574-6968.2001.tb10556.x
Fraga-Corral M, Carpena M, Garcia-Oliveira P et al (2022) Analytical metabolomics and applications in health, environmental and food science. Crit Rev Anal Chem 52:712–734. https://doi.org/10.1080/10408347.2020.1823811
Gama S, Hermenau R, Frontauria M et al (2021) Iron coordination properties of gramibactin as model for the new class of diazeniumdiolate based siderophores. Chemistry 27:2724–2733. https://doi.org/10.1002/chem.202003842
Gao H, Bian X (2022) Microbial siderophores: biosynthesis, regulation, and physiological and ecological impacts. Front Microbiol 13:1377. https://doi.org/10.3389/fmicb.2022.892485
Gupta R, Khan F, Alqahtani FM et al (2023) Plant growth-promoting rhizobacteria (PGPR) assisted bioremediation of heavy metal toxicity. Appl Biochem Biotechnol. https://doi.org/10.1007/s12010-023-04545-3
Handore AV, Khandelwal SR, Karmakar R, Handore DV (2022) Exploration of bacterial siderophores for sustainable future. In: Climate change and microbial diversity. Apple Academic Press, pp 163–189.
Hofmann M, Retamal-Morales G, Tischler D (2020) Metal binding ability of microbial natural metal chelators and potential applications. Nat Prod Rep 37:1262–1283. https://doi.org/10.1039/C9NP00058E
Jaremko MJ, Davis TD, Corpuz JC, Burkart MD (2020) Type II non-ribosomal peptide synthetase proteins: structure, mechanism, and protein–protein interactions. Nat Prod Rep 37:355–379. https://doi.org/10.1039/C9NP00047J
Jelowicki AM, Butler A (2022) On the origin of amphi-enterobactin fragments produced by Vibrio campbellii species. J Biol Inorg Chem 27:565–572. https://doi.org/10.1007/s00775-022-01949-0
Kalyan VSRK, Meena S, Karthikeyan S, Jawahar D (2022) Isolation, screening, characterization, and optimization of bacteria isolated from calcareous soils for siderophore production. Arch Microbiol 204:721. https://doi.org/10.1007/s00203-022-03322-w
Kaur D, Zhang Y, Reiss KM et al (2021) Proton exit pathways surrounding the oxygen evolving complex of photosystem II. Biochimica Et Biophysica Acta (BBA) 1862:148446. https://doi.org/10.1016/j.bbabio.2021.148446
Khasheii B, Mahmoodi P, Mohammadzadeh A (2021) Siderophores: Importance in bacterial pathogenesis and applications in medicine and industry. Microbiol Res 250:126790. https://doi.org/10.1016/j.micres.2021.126790
Khazaal MT, El-Hendawy HH, Mabrouk MI et al (2022) Antibiotic resistance and siderophores production by clinical Escherichia coli strains. Biotechnologia 103:169–184. https://doi.org/10.5114/bta.2022.116211
Korany SM, El-Hendawy HH, Sonbol H, Hamada MA (2021) Partial characterization of levan polymer from Pseudomonas fluorescens with significant cytotoxic and antioxidant activity. Saudi J Biol Sci 28:6679–6689. https://doi.org/10.1016/j.sjbs.2021.08.008
Kramer J, Özkaya Ö, Kümmerli R (2020) Bacterial siderophores in community and host interactions. Nat Rev Microbiol 18:152–163. https://doi.org/10.1038/s41579-019-0284-4
Leela K, Anchana DC (2017) A study on the applications of silver nanoparticle synthesized using the aqueous extract and the purified secondary metabolites of lichen Parmelia perlata. Int J Pharm Sci Invent 6:42–59
Liu Z, Huang T, Shi Q et al (2023) Catechol siderophores framed on 2, 3-dihydroxybenzoyl-l-serine from Streptomyces varsoviensis. Front Microbiol. https://doi.org/10.3389/fmicb.2023.1182449
Molnár Z, Solomon W, Mutum L, Janda T (2023) Understanding the mechanisms of Fe deficiency in the rhizosphere to promote plant resilience. Plants 12:1945. https://doi.org/10.3390/plants12101945
Moreno M, Zacarias A, Porzel A et al (2018) IR and NMR spectroscopic correlation of enterobactin by DFT. Spectrochim Acta A 198:264–277. https://doi.org/10.1016/j.saa.2018.02.060
Mushtaq Z, Nazir A, Asghar HN, Zahir ZA (2022) Interactive effect of siderophore-producing bacteria and l-tryptophan on physiology, tuber characteristics, yield, and iron concentration of potato. Potato Res 65:1015–1027. https://doi.org/10.1007/s11540-022-09565-w
Panda SH, Goli JK, Das S, Mohanty N (2017) Production, optimization and probiotic characterization of potential lactic acid bacteria producing siderophores. AIMS Microbiol 3:88. https://doi.org/10.3934/microbiol.2017.1.88
Peralta DR, Farizano JV, Bulacio Gil N et al (2022) Less is more: enterobactin concentration dependency in copper tolerance and toxicity. Front Mol Biosci 9:961917. https://doi.org/10.3389/fmolb.2022.961917
Pham TN, Tran BP, Tran TH, et al (2019) Response surface modeling and optimizing conditions for anthocyanins extraction from purple sweet potato (Ipomoea batatas (L.) Lam) grown in Lam Dong province, Vietnam. In: IOP Conference Series: Materials Science and Engineering. IOP Conf. Ser. 479: 012012. Doi: https://doi.org/10.1088/1757-899X/479/1/012012.
