Abstract
A genetic analysis of unexplained mild-moderate intellectual disability and co-morbid psychiatric or behavioural disorders is not systematically conducted in adults. A cohort of 100 adult patients affected by both phenotypes were analysed in order to identify the presence of copy number variants (CNVs) responsible for their condition identifying a yield of 12.8% of pathogenic CNVs (19% when including clinically recognizable microdeletion syndromes). Moreover, there is a detailed clinical description of an additional 11% of the patients harbouring possible pathogenic CNVs—including a 7q31 deletion (IMMP2L) in two unrelated patients and duplications in 3q29, 9p24.2p24.1 and 15q14q15.1—providing new evidence of its contribution to the phenotype. This study adds further proof of including chromosomal microarray analysis (CMA) as a mandatory test to improve the diagnosis in the adult patients in psychiatric services.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Intellectual disability (ID) is a complex and multifactorial disorder that includes both intellectual and adaptive functioning deficits in the conceptual, social and practical domains with onset during the developmental period. This disorder affects approximately 1–3% of the general population, and between 10 and 40% of people with ID also present with mental illness or behavioural disorders (Cooper et al. 2007; Lowe et al. 2007; Morgan et al. 2008). The diagnostic categories of these mental disorders are based on the symptoms (Stein et al. 2013), but there is considerable clinical heterogeneity and overlap with different psychiatric categories (Burmeister et al. 2008). Indeed, the boundaries of the diagnostic categories can be blurred when the patients’ symptoms are not clearly expressed. The diagnosis of a psychiatric disorder in subjects with ID can be difficult, and most symptoms tend to be attributed to the ID. For this reason, the co-occurrence of both entities is usually overlooked (Costello and Bouras 2006).
Copy number variants (CNVs) are a source of human genetic variation and have been described as an important genomic cause of human disease (Iafrate et al. 2004; Sebat et al. 2004). Screening of ID patient cohorts via chromosomal microarray analysis (CMA) has led to the characterization of new syndromes, such as 8q21.11 deletion syndrome (OMIM: 614230) and 19p13.3 microdeletion/microduplication syndrome (Dolan et al. 2010; Orellana et al. 2015). Additionally, there is evidence that CNVs can predispose individuals to the development of psychiatric disorders, such as the autism spectrum disorders (ASDs) (Marshall et al. 2008; Hedges et al. 2012), schizophrenia (SQZ) (Kirov et al. 2012; Xu et al. 2008), bipolar disorder (Green et al. 2015) and attention-deficit/hyperactive disorder (ADHD) (Jarick et al. 2014; Ramos-Quiroga et al. 2014). Numerous CNV loci have been recurrently observed across ID and various neuropsychiatric phenotypes, such as the16p11.2 and NRXN1 deletions, both of which are associated with ID, SQZ and ASD. These findings suggest that ID and psychiatric disorders may share genetic susceptibility factors (Guilmatre et al. 2009).
A large proportion of the adult population affected by ID lacks a genetic diagnosis. Some of these adult patients have never received a diagnostic assessment; alternatively, in some cases the assessment is completed without finding an explanation for the ID possibly due to the use of less advanced technologies than are currently available. At present, there is little knowledge of the genetics of ID and co-morbid psychiatric disorder in adults. Nevertheless, CMA and whole exome sequencing could shed light on the genetic diagnoses in adults with idiopathic ID (Baker et al. 2012; Posey et al. 2016; Taylor et al. 2010; Wolfe et al. 2016). Here, we report the genetic analysis of 100 adult patients affected by ID and psychiatric and/or behavioural disorders. The main purpose of this study is to investigate the contribution of putative pathogenic CNVs among patients with ID and comorbid psychiatric/behavioural disorders.
Materials and methods
Participants
This study was designed prospectively. Cognitive, psychiatric and behavioural evaluation was performed by psychiatric specialists at the Mental Health ID Service (“Parc Hospitalari Martí i Julià”, Girona, Catalonia, Spain) while clinical-dysmorphic evaluation and genetic assessment was performed by a clinical geneticist at the Clinical Genetics Department (“Parc Taulí Hospital Universitari”, Sabadell, Catalonia, Spain). This study was approved by the institutional ethics committee (CEIC 2009/582). A legal guardian or family member that legally represented the participant signed the informed consent form. Adult patients over the age of 18 years were consecutively recruited using the following inclusion criteria: mild (IQ = 75 − 50) or moderate (IQ = 50 − 35) ID according to the Diagnostic and Statistical Manual of Mental Disorders (DSM-5) criteria and a defined psychiatric disorder or behavioural disorder according to the measures listed in Table 1. The exclusion criteria were having severe ID or sensory impairment that precluded a proper examination, having suffered alterations in the central nervous system unrelated to the ID (i.e., head injury, stroke or brain tumours), the presence of untreated diseases with associated cognitive deficits (i.e., hypothyroidism, vitamin B12 deficiency or diabetes mellitus) and substance abuse. This recruitment led to 100 eligible patients for the analysis, including five sibling pairs and a sibling trio.
