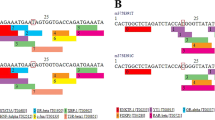
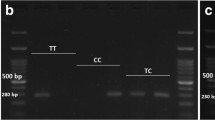
Abstract
Bangladesh has the second largest number of adults with diabetes in South Asia. Compelling evidence suggest that miRNAs contribute to the etiology of Type 2 diabetes mellitus (T2DM) by regulating many aspects of glucose homeostasis. Hence, we hypothesized that genetic polymorphisms in the diabetes-related miRNA target-binding sites could be associated with the risk of T2DM in Bangladesh. The reference Single nucleotide polymorphism (SNP) data from the Insulin Receptor (INSR) gene were downloaded from the ENSEMBL genome browser release 88 and further analyzed in silico for identifying SNPs with deleterious effect and clinical relationships. Further, case–control study using the microRNA-binding site polymorphism rs1366600 (T > C) located at the 3′ UTR of the INSR gene was carried out in 217 T2DM patients and 237 healthy controls from Bangladesh. Genotyping was performed using the real time PCR based allele discrimination method. The results showed that the minor allele ‘C’ is associated with increased risk of T2DM [Odds ratio (OR) 1.87; 95% confidence intervals (CI) 1.28–2.74; P = 0.0010]. When we dissected our analysis to include the dominant model (CC + TC genotype against the TT genotype), we found that the CC and TC genotypes were associated with increased risk of T2DM in Bangladeshi population (OR 2.01; 95% CI 1.31–3.07; P = 0.0012). However, in recessive model (CC vs TT + TC); the effect was not statistically significant (OR 2.23; 95% CI 0.66–7.51; P = 0.1848). Stratification of our data based on the gender of the cases and controls showed similar degree of risk association with respect to different genotypes and alleles. Our study showed that the miRNA binding site polymorphism rs1366600 located at the 3′-UTR region of the INSR gene is associated with increased risk of T2DM in Bangladeshi individuals.

Similar content being viewed by others
References
Afruza R, Islam LN, Banerjee S, Hassan MM, Suzuki F, Nabi AN (2014) Renin gene polymorphisms in bangladeshi hypertensive population. J Genomics 2:45–53
Aken BL, Ayling S, Barrell D, Clarke L, Curwen V, Fairley S, Fernandez Banet J, Billis K, García Girón C, Hourlier T, Howe K, Kähäri A, Kokocinski F, Martin FJ, Murphy DN, Nag R, Ruffier M, Schuster M, Tang YA, Vogel J-H, White S, Zadissa A, Flicek P, Searle SMJ (2016) The Ensembl gene annotation system. Database, baw093
Akter S, Rahman MM, Abe SK, Sultana P (2014) Prevalence of diabetes and prediabetes and their risk factors among Bangladeshi adults: a nationwide survey. Bull World Health Organ 92(204–13):213a
Bhattacharya A, Ziebarth JD, Cui Y (2014) PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res 42:D86–D91
Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM, Snyder M (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 22:1790–1797
Brennecke J, Stark A, Russell RB, Cohen SM (2005) Principles of microRNA-target recognition. PLoS Biol 3:e85
Brodersen P, Voinnet O (2009) Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol 10:141–148
Chen K, Song F, Calin GA, Wei Q, Hao X, Zhang W (2008) Polymorphisms in microRNA targets: a gold mine for molecular epidemiology. Carcinogenesis 29:1306–1311
Chen B, Wilkening S, Drechsel M, Hemminki K (2009) SNP_tools: a compact tool package for analysis and conversion of genotype data for MS-Excel. BMC Res Notes 2:214
Chowdhury MA, Uddin MJ, Khan HM, Haque MR (2015) Type 2 diabetes and its correlates among adults in Bangladesh: a population based study. BMC Public Health 15:1070
De Silva NM, Frayling TM (2010) Novel biological insights emerging from genetic studies of type 2 diabetes and related metabolic traits. Curr Opin Lipidol 21:44–50
Elek Z, Nemeth N, Nagy G, Nemeth H, Somogyi A, Hosszufalusi N, Sasvari-Szekely M, Ronai Z (2015) Micro-RNA binding site polymorphisms in the WFS1 gene are risk factors of diabetes mellitus. PLoS ONE 10:e0139519
Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, Asplund A, Sjostedt E, Lundberg E, Szigyarto CA, Skogs M, Takanen JO, Berling H, Tegel H, Mulder J, Nilsson P, Schwenk JM, Lindskog C, Danielsson F, Mardinoglu A, Sivertsson A, von Feilitzen K, Forsberg M, Zwahlen M, Olsson I, Navani S, Huss M, Nielsen J, Ponten F, Uhlen M (2014) Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics 13:397–406
Goda N, Murase H, Kasezawa N, Goda T, Yamakawa-Kobayashi K (2015a) Polymorphism in microRNA-binding site in HNF1B influences the susceptibility of type 2 diabetes mellitus: a population based case-control study. BMC Med Genet 16:75