Pita-Grisanti V, Chasser K, Sobol T, Cruz-Monserrate Z (2022) Understanding the potential and risk of bacterial siderophores in cancer. Front Oncol 12:1885. https://doi.org/10.3389/fonc.2022.867271
RajeNimbalkar RU, Barge NS, Marathe RJ et al (2022) Application of response surface methodology for optimization of siderophore production. Indian J Agric Res 56:230–237. https://doi.org/10.3389/fpls.2022.984522
Rayner B, Verderosa AD, Ferro V, Blaskovich MAT (2023) Siderophore conjugates to combat antibiotic-resistant bacteria. RSC Med Chem 14:800–822. https://doi.org/10.1039/D2MD00465H
Reitz ZL, Sandy M, Butler A (2017) Biosynthetic considerations of triscatechol siderophores framed on serine and threonine macrolactone scaffolds. Metallomics 9:824–839. https://doi.org/10.1039/c7mt00111h
Ribeiro M, Sousa CA, Simões M (2022) Harnessing microbial iron chelators to develop innovative therapeutic agents. J Adv Res 39:89–101. https://doi.org/10.1016/j.jare.2021.10.010
Saha P, San Yeoh B, Xiao X et al (2019) Enterobactin, an iron chelating bacterial siderophore, arrests cancer cell proliferation. Biochem Pharmacol 168:71–81. https://doi.org/10.1016/j.bcp.2019.06.017
Sahu S, Prakash A (2021) Siderophore from Talaromyces trachyspermus: augmentation and characterization. bioRxiv. https://doi.org/10.1101/2021.04.13.439607
Sayyed RZ, Seifi S, Patel PR et al (2019) Siderophore production in groundnut rhizosphere isolate, Achromobacter sp. RZS2 influenced by physicochemical factors and metal ions. Environ Sustain 2:117–124. https://doi.org/10.1007/s42398-019-00070-4
Shaikh SS, Wani SJ, Sayyed RZ (2016) Statistical-based optimization and scale-up of siderophore production process on laboratory bioreactor. 3 Biotech 6:1–10. https://doi.org/10.1007/s13205-016-0365-2
Soares EV (2022) Perspective on the biotechnological production of bacterial siderophores and their use. Appl Microbiol Biotechnol 106:3985–4004. https://doi.org/10.1007/s00253-022-11995-y
Srivastava P, Sahgal M, Sharma K et al (2022) Optimization and identification of siderophores produced by Pseudomonas monteilii strain MN759447 and its antagonism toward fungi associated with mortality in Dalbergia sissoo plantation forests. Front Plant Sci 13:984522. https://doi.org/10.3389/fpls.2022.984522
Subi B, Dhas DA, Joe IH, Balachandran S (2022) Synthesis, spectroscopic (FTIR, FT-Raman and UV-Vis), structural investigation, Hirshfeld, AIM, NBO, chemical reactivity, in-vitro and in-silico analysis of N-(2-Hydroxyphenyl)-4-Toluenesulfonamide. Polycycl Aromat Compd. https://doi.org/10.1080/10406638.2022.2144916
Vindeirinho JM, Soares HMVM, Soares EV (2021) Modulation of siderophore production by Pseudomonas fluorescens through the manipulation of the culture medium composition. Appl Biochem Biotechnol 193:607–618. https://doi.org/10.1007/s12010-020-03349-z
Wang T-T, Wang S, Shao S et al (2022) Perfluorooctanoic acid (PFOA)-induced alterations of biomolecules in the wetland plant Alisma orientale. Sci Total Environ 820:153302. https://doi.org/10.1016/j.scitotenv.2022.153302
Wellawa DH, Lam P-KS, White AP et al (2022) High affinity iron acquisition systems facilitate but are not essential for colonization of chickens by Salmonella Enteritidis. Front Microbiol 13:280. https://doi.org/10.3389/fmicb.2022.824052
Workman J Jr, Weyer L (2012) Practical guide and spectral atlas for interpretive near-infrared spectroscopy. CRC Press, Boca Raton
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). Open access funding was provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). The authors have not disclosed any funding.
Author information
Authors and Affiliations
Contributions
MK contributed to the study conception, performed experiments, data analysis, and wrote the original draft. AF contributed to the study conception, design, and to laboratory experiments. MH performed the statistical work up to writing in the manuscript and assisted in TLC laboratory work. HHE conceptualized the study, supervised, reviewed, and edited the manuscript. All authors have read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Khazaal, M.T., Faraag, A.H.I., Hamada, M.A. et al. Characterization and Statistical Optimization of Enterobatin Synthesized by Escherichia coli OQ866153. Biochem Genet (2024). https://doi.org/10.1007/s10528-023-10626-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10528-023-10626-z