Clinical evaluation: cognitive, behavioural, psychiatric and dysmorphic measures
Different tests were administered to all participants to identify the presence of ID and establish its severity level, as well as to identify the presence of a psychiatric and/or behavioural disorder (Table 1). The presence of a behavioural disorder not necessarily related to a mental disorder was defined according to (Emerson 1995) as “culturally abnormal behaviour of such intensity, frequency or duration that the physical safety of the person or others is placed in serious jeopardy, or behaviour which is likely to seriously limit or deny them access to ordinary community facilities”. A family history of ID, psychiatric or behavioural disorders was also recorded.
Dysmorphic features were classified into five categories as follows: craniofacial, limbs, cutaneous, genital and body (all other dysmorphisms). A category was considered dysmorphic if at least one feature was abnormal.
Genetic analysis
The cohort was first analysed by G-banded karyotyping to determine the presence of unbalanced and balanced rearrangements. FMR1 screening and other specific molecular technologies were applied to subjects who were clinically suspected of having a syndrome.
The CMA analysis was performed with the 400K Agilent platform (Agilent Technologies, Santa Cruz, CA, USA) on all patients without a clinically recognized syndrome (including subjects known to possess a chromosomal rearrangement). This oligonucleotide-based comparative genomic hybridization array covered the entire genome with an average resolution of 5.3 kb. The microarrays were processed according to the manufacturer’s specifications, and the Agilent Workbench 5.0, Feature Extraction and Cytogenomics softwares (Agilent Technologies, Santa Cruz, CA, USA) were used to render the image analysis with the manufacturer’s recommended settings and human genome assembly hg19. We called CNVs when there were at least five consecutive probes with a minimum log2 ratio of ± 0.25.This low rate is capable to detect mosaicisms and using five consecutive probes avoid false positives.
The identified CNVs were cross-referenced with the Database of Genomic Variants (DGV, http://projects.tcag.ca/variation); those variants completely overlapped with common CNVs (prevalence > 1% in the general population) were excluded from further analysis. All rare CNVs (prevalence < 1% in the general population) were interpreted individually by comparing each genomic region to information available in public databases [University of California, Santa Cruz Genome Browser (http://genome.ucsc.edu), National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), Ensembl (http://www.ensembl.org/index.html), Decipher (https://decipher.sanger.ac.uk/), Clinical Genome Resource (https://www.clinicalgenome.org)] and Online Mendelian Inheritance in Man database (https://www.omim.org/) as well as literature, and classified into four categories as follows: (1) Pathogenic CNVs (pCNV), which overlap with known causative findings previously associated with ID or psychiatric disorders (from databases and literature). (2) Variants of unknown significance (VOUS) that were likely pathogenic (pVOUS) when at least two of the following conditions are met: (a) Partially overlap with a pathogenic susceptibility locus; (b) It is not reported in control population from (Coe et al. 2014); (c) Include genes enriched for deletions/duplications at nominal level of significance according to (Coe et al. 2014); (d) Include developmental delay (DD) genes from (Deciphering Developmental Disorders Study 2017); (e) Include genes with relevant function in the nervous system. (3) VOUS that were likely benign (bVOUS), which included only intronic regions of genes with a function in the nervous system not yet described in the patients or CNVs that included genes with unknown functions or functions not related to the central nervous system. (4) Benign CNVs (bCNVs), which were without genes or devoid of known regulatory elements. We focused on pCNVs and pVOUSs, both of which are likely associated with the affected phenotype. Customized multiplex ligation-dependent probe amplification (MLPA) and fluorescent in situ hybridization (FISH) were performed according to standard protocols to validate and determine the inheritance of CNVs. Custom MLPA probes were designed according to protocols and guidelines from MRC-Holland (Amsterdam, the Netherlands) and the ProSeek web server created by Estivill et al. (Pantano et al. 2008), and specific bacterial artificial chromosome clones were selected for the aberration regions.
Finally, since the cohort of 100 patients results from a 50-patient set which was subsequently increased with a second 50-patient set, we selected seven CNVs (pCNVs and pVOUS) identified in the first patient-set analysed by CMA to evaluate their recurrence. Two pCNVs associated with ID and psychiatric disorders (2p16.3 and 12p12.1) and five pVOUSs (2p12, 3q29, 15q14q15, 15q26.2 and 17q24) were analysed in a new set of 161 adult patients affected by mild/moderate ID and 189 controls using a custom MLPA.
Data analysis
The potential associations between categorical variables were tested using the χ2 test. When one or more of the expected values for the χ2 computation was lower than 5, the p value was computed using Fisher’s exact test. When a result was significant, the odds ratio was indicated as a measure of the effect size. The Kruskal–Wallis and Mann–Whitney U tests were performed for dependent continuous variables that showed non-normal distributions (as determined by the Shapiro–Wilk test and visual inspection). A threshold of p < 0.05 was set to indicate statistical significance, and the Bonferroni correction was applied for post hoc comparisons. The statistical analyses were performed using the Statistical Package for Social Sciences (SPSS for Windows, Version 16.0., SPSS Inc., Chicago, IL, USA).
