Abstract
The association of the rs4420638 polymorphism, near the APOC1 gene, was examined with the risk of obesity among Portuguese children. A sample of 446 Portuguese individuals (231 boys and 215 girls) of European descent, aged 3.2 to 13.7 years old (mean age: 7.98 years), were selected to conduct a case-control study. Body mass index (BMI), BMI Z-scores, and waist circumference were calculated. Genotyping was performed by real time PCR using a pre-designed TaqMan probe. Logistic regression and the nonparametric Mann-Whitney test were used to test the associations. The association results revealed a significant protective effect from the minor G-allele of SNP rs4420638 against obesity, with an odds ratio (OR) of 0.619 (95% CI 0.421–0.913; p = 0.0155) in the additive model, and OR of 0.587 (95% CI 0.383-0.9; p = 0.0145) in the dominant model. Moreover, comparing genotype groups (AA vs. AG + GG), significantly lower values (p < 0.05) for the anthropometric traits weight, height, BMI, BMI Z-score and waist circumference, were observed in the carriers of allele G. The present study provides further evidence for the APOE/APOC1 candidate-region association with the risk of obesity. This was the first study to describe the protective association of the rs4420638 minor G-allele against obesity in childhood exclusively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The prevalence of overweight and obesity in children and adolescents has increased enormously in the last few decades, both in girls and boys (Ng et al. 2014). This epidemic proportion reached by obesity in the pediatric population suggests the importance of environmental factors, mostly the consumption of high-calorie foods and a sedentary lifestyle (Vilchis-Gil et al. 2015). However, it is well established that childhood obesity results from a complex interplay between genetic and these non-genetic environmental factors (Mason et al. 2020; Loos and Yeo 2022). Furthermore, some studies have elucidated epigenetic modifications with a role in weight gain (Alfano et al. 2022).
The SNP rs4420638 (NM_001645.3, c.*459A > G) located approximately 340 bp from the 3’ end of the apolipoprotein C-I (APOC1) gene, in the chromosomal region 19q13.32, tags a linkage disequilibrium (LD) block involving the TOMM40, APOE, and APOC1 genes (Suchindran et al. 2010; Beekman et al. 2013), and has been implicated in dyslipidemia by numerous genetic association studies [GWAS Catalog EMBL-EBI (http://www.ebi.ac.uk/gwas; accessed on January 24, 2023)] (Sollis et al. 2023). The rs4420638 A > G polymorphism was significantly related with activity of lipoprotein-associated phospholipase A2 (Lp-PLA2) and serum levels of total cholesterol (TC), triglyceride (TG), low-density-lipoprotein (LDL) cholesterol or high-density lipoprotein (HDL) cholesterol (Kathiresan et al. 2008; Drenos et al. 2009; Suchindran et al. 2010; Liu et al. 2011; Grallert et al. 2012; Breitling et al. 2015; Zhang et al. 2018; Wang et al. 2022). Furthermore, rs4420638 was associated with risks of metabolic syndrome, type 2 diabetes, and coronary heart disease (Zhang et al. 2018). However, the mechanism by which the rs4420638 A > G variant influences lipid metabolism and risk of diseases or whether these associations are related with the LD observed with APOE polymorphisms remain to be clarified (Zhang et al. 2018).
Additionally, while a few studies have identified rs4420638 as an obesity-linked genetic variant (Winkler et al. 2015; Lotta et al. 2018; Boulenouar et al. 2019), some did not reveal this association (Wang et al. 2022). Moreover, to our knowledge, there is no studies on the relationship between obesity and the rs4420638 variant in childhood exclusively. Considering these conflicting results for rs4420638 as an obesity-associated variant, and the total lack of studies exploring the association of this genetic variant with the risk of obesity in the Portuguese population, we conducted the present case-control study in a sample of Portuguese children.
Materials and Methods
Study Population
Participants were randomly recruited from several pre- and primary public schools in the Portuguese Midlands between 2013 and 2018 in two independent studies carried out (Albuquerque et al. 2013; Manco et al. 2019). Both projects were approved by the Direcção Geral de Inovação e de Desenvolvimento Curricular of the Ministry of Education, and they were conducted anonymously, and according to the guidelines laid down in the Declaration of Helsinki. Written informed consent was obtained from all children’s parents or tutors. From a total of 1170 Portuguese children, 446 of them (231 boys and 215 girls) from European descent, aged 3.2 to 13.7 years-old (mean 7.98 years), were selected to conduct the present case-control study.