Goda N, Murase H, Kasezawa N, Goda T, Yamakawa-Kobayashi K (2015b) Polymorphism in microRNA-binding site in HNF1B influences the susceptibility of type 2 diabetes mellitus: a population based case–control study. BMC Med Genet 16:75
Gong W, Xiao D, Ming G, Yin J, Zhou H, Liu Z (2014) Type 2 diabetes mellitus-related genetic polymorphisms in microRNAs and microRNA target sites. J Diabetes 6:279–289
Grarup N, Sandholt CH, Hansen T, Pedersen O (2014) Genetic susceptibility to type 2 diabetes and obesity: from genome-wide association studies to rare variants and beyond. Diabetologia 57:1528–1541
Guariguata L, Whiting DR, Hambleton I, Beagley J, Linnenkamp U, Shaw JE (2014) Global estimates of diabetes prevalence for 2013 and projections for 2035. Diabetes Res Clin Pract 103:137–149
He Y, Ding Y, Liang B, Lin J, Kim TK, Yu H, Hang H, Wang K (2017) A systematic study of dysregulated microRNA in type 2 diabetes mellitus. Int J Mol Sci 18:456
Kahn SE, Cooper ME, Del Prato S (2014) Pathophysiology and treatment of type 2 diabetes: perspectives on the past, present, and future. Lancet 383:1068–1083
Keen JC, Moore HM (2015) The genotype-tissue expression (GTEx) project: linking clinical data with molecular analysis to advance personalized medicine. J Pers Med 5:22–29
Lyssenko V, Groop L (2009) Genome-wide association study for type 2 diabetes: clinical applications. Curr Opin Lipidol 20:87–91
Moszyńska A, Gebert M, Collawn JF, Bartoszewski R (2017) SNPs in microRNA target sites and their potential role in human disease. Open Biology 7:170019
Pradeepa R, Mohan V (2017) Prevalence of type 2 diabetes and its complications in India and economic costs to the nation. Eur J Clin Nutr 71:816–824
Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29:308–311
Skol AD, Scott LJ, Abecasis GR, Boehnke M (2006) Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet 38:209
The Genomes Project, C. (2015) A global reference for human genetic variation. Nature 526:68
Wang X, Li W, Ma L, Ping F, Liu J, Wu X, Mao J, Wang X, Nie M (2017) Investigation of miRNA-binding site variants and risk of gestational diabetes mellitus in Chinese pregnant women. Acta Diabetol 54:309–316
Whiting DR, Guariguata L, Weil C, Shaw J (2011) IDF diabetes atlas: global estimates of the prevalence of diabetes for 2011 and 2030. Diabetes Res Clin Pract 94:311–321
Wu B, Miller D (2017) Involvement of microRNAs in diabetes and its complications. Methods Mol Biol 1617:225–239
Wu Y, Ding Y, Tanaka Y, Zhang W (2014) Risk factors contributing to type 2 diabetes and recent advances in the treatment and prevention. Int J Med Sci 11:1185–1200
Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, Gil L, Gordon L, Haggerty L, Haskell E, Hourlier T, Izuogu OG, Janacek SH, Juettemann T, To JK, Laird MR, Lavidas I, Liu Z, Loveland JE, Maurel T, McLaren W, Moore B, Mudge J, Murphy DN, Newman V, Nuhn M, Ogeh D, Ong CK, Parker A, Patricio M, Riat HS, Schuilenburg H, Sheppard D, Sparrow H, Taylor K, Thormann A, Vullo A, Walts B, Zadissa A, Frankish A, Hunt SE, Kostadima M, Langridge N, Martin FJ, Muffato M, Perry E, Ruffier M, Staines DM, Trevanion SJ, Aken BL, Cunningham F, Yates A, Flicek P (2018) Ensembl 2018. Nucleic Acids Res 46:D754–D761
Zhao X, Ye Q, Xu K, Cheng J, Gao Y, Li Q, Du J, Shi H, Zhou L (2013) Single-nucleotide polymorphisms inside microRNA target sites influence the susceptibility to type 2 diabetes. J Hum Genet 58:135–141
Zhou X, Yang PC (2012) MicroRNA: a small molecule with a big biological impact. Microrna 1:1
Acknowledgement
We would like to acknowledge the generosity of the individuals who participated in this study. We would also like to thank Professor Dr Haseena Khan, Department of Biochemistry and Molecular Biology, University of Dhaka for providing us the facilities for performing RT-PCR based allelic discrimination assay.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no conflict of interest.
Additional information
The original version of this article was revised: The co-author name should be Farhana Jahan instead of Farhan Jahan. This has been corrected in this version.
Rights and permissions
About this article
Cite this article
Parvin, M., Jahan, F., Sarkar, P.K. et al. Functional Polymorphism Located in the microRNA Binding Site of the Insulin Receptor (INSR) Gene Confers Risk for Type 2 Diabetes Mellitus in the Bangladeshi Population. Biochem Genet 57, 20–33 (2019). https://doi.org/10.1007/s10528-018-9872-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-018-9872-7