Results
Description of the patient cohort
A patient cohort of 100 adults affected by ID and co-morbid psychiatric/behavioural disorders without a genetic diagnosis was recruited with the main purpose of identifying CNVs responsible for their conditions. The cohort comprised 50 men and 50 women with an average age of 31.28 years (18–56 years, SD = 10.14), of whom 60% had mild ID and 40% had moderate ID. Out of the 100 patients, 50 had both a psychiatric and a behavioural disorder, 37 had only a psychiatric disorder and 13 had only a behavioural disorder. Sixteen patients had a diagnosis of two different psychiatric disorders, nine of whom also presented with a behavioural disorder. Table 2 shows the distribution of the psychiatric disorders in our cohort according to ID severity level and the presence or absence of behavioural disorders. The χ2 test did not show a significant difference in the presence of psychiatric or behavioural disorders between the mild and moderate ID groups. Mild dysmorphic features were present in all patients and were identified via minor facial or cranial dysmorphologies (98%) and abnormalities in the limbs (44%), cutaneous tissue (52), genitals (16%) and other (60%).
Genetic analysis of the patient cohort
A preliminary karyotype identified four rearrangements and the specific molecular technologies confirmed the presence of a clinically recognized syndromes in fourteen individuals (Table 3). The CMA performed in the 86 patients with no clinically recognized syndromes identified a total of 216 rare CNVs (additional file 1) with an average of 2.5 CNVs/patient and range of 0–8 CNVs/patient. According to the classification criteria, 13 pCNVs were the genetic cause of the phenotype and 11 pVOUSs were the putative cause of the phenotype (additional file 2) while 192 CNVs (88.9%) were non-pathogenic (139bVOUSs and 53 bCNVs). The 13pCNVs, nine deletions and four duplications, were identified in 11 of the 86 patients (12.8%) (Table 4), given that two patients presented two CNVs—in one case the 2 pCNVs arose from a maternal inversion (patient 10) and in the other case the 2 pCNVs derived from an unbalanced translocation (patient 26) according to the FISH performed afterwards. The 11pVOUS, five deletions and six duplications, were identified in 11 of the 86 patients (12.8%) (Table 4), but if we consider only one patient of each sibling set (given that we include four set of siblings in the CMA population), pVOUS are the putative cause of disease in nine of 82 patients (11%). The analysis of parental samples (when available) revealed that the pCNVs were de novo in seven patients and maternally inherited in two cases (one X-linked). In contrast, of the eight cases with pVOUSs with available parental samples, there were no de novo pVOUSs (Table 4).
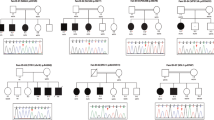
Two shared CNV regions were present in unrelated patients. The first CNV region was the pathogenic 2p16.3 deletion in patients 55 and 94, which partially included the NRXN1 gene (Table 4). The shared phenotype between these patients and the neuropsychological evaluation of deletion family carriers was previously reported (Vinas-Jornet et al. 2014). The second shared CNV was a 7q31.1 deletion that disrupted the IMMP2L gene, which encodes a catalytic subunit of the mitochondrial inner membrane peptidase (IMP) complex. This CNV was identified in two males (patients 32 and 151 from Table 4) affected by moderate ID and psychiatric disorders [a post-traumatic stress disorder in one patient and obsessive–compulsive disorder (OCD) with childhood autism in the other patient]. The deletion was maternally inherited in these two unrelated patients, and both patients had a registered familial history: patient 32’s mother was diagnosed with early Alzheimer’s disease and patient 151’s maternal aunt was diagnosed with a psychiatric disorder.
Family studies may help to understand the pathogenicity of CNVs and delineate genotype-phenotype correlations. Of the five sibling pairs included in the cohort, we identified a putative genetic cause that was shared between siblings in two pairs. A 9p24.2p24.1 duplication was identified in two brothers affected by moderate ID and behavioural disorders (patients 122 and 123), but a generalized anxiety disorder was diagnosed in only one patient (Table 4). This duplication overlaps two duplications described in DECIPHER in patients affected by cognitive and behavioural disorders (295,026 and 254,714, respectively). The second sibling pair was a female and her brother (patients 59 and 60) who were both affected by moderate ID, hyperkinetic conduct disorder and minor facial/cranial dysmorphology; the siblings shared a 2.7 Mb duplication in 15q14q15.1 (Table 4). There are overlapping duplications in public databases (ClinVar) with unknown clinical significance in patients with global developmental delay (nssv580863 and nssv1609978), the first of whom also presented with microcephaly and upslanted palpebral fissures.