The anthropometric measurements of height, weight and waist circumference were taken by trained researchers according to established protocols, as described elsewhere (Albuquerque et al. 2013; Manco et al. 2019). Body mass index (BMI) was calculated. Overweight and obesity were classified using age and sex specific BMI cut-offs adopted by the International Obesity Task Force (IOTF) (Cole et al. 2000). A child was considered with overweight or obesity when the BMI was above the 85th percentile (i.e., overweight if 95th > BMI ≥ 85th percentile, corresponding to Z-scores between + 1 and < + 2 SD; obesity if BMI ≥ 95th percentile, corresponding to Z-scores ≥ + 2 SD) (de Onis et al. 2007). Within the 446 participants, 234 were classified has having normal-weight, and 212 had overweight or obesity.
Genotyping
Buccal swabs were collected from each participant and the genomic DNA was extracted using the FavorPrep™ Genomic DNA Mini Kit (Favorgen® Biotech Corp, Taiwan). Polymorphism rs4420638 was genotyped by real-time polymerase chain reaction (qPCR) using the pre-designed TaqMan SNP® Genotyping assay C__26682080_10 (Applied Biosystems, Foster City, CA) in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories, CA). The PCR amplification was carried out in 10 µl of a total reaction volume containing 1 µl (~ 10 ng) of DNA, 0.25 µl (20x) of TaqMan SNP assay in 1x of SsoFast™ Probes Supermix (Bio-Rad, Hercules, CA, USA). PCR conditions were an initial denature step at 95 °C for 10 min, followed by 40 cycles of 1 min at 62 °C and 15 s at 95 °C. To assess genotyping reproducibility, a 10% random selection of samples were re-genotyped.
Statistical Analysis
Genotype and allele frequencies were estimated by direct counting and the Hardy-Weinberg equilibrium (HWE) probability value was achieved using an exact test. Differences in the genotype distribution between cases and controls were tested using logistic regression, under the additive (AA vs. AG vs. GG) and dominant (AA vs. AG/GG) models, to determine odds ratio (OR), 95% confidence intervals (CI) and p-values. These statistical analyses were performed using the PLINK software v.1.07 (http://pngu.mgh.harvard.edu/purcell/plink/) (Purcell et al. 2007).
The association of the rs4420638 polymorphism with obesity-related quantitative traits (e.g., weight, height, BMI, BMI Z-score and waist circumference), by comparing genotypes AA against AG + GG, were tested using the nonparametric Mann-Whitney test. These statistical analyses were performed using the IBM SPSS Statistics software for Windows, version 27 (SPSS, Chicago, IL). A statistically significant association or difference was set at 5%.
Results
The characteristics of the participants are shown in Table 1 and supplementary Table S1 stratified by sex. The study included a total of 446 children (231 boys and 215 girls): 234 normal-weight and 212 with overweight (including obesity). The obtained genotypes and minor allele frequencies (MAFs) in the total population and the association results with the risk of obesity in a case-control outcome are summarized in Table 2. Minor allele frequency (MAF) for the rs4420638 SNP was 0.146 (G-allele) in the total population, and genotype frequencies were AA 72.87%, AG 25.11% and GG 2.02%. The MAF were 0.145 in boys and 0.147 in girls, and genotype frequencies were AA 73.16%, AG 24.68% and GG 2.16% for boys, and AA 72.56%, AG 25.58% and GG 1.86% for girls (supplementary Table S2). The obtained genotype frequencies were according to the Hardy-Weinberg equilibrium (p = 1) in the total population as also in boys and girls (Table 2 and supplementary Table S2).
The frequency of the minor allele was higher in normal-weight individuals in comparison with children with overweight (including obesity), in the total population (0.173 vs. 0.116) as also in boys (0.177 vs. 0.111) and girls (0.169 vs. 0.12). In according to these dissimilar frequencies, logistic regression, comparing children with normal-weight vs. children with overweight and obesity, revealed a significant association with risk of obesity, with the rs4420638 minor G-allele providing protection against obesity in both the additive (OR 0.619; 95% CI 0.421–0.913; p = 0.016) and dominant (OR 0.5875; 95% CI 0.383-0.9; p = 0.015) models (Table 2). Also, stratifying data by sex, logistic regression revealed near significant protective associations against overweight/obesity for the G-allele, both in boys (OR 0.581; 95% CI 0.321–1.052; p = 0.073) and girls (OR 0.595; 95% CI 0.322–1.099; p = 0.097) (dominant model) (supplementary Table S2).
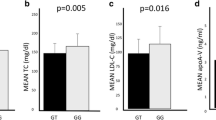
The associations between SNP rs4420638 and obesity-related anthropometric traits are detailed in Table 3. Consistently with the case-control results, children homozygous for the major allele (genotype AA) present higher BMI Z-score (mean 0.98) when compared to those carriers of the minor allele (genotypes AG + GG) (mean 0.75) (p = 0.049). Moreover, the homozygous individuals for the major allele (genotype AA) revealed significantly higher values for the anthropometric traits weight (p = 0.008), height (p = 0.023), BMI (p = 0.022), and waist circumference (p = 0.005), in comparison with carriers of the minor allele (genotypes AG + GG) (Table 3). This same pattern was observed when splitting the sample by sex, although only two significant values were observed, for weight and waist circumference in girls (supplementary Table S3).
Discussion
In this study, we examined the association of the polymorphism rs4420638 A > G, tagging the LD block harboring the APOE and APOC1 genes, with the risk of obesity using BMI case-control groups of Portuguese children (normal-weight vs. overweight, including obesity). The present study revealed a significant protective effect from the rs4420638 minor G-allele against the risk of obesity in the total population (OR 0.583; 95% CI 0.383-0.9; p = 0.015, in the dominant model). Also, near significant protective associations with obesity risk were observed both in boys (p = 0.073) and in girls (p = 0.097) (dominant model). Moreover, the association with anthropometric variables, by comparing genotype groups (AA vs. AG + GG), revealed the same protective association with lower values for the anthropometric traits of weight, BMI, BMI Z-score, and waist circumference in the carriers of G-allele in the total population. The same pattern was observed in boys and girls when splitting the whole population by sex. These results suggest no sex-specific associations with obesity risk for the rs4420638 variant, in contrast with other genetic loci where sex-dependent effects on adiposity were identified (Jacobsson et al. 2008; Al Asoom et al. 2020).
Numerous association studies have correlated SNP rs4420638 with lipid metabolism by revealing strong associations between the minor G-allele and elevated activity of Lp-PLA2 and high serum levels of TC, TG, LDL cholesterol or HDL cholesterol, thus providing evidence for a significant function in dyslipidemia (Kathiresan et al. 2008; Drenos et al. 2009; Suchindran et al. 2010; Liu et al. 2011; Grallert et al. 2012; Breitling et al. 2015; Zhang et al. 2018; Wang et al. 2022). Curiously, some studies also revealed opposite effects for this locus, showing the rs4420638 A-allele associated with these same high levels of serum lipids (Teslovich et al. 2010; Hoffmann et al. 2018; Bentley et al. 2019; Richardson et al. 2022). The GWAS Catalog EMBL-EBI (http://www.ebi.ac.uk/gwas, accessed on January 24, 2023) (Sollis et al. 2023) provides 156 studies for the variant rs4420638, showing multiple associations with serum lipid traits for both the G and A alleles.
Additionally, while the rs4420638 G-allele was identified conferring protection against obesity (OR 0.48; 95% CI 0.29–0.81; p = 0.004) in an Algerian population sample (mean age 44.0) (Boulenouar et al. 2019), a recent study in Chinese children and adolescents (6–18 years old) has not identified rs4420638 as an obesity-linked variant (Wang et al. 2022). Therefore, the observed protective association of the rs4420638 G-allele against obesity risk in Portuguese children is consistent with the study by Boulenouar et al. (2019).
For non-Mendelian diseases, significant protective associations (i.e., the major allele of an associated locus being the risk allele) can be explained by genetic drift, overdominance, frequency-dependent selection, and gene–gene or gene–environment interactions (Kido et al. 2018). However, the mechanism by which the rs4420638A/G variant influences plasma lipid levels and the risk of obesity is unclear. This variant, near the APOC1 gene, have been reported to exist in LD with APOE ε2/ε3/ε4 polymorphisms (Zhang et al. 2018), and it is unknown whether these associations are independent of the APOE alleles that are known to increase the risk of obesity (Galal et al. 2021; Zeljko et al. 2011).