Finally, a homozygous 3q29 duplication was identified in patient 34, who was affected by mild ID, post-traumatic stress disorder and behavioural disorders (Table 4). The patient was the third child of a consanguineous couple; both parents had borderline IQs and a heterozygous 3q29 duplication. The patient had a younger brother with a severe ID and ASD phenotype who also presented with the duplication in homozygosity.
Effect of CNVs on dysmorphic and neurodevelopmental traits
Demographic and clinical variables (gender, ID severity level, dysmorphology, psychiatric disorders, behavioural disorders and psychiatric co-morbidity) from the group of patients with an identified putative genetic cause (pCNV and pVOUS) were compared to the patients with an unknown possibly genetic cause (bVOUS, bCNVs and absence of rare CNVs). The comparison of the number of dysmorphic features, the ID severity level, psychiatric disorders or behavioural disorders between the two groups did not show any significant difference. Interestingly, the odds of having two psychiatric disorders diagnosed in the same patient were 4.22 times higher in the genetic cause group than in the unknown possible genetic cause group (95% CI 1.21–14.74, χ2 (1) = 5.56, p = 0.035).
Analysis of specific CNVs in an additional cohort
In order to evaluate the recurrence of seven pCNVs/pVOUS, an independent population of 161 patients affected by mild/moderate ID and 189 controls were analysed. None of the selected seven CNVs were detected either in the patients or the control individuals indicating a very low frequency of these pCNVs/pVOUS.
Discussion
A genetic cause of the ID and psychiatric phenotypes was identified in 25 patients of our adult cohort. This incidence is due to the diagnosis of clinically recognized syndromes such as fragile X, Velocardiofacial, Prader Willi, Smith Magenis and Williams not recognised at the adult psychiatric service. The application of CMA test in those patients without a recognised syndrome allows the genetic diagnosis in 12.8% in agreement to a similar adult population affected by ID and co-morbid psychiatric disorders (Wolfe et al. 2016). This rate would have increased to 19% if the CMA had been performed in all patients being comparable to an adult population with ID and mild-severe congenital malformation anomalies (Ho et al. 2016).
Interestingly, in our series we found a NRXN1 deletion in two cases responsible for bipolar disorder, persistent delusional disorders and behavioural phenotype (Vinas-Jornet et al. 2014) in keeping with (Lowther et al. 2017).
An additional 11% of the patients present pVOUS that may contribute to the phenotype despite there not being strong evidence for their pathogenicity. Given the low frequency of each individual CNV it is important to report them to increase the knowledge and clarify their possible association with the phenotype.
The 7q31 deletion identified in two unrelated patients (Table 4) disrupted the IMMP2L gene (NM_001244606). Although deletions in this region are reported as benign loss in ISCA database and are identified in control population, they are considered rare CNVs because their frequency is lower than 1% considering the DGV Gold Standard Variants (additional file 3). The 7q31 deletion was considered a risk factor in several neuropsychiatric disorders, including ASD (Maestrini et al. 2010; Pagnamenta et al. 2010; Casey et al. 2012), ADHD (Elia et al. 2010) and language disorder (Lai et al. 2001) and partial deletions of the IMMP2L gene in particular has been described as risk factors for neurological diseases with an incomplete penetrance (Gimelli et al. 2014). The history of Alzheimer’s disease in the carrier mother is interesting, particularly because the IMMP2L gene encodes a mitochondrial protein that regulates the levels of reactive oxygen species (George et al. 2011), and has been implicated in Alzheimer’s disease susceptibility (Swaminathan et al. 2012). This evidence suggests that IMMP2L may contribute to the ID and psychiatric disorders in these patients.
Little is known about the clinical effects of duplications in 3q29, 9p24.2p24.1 and 15q14q15.1 in contrast to the deletions in these regions that have been previously associated with neurodevelopmental disorders (Myles-Worsley et al. 2013; Bianchi et al. 2014; Spencer et al. 2011; Willatt et al. 2005). However, patients presented here suggest that these duplications could be pathogenic. The 3q29 duplication not only could disrupt the PAK2 gene, which codifies a serine/threonine protein kinase involved in the dendritic development of early cortical neurons, but also includes the FBXO45 gene. This gene, which is a component of an E3 ubiquitin ligase complex, is evolutionarily conserved and selectively expressed in the nervous system, plays an important role in the regulation of neurotransmission (Tada et al. 2010) and has been described as a candidate gene for SQZ (Wang et al. 2014). This CNV partially overlaps the 3.5 Mb critical region in 3q29 present in five members of a family affected by ID and microcephaly (Lisi et al. 2008) and spans some smaller duplications described in patients affected by ID and a wide range of minor dysmorphic features (Ballif et al. 2008). In our case, phenotypic severity correlated with the copy number of the 3q29 region in the proband, who harbours four copies of the 3q29 material and was affected by ID and a psychiatric disorder, and the parents, both of whom harbour three copies of 3q29 and had borderline IQs. A second putative pathogenic duplication identified in our cohort is located in 9p24.2p24.1 and includes the SLC1A1 gene. This gene encodes a member of the high-affinity glutamate transporters, which are crucial for the termination of the postsynaptic action of the neurotransmitter glutamate and maintenance of extracellular glutamate concentrations below the neurotoxic levels. Changes in its expression are associated with neuropsychiatric diseases, such as OCD and SQZ (Porton et al. 2013; Bauer et al. 2008), and overexpression of SLC1A1 has been demonstrated to increase the expression level of the two glial members of the glutamate transporter family (SLC1A2 and SLC1A3), which are associated with SQZ (Afshari et al. 2015). Finally, although pathogenicity of the 15q14q15.1 duplication has not been demonstrated, this duplication includes three genes (SPRED1, RASGRP1 and PAK6) that have been previously related to neuropsychiatric diseases (Brems et al. 2007; Denayer et al. 2008; Kato et al. 2011; Furnari et al. 2013). The presence of SPRED1 is particularly interesting given that deletions and point mutations in this gene are responsible for Legius syndrome, which is a genetic skin pigmentation disorder that is sometimes accompanied by other common manifestations, including moderate ID, ADHD, hypotelorism and pectus excavatum; these symptoms were present in the two patients with the 15q14q15.1 duplication. This evidence suggests that the SPRED1 gene may be responsible for the ID and neuropsychiatric disorders in our patients and that increased dosage in this region is capable of yielding a similar phenotype as decreased dosage.
Of the 13pCNVs and 11 pVOUSs, ten genes (NRXN1, IMMP2L, MSRA, SLC1A1, SOX5, UBE3A, CHRNA7, SPRED1, PRKCA, and SHANK3) have each been associated with more than one psychiatric phenotype (Table 5) and neurodevelopmental disorders based on the hypothesis that perturbation of the same molecular pathway can result in different psychiatric diagnoses (Plummer et al. 2016); for instance, SHANK3 and SLC1A1 participate in the glutamatergic pathway and UBE3A and FBXO45 in the ubiquitin pathway (Javitt 2007, Tebartz van Elst et al. 2014; Glessner et al. 2009; Plummer et al. 2016). Other genes involved in synaptic formation and function may contribute to behaviour impairments and a brain malfunction (Mehregan et al. 2016). Interestingly, we found that the presence of two psychiatric disorders increases the likelihood of detecting a pathogenic or possibly pathogenic CNV supporting the fact that different psychiatric disorders share common genetic aetiologies (Moreno-De-Luca et al. 2013).
Our cohort has been clinically examined in great detail for psychiatric and behavioural disorders as well as a dysmorphological evaluation was performed by a clinical geneticist. Almost all patients in our cohort present mild cranial or facial dysmorphic features suggesting that having multiple mild dysmorphic features may be a clue to an underlying genetic cause, despite specific comparison was not possible. We suggest that adults with mild or moderate ID, psychiatric/behavioural disorders and mild dysmorphic signs are an especially CNV enriched group as shown in the present study.
We highlight there is a high familial burden of ID and neuropsychiatric disorders in all individuals with an inherited genetic cause mainly gathered in the pVOUS group. Inherited variants must be taken into account because they can act as susceptibility factors having an additive or synergistic effect (Pinto et al. 2010; Girirajan and Eichler 2010). The identification of a familial history in individuals with ID and neuropsychiatric disorders is challenging due to the continuous spectrum of the phenotype that could explain the discrepancy between family members. Therefore, pVOUS should be considered in larger studies to reinforce their pathogenicity for ID and co-morbid psychiatric disorders.
The data provided here from an adult cohort with mild-moderate ID and co-morbid psychiatric and behavioural disorders is essential to advance our knowledge of these pathologies and useful for genotype-phenotype correlations as well as contribute to the prognosis of the behavioural phenotype in children and adolescents with the same diagnoses. Most behaviours and organic/mental health problems are easier to work with and to understand when an aetiological diagnosis is delivered, which enables the planning of better medical intervention strategies. Furthermore, having a genetic diagnosis provides relevant information for families in terms of genetic counselling, allows improved care of all family members and provides an early diagnosis of related diseases, which is a significant issue to take into account when governments and authorities plan local and national health strategies. We propose that CMA testing together with a clinical genetics assessment would help to achieve more aetiological diagnoses in adult patients with ID and psychiatric disorders.
References
Afshari P, Myles-Worsley M, Cohen OS, Tiobech J, Faraone SV, Byerley W, Middleton FA (2015) Characterization of a novel mutation in SLC1A1 associated with schizophrenia. Mol Neuropsychiatr 1:125–144
Bacchelli E, Ceroni F, Pinto D, Lomartire S, Giannandrea M, D’Adamo P, Bonora E, Parchi P, Tancredi R, Battaglia A, Maestrini E (2014) A CTNNA3 compound heterozygous deletion implicates a role for alphaT-catenin in susceptibility to autism spectrum disorder. J Neurodev Disord 6(1):17
Baker K, Raymond FL, Bass N (2012) Genetic investigation for adults with intellectual disability: opportunities and challenges. Curr Opin Neurol 25:150–158
Ballif BC, Theisen A, Coppinger J, Gowans GC, Hersh JH, Madan-Khetarpal S, Schmidt KR, Tervo R, Escobar LF, Friedrich CA, Mcdonald M, Campbell L, Ming JE, Zackai EH, Bejjani BA, Shaffer LG (2008) Expanding the clinical phenotype of the 3q29 microdeletion syndrome and characterization of the reciprocal microduplication. Mol Cytogenet 1:8
Bauer D, Gupta D, Harotunian V, Meador-Woodruff JH, Mccullumsmith RE (2008) Abnormal expression of glutamate transporter and transporter interacting molecules in prefrontal cortex in elderly patients with schizophrenia. Schizophr Res 104:108–120
Bianchi MG, Bardelli D, Chiu M, Bussolati O (2014) Changes in the expression of the glutamate transporter EAAT3/EAAC1 in health and disease. Cell Mol Life Sci 71:2001–2015
Brems H, Chmara M, Sahbatou M, Denayer E, Taniguchi K, Kato R, Somers R, Messiaen L, de Schepper S, Fryns JP, Cools J, Marynen P, Thomas G, Yoshimura A, Legius E (2007) Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat Genet 39:1120–1126
Burmeister M, Mcinnis MG, Zollner S (2008) Psychiatric genetics: progress amid controversy. Nat Rev Genet 9:527–540
Carroll LS, Williams NM, Moskvina V, Russell E, Norton N, Williams HJ, Peirce T, Georgieva L, Dwyer S, Grozeva D, Greene E, Farmer A, McGuffin P, Morris DW, Corvin A, Gill M, Rujescu D, Sham P, Holmans P, Jones I et al (2010) Evidence for rare and common genetic risk variants for schizophrenia at protein kinase C, alpha. Mol Psychiatry 15(11):1101–1111
Casey JP, Magalhaes T, Conroy JM, Regan R, Shah N, Anney R, Shields DC, Abrahams BS, Almeida J, Bacchelli E, Bailey AJ, Baird G, Battaglia A, Berney T, Bolshakova N, Bolton PF, Bourgeron T, Brennan S, Cali P, Correia C, Corsello C, Coutanche M, Dawson G, de Jonge M, Delorme R, Duketis E, Duque F, Estes A, Farrar P, Fernandez BA, Folstein SE, Foley S, Fombonne E, Freitag CM, Gilbert J, Gillberg C, Glessner JT, Green J, Guter SJ, Hakonarson H, Holt R, Hughes G, Hus V, Igliozzi R, Kim C, Klauck SM, Kolevzon A, Lamb JA, Leboyer M, Le Couteur A, Leventhal BL, Lord C, Lund SC, Maestrini E, Mantoulan C, Marshall CR, Mcconachie H, Mcdougle CJ, Mcgrath J, Mcmahon WM, Merikangas A, Miller J, Minopoli F, Mirza GK, Munson J, Nelson SF, Nygren G, Oliveira G, Pagnamenta AT, Papanikolaou K, Parr JR, Parrini B, Pickles A, Pinto D, Piven J, Posey DJ, Poustka A, Poustka F, Ragoussis J, Roge B, Rutter ML, Sequeira AF, Soorya L, Sousa I, Sykes N, Stoppioni V, Tancredi R, Tauber M, Thompson AP, Thomson S, Tsiantis J, van Engeland H, Vincent JB, Volkmar F, Vorstman JA, Wallace S, Wang K, Wassink TH, White K, Wing K et al (2012) A novel approach of homozygous haplotype sharing identifies candidate genes in autism spectrum disorder. Hum Genet 131:565–579
Coe BP, Witherspoon K, Rosenfeld JA, van Bon BW, Vulto-van Silfhout AT, Bosco P, Friend KL, Baker C, Buono S, Vissers LE, Schuurs-Hoeijmakers JH, Hoischen A, Pfundt R, Krumm N, Carvill GL, Li D, Amaral D, Brown N, Lockhart PJ, Scheffer IE, Alberti A, Shaw M, Pettinato R, Tervo R, de Leeuw N, Reijnders MR, Torchia BS, Peeters H, O’roak BJ, Fichera M, Hehir-Kwa JY, Shendure J, Mefford HC, Haan E, Gecz J, de Vries BB, Romano C, Eichler EE (2014) Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet 46:1063–1071
Cooper SA, Smiley E, Morrison J, Williamson A, Allan L (2007) Mental ill-health in adults with intellectual disabilities: prevalence and associated factors. Br J Psychiatry 190:27–35
Costello H, Bouras N (2006) Assessment of mental health problems in people with intellectual disabilities. Isr J Psychiatry Relat Sci 43:241–251
Deciphering Developmental Disorders Study (2017) Prevalence and architecture of de novo mutations in developmental disorders. Nature 542:433–438
Denayer E, Ahmed T, Brems H, van Woerden G, Borgesius NZ, Callaerts-Vegh Z, Yoshimura A, Hartmann D, Elgersma Y, D’hooge R, Legius E, Balschun D (2008) Spred1 is required for synaptic plasticity and hippocampus-dependent learning. J Neurosci 28:14443–14449
Djurovic S, Le Hellard S, Kahler AK, Jonsson EG, Agartz I, Steen VM, Hall H, Wang AG, Rasmussen HB, Melle I, Werge T, Andreassen OA (2009) Association of MCTP2 gene variants with schizophrenia in three independent samples of Scandinavian origin (SCOPE). Psychiatry Res 168(3):256–258
Dolan M, Mendelsohn NJ, Pierpont ME, Schimmenti LA, Berry SA, Hirsch B 2010. A novel microdeletion/microduplication syndrome of 19p13.13. Genet Med 12:503–511
Elia J, Gai X, Xie HM, Perin JC, Geiger E, Glessner JT, D’arcy M, Deberardinis R, Frackelton E, Kim C, Lantieri F, Muganga BM, Wang L, Takeda T, Rappaport EF, Grant SF, Berrettini W, Devoto M, Shaikh TH, Hakonarson H, White PS 2010. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol Psychiatry 15:637–646
Emerson E (1995) Challenging behaviour: analysis and intervention in people with intellectual disabilities. Cambridge University Press, Cambridge
Furnari MA, Jobes ML, Nekrasova T, Minden A, Wagner GC (2013) Functional deficits in PAK5, PAK6 and PAK5/PAK6 knockout mice. PLoS ONE 8:e61321
Gauthier J, Champagne N, Lafreniere RG, Xiong L, Spiegelman D, Brustein E, Lapointe M, Peng H, Cote M, Noreau A, Hamdan FF, Addington AM, Rapoport JL, Delisi LE, Krebs MO, Joober R, Fathalli F, Mouaffak F, Haghighi AP, Neri C et al (2010) De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proc Natl Acad Sci USA 107(17):7863–7868
George SK, Jiao Y, Bishop CE, Lu B 2011. Mitochondrial peptidase IMMP2L mutation causes early onset of age-associated disorders and impairs adult stem cell self-renewal. Aging Cell 10:584–594
Gillentine MA, Schaaf CP (2015) The human clinical phenotypes of altered CHRNA7 copy number. Biochem Pharmacol 97(4):352–362
Gimelli S, Capra V, Di Rocco M, Leoni M, Mirabelli-Badenier M, Schiaffino MC, Fiorio P, Cuoco C, Gimelli G, Tassano E (2014) Interstitial 7q31.1 copy number variations disrupting IMMP2L gene are associated with a wide spectrum of neurodevelopmental disorders. Mol Cytogenet 7:54
Girirajan S, Brkanac Z, Coe BP, Baker C, Vives L, Vu TH, Shafer N, Bernier R, Ferrero GB, Silengo M, Warren ST, Moreno CS, Fichera M, Romano C, Raskind WH, Eichler EE (2011) Relative burden of large CNVs on a range of neurodevelopmental phenotypes. PLoS Genet 7(11):e1002334
Girirajan S, Eichler EE (2010) Phenotypic variability and genetic susceptibility to genomic disorders. Hum Mol Genet 19:R176–R187
Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, Goldstein A, Filipink RA, McConnell JS, Angle B, Meschino WS, Nezarati MM, Asamoah A, Jackson KE, Gowans GC, Martin JA, Carmany EP, Stockton DW, Schnur RE, Penney LS, Martin DM et al (2012) Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N Engl J Med 367(14):1321–1331
Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, Imielinski M, Frackelton EC, Reichert J, Crawford EL, Munson J, Sleiman PM, Chiavacci R, Annaiah K, Thomas K, Hou C, Glaberson W, Flory J, Otieno F, Garris M, Soorya L, Klei L, Piven J, Meyer KJ, Anagnostou E, Sakurai T, Game RM, Rudd DS, Zurawiecki D, Mcdougle CJ, Davis LK, Miller J, Posey DJ, Michaels S, Kolevzon A, Silverman JM, Bernier R, Levy SE, Schultz RT, Dawson G, Owley T, Mcmahon WM, Wassink TH, Sweeney JA, Nurnberger JI, Coon H, Sutcliffe JS, Minshew NJ, Grant SF, Bucan M, Cook EH, Buxbaum JD, Devlin B, Schellenberg GD, Hakonarson H (2009) Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 459:569–573
Grayton HM, Missler M, Collier DA, Fernandes C (2013) Altered social behaviours in neurexin 1alpha knockout mice resemble core symptoms in neurodevelopmental disorders. PLoS ONE 8(6):e67114
Green EK, Rees E, Walters JT, Smith KG, Forty L, Grozeva D, Moran JL, Sklar P, Ripke S, Chambert KD, Genovese G, Mccarroll SA, Jones I, Jones L, Owen MJ, O’donovan MC, Craddock N, Kirov G (2015) Copy number variation in bipolar disorder. Mol Psychiatry 21(1):89–93
Guilmatre A, Dubourg C, Mosca AL, Legallic S, Goldenberg A, Drouin-Garraud V, Layet V, Rosier A, Briault S, Bonnet-Brilhault F, Laumonnier F, Odent S, Le Vacon G, Joly-Helas G, David V, Bendavid C, Pinoit JM, Henry C, Impallomeni C, Germano E, Tortorella G, Di Rosa G, Barthelemy C, Andres C, Faivre L, Frebourg T, Saugier Veber P, Campion D (2009) Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry 66:947–956
Hahn CG, Friedman E (1999) Abnormalities in protein kinase C signaling and the pathophysiology of bipolar disorder. Bipolar Disord 1(2):81–86
Hedges DJ, Hamilton-Nelson KL, Sacharow SJ, Nations L, Beecham GW, Kozhekbaeva ZM, Butler BL, Cukier HN, Whitehead PL, Ma D, Jaworski JM, Nathanson L, Lee JM, Hauser SL, Oksenberg JR, Cuccaro ML, Haines JL, Gilbert JR, Pericak-Vance MA (2012) Evidence of novel fine-scale structural variation at autism spectrum disorder candidate loci. Mol Autism 3:2
Ho KS, Wassman ER, Baxter AL, Hensel CH, Martin MM, Prasad A, Twede H, Vanzo RJ, Butler MG (2016) Chromosomal microarray analysis of consecutive individuals with autism spectrum disorders using an ultra-high resolution chromosomal microarray optimized for neurodevelopmental disorders. Int J Mol Sci 17:2070
Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C (2004) Detection of large-scale variation in the human genome. Nat Genet 36:949–951
Jarick I, Volckmar AL, Putter C, Pechlivanis S, Nguyen TT, Dauvermann MR, Beck S, Albayrak O, Scherag S, Gilsbach S, Cichon S, Hoffmann P, Degenhardt F, Nothen MM, Schreiber S, Wichmann HE, Jockel KH, Heinrich J, Tiesler CM, Faraone SV, Walitza S, Sinzig J, Freitag C, Meyer J, Herpertz-Dahlmann B, Lehmkuhl G, Renner TJ, Warnke A, Romanos M, Lesch KP, Reif A, Schimmelmann BG, Hebebrand J, Scherag A, Hinney A (2014) Genome-wide analysis of rare copy number variations reveals PARK2 as a candidate gene for attention-deficit/hyperactivity disorder. Mol Psychiatry 19:115–121
Javitt DC (2007) Glutamate and schizophrenia: phencyclidine, N-methyl-D-aspartate receptors, and dopamine-glutamate interactions. Int Rev Neurobiol 78:69–108
Kato T, Hayashi-Takagi A, Toyota T, Yoshikawa T, Iwamoto K (2011) Gene expression analysis in lymphoblastoid cells as a potential biomarker of bipolar disorder. J Hum Genet 56:779–783
Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, Moran J, Chambert K, Toncheva D, Georgieva L, Grozeva D, Fjodorova M, Wollerton R, Rees E, Nikolov I, Van De Lagemaat LN, Bayes A, Fernandez E, Olason PI, Bottcher Y, Komiyama NH, Collins MO, Choudhary J, Stefansson K, Stefansson H, Grant SG, Purcell S, Sklar P, O’donovan MC, Owen MJ (2012) De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry 17:142–153
Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP (2001) A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 413:519–523
Lamb AN, Rosenfeld JA, Neill NJ, Talkowski ME, Blumenthal I, Girirajan S, Keelean-Fuller D, Fan Z, Pouncey J, Stevens C, Mackay-Loder L, Terespolsky D, Bader PI, Rosenbaum K, Vallee SE, Moeschler JB, Ladda R, Sell S, Martin J, Ryan S et al (2012) Haploinsufficiency of SOX5 at 12p12.1 is associated with developmental delays with prominent language delay, behavior problems, and mild dysmorphic features. Hum Mutat 33(4):728–740
Levy D, Ronemus M, Yamrom B, Lee YH, Leotta A, Kendall J, Marks S, Lakshmi B, Pai D, Ye K, Buja A, Krieger A, Yoon S, Troge J, Rodgers L, Iossifov I, Wigler M (2011) Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron 70(5):886–897
Lisi EC, Hamosh A, Doheny KF, Squibb E, Jackson B, Galczynski R, Thomas GH, Batista DA (2008) 3q29 interstitial microduplication: a new syndrome in a three-generation family. Am J Med Genet A 146A:601–609