In conclusion, the present study provides further evidence for the APOE/APOC1 candidate-region association with the risk of obesity, in particular in children. Findings revealed the rs4420638 SNP minor G-allele is associated with a lower risk of obesity. This was the first study to describe this significant protective association for the rs4420638 G-allele against obesity risk in childhood exclusively. As the small population sample could be considered a limitation of this study, mainly when stratifying the total population by sex, further investigations with large sample sizes are required to confirm the obtained results.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Al Asoom LI, Al Afandi DT, Al Abdulhadi AS, Rafique N, Chathoth S, Al Sunni AA (2020) Protective Association of single nucleotide polymorphisms rs1861868-FTO and rs7975232-VDR and obesity in saudi females. Int J Gen Med 13:235–241. https://doi.org/10.2147/IJGM.S251466
Albuquerque D, Nóbrega C, Manco L (2013) Association of FTO polymorphisms with obesity and obesity-related outcomes in portuguese children. PLoS ONE 8:e54370
Alfano R, Robinson O, Handakas E, Nawrot TS, Vineis P, Plusquin M (2022) Perspectives and challenges of epigenetic determinants of childhood obesity: a systematic review. Obes Rev 23 Suppl 1e13389. https://doi.org/10.1111/obr.13389
Beekman M, Blanché H, Perola M et al (2013) Genome-wide linkage analysis for human longevity: Genetics of Healthy Aging Study. Aging Cell 12:184–193. https://doi.org/10.1111/acel.12039
Bentley AR, Sung YJ, Brown MR et al (2019) Multi-ancestry genome-wide gene-smoking interaction study of 387,272 individuals identifies new loci associated with serum lipids. Nat Genet 51:636–648. https://doi.org/10.1038/s41588-019-0378-y
Boulenouar H, Mediene Benchekor S, Ouhaibi Djellouli H et al (2019) Association study of APOE gene polymorphisms with diabetes and the main cardiometabolic risk factors, in the algerian population. Egypt J Med Hum Genet 20:5. https://doi.org/10.1186/s43042-019-0013-6
Breitling C, Gross A, Büttner P, Weise S, Schleinitz D, Kiess W, Scholz M, Kovacs P, Körner A (2015) Genetic contribution of Variants near SORT1 and APOE on LDL cholesterol independent of obesity in children. PLoS ONE 10:e0138064. https://doi.org/10.1371/journal.pone.0138064
Cole TJ, Bellizzi MC, Flegal KM, Dietz WH (2000) Establishing a standard definition for child overweight and obesity worldwide: international survey. BMJ 320:1240–1243. https://doi.org/10.1136/bmj.320.7244.1240
de Onis M, Onyango AW, Borghi E, Siyam A, Nishida C, Siekmann J (2007) Development of a WHO growth reference for school-aged children and adolescents. Bull World Health Organ 85:660–667. https://doi.org/10.2471/blt.07.043497
Drenos F, Talmud PJ, Casas JP, Smeeth L, Palmen J, Humphries SE, Hingorani AD (2009) Integrated associations of genotypes with multiple blood biomarkers linked to coronary heart disease risk. Hum Mol Genet 18:2305–2316. https://doi.org/10.1093/hmg/ddp159
Galal AA, Abd Elmajeed AA, Elbaz RA, Wafa AM, Elshazli RM (2021) Association of Apolipoprotein E gene polymorphism with the risk of T2DM and obesity among egyptian subjects. Gene 769:145223. https://doi.org/10.1016/j.gene.2020.145223
Grallert H, Dupuis J, Bis JC et al (2012) Eight genetic loci associated with variation in lipoprotein-associated phospholipase A2 mass and activity and coronary heart disease: meta-analysis of genome-wide association studies from five community-based studies. Eur Heart J 33:238–251. https://doi.org/10.1093/eurheartj/ehr372
Hoffmann TJ, Theusch E, Haldar T et al (2018) A large electronic-health-record-based genome-wide study of serum lipids. Nat Genet 50:401–413. https://doi.org/10.1038/s41588-018-0064-5
Jacobsson JA, Danielsson P, Svensson V et al (2008) Major gender difference in association of FTO gene variant among severely obese children with obesity and obesity related phenotypes. Biochem Biophys Res Commun 368:476–482. https://doi.org/10.1016/j.bbrc.2008.01.087
Kathiresan S, Melander O, Guiducci C et al (2008) Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet 40:189–197. https://doi.org/10.1038/ng.75
Kido T, Sikora-Wohlfeld W, Kawashima M, Kikuchi S, Kamatani N, Patwardhan A, Chen R, Sirota M, Kodama K, Hadley D, Butte AJ (2018) Are minor alleles more likely to be risk alleles? BMC Med. Genomics 11:3. https://doi.org/10.1186/s12920-018-0322-5
Liu Y, Zhou D, Zhang Z, Song Y, Zhang D, Zhao T, Chen Z, Sun Y, Zhang D, Yang Y, Xing Q, Zhao X, Xu H, He L (2011) Effects of genetic variants on lipid parameters and dyslipidemia in a chinese population. J Lipid Res 52:354–360. https://doi.org/10.1194/jlr.P007476
Loos RJF, Yeo GSH (2022) The genetics of obesity: from discovery to biology. Nat Rev Genet 23:120–133
Lotta LA, Wittemans LBL, Zuber V et al (2018) Association of genetic variants related to Gluteofemoral vs Abdominal Fat distribution with type 2 diabetes, Coronary Disease, and Cardiovascular Risk factors. JAMA 320:2553–2563. https://doi.org/10.1001/jama.2018.19329
Manco L, Pinho S, Albuquerque D, Machado-Rodrigues AM, Padez C (2019) Physical activity and the association between the FTO rs9939609 polymorphism and obesity in portuguese children aged 3 to 11 years. Am J Hum Biol 31:e23312. https://doi.org/10.1002/ajhb.23312
Mason KE, Palla L, Pearce N, Phelan J, Cummins S (2020) Genetic risk of obesity as a modifier of associations between neighbourhood environment and body mass index: an observational study of 335 046 UK Biobank participants. BMJ Nutr Prev Health 3:247–255. https://doi.org/10.1136/bmjnph-2020-000107
Ng M, Fleming T, Robinson M et al (2014) Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the global burden of Disease Study 2013. Lancet 384:766–781. https://doi.org/10.1016/S0140-6736(14)60460-8
Purcell S, Neale B, Todd-Brown K et al (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. https://doi.org/10.1086/519795
Richardson TG, Leyden GM, Wang Q, Bell JA, Elsworth B, Davey Smith G, Holmes MV (2022) Characterising metabolomic signatures of lipid-modifying therapies through drug target mendelian randomisation. PLoS Biol 20:e3001547. https://doi.org/10.1371/journal.pbio.3001547
Sollis E, Mosaku A, Abid A et al (2023) The NHGRI-EBI GWAS catalog: knowledgebase and deposition resource. Nucleic Acids Res 51(D1):D977–D985. https://doi.org/10.1093/nar/gkac1010
Suchindran S, Rivedal D, Guyton JR et al (2010) Genome-wide association study of Lp-PLA(2) activity and mass in the Framingham Heart Study. PLoS Genet 6:e1000928. https://doi.org/10.1371/journal.pgen.1000928
Teslovich TM, Musunuru K, Smith AV et al (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466:707–713. https://doi.org/10.1038/nature09270
Vilchis-Gil J, Galván-Portillo M, Klünder-Klünder M, Cruz M, Flores-Huerta S (2015) Food habits, physical activities and sedentary lifestyles of eutrophic and obese school children: a case-control study. BMC Public Health 15:124. https://doi.org/10.1186/s12889-015-1491-1
Wang H, Wang Y, Song JY et al (2022) Associations of genetic variants of lysophosphatidylcholine metabolic enzymes with levels of serum lipids. Pediatr Res 91:1595–1599. https://doi.org/10.1038/s41390-021-01549-9
Winkler TW, Justice AE, Graff M et al (2015) The influence of Age and Sex on Genetic Associations with adult body size and shape: a large-scale genome-wide Interaction Study. PLoS Genet 11:e1005378. https://doi.org/10.1371/journal.pgen.1005378
Zeljko HM, Škarić-Jurić T, Narančić NS et al (2011) E2 allele of the apolipoprotein E gene polymorphism is predictive for obesity status in Roma minority population of Croatia. Lipids Health Dis 10:9. https://doi.org/10.1186/1476-511X-10-9
Zhang R, Liu Q, Liu H et al (2018) Effects of apoC1 genotypes on the hormonal levels, metabolic profile and PAF-AH activity in chinese women with polycystic ovary syndrome. Lipids Health Dis 17:77. https://doi.org/10.1186/s12944-018-0725-5
Acknowledgements
The authors would like to thank children and their parents for participation in the study. We would also like to thank the staff in the schools.
Funding
This work was supported by Fundação para a Ciência e a Tecnologia (FCT), co-financed by COMPETE 2020 and Portugal 2020 (Grant Number: PTDC/DTP-SAP/1520/2014) and the institutional FCT Grant Number: UIDB/00283/2020).
Open access funding provided by FCT|FCCN (b-on).
Author information
Authors and Affiliations
Contributions
LM: conceptualization, investigation, statistical analysis, original draft preparation. DA, DR, AM-R: collected samples and data, statistical analysis, reviewed the manuscript. CP: Funding acquisition, reviewed the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Direcção Geral de Inovação e de Desenvolvimento Curricular of the Ministry of Education – Portugal, under the Grant Number PTDC/DTP-SAP/1520/2014.
Consent to Participate
The informed consent was required for the present study and all the informed consent were obtained from children’s parents or tutors.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Manco, L., Albuquerque, D., Rodrigues, D. et al. Protective Association of APOC1/rs4420638 with Risk of Obesity: A case-control Study in Portuguese Children. Biochem Genet 62, 254–263 (2024). https://doi.org/10.1007/s10528-023-10427-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-023-10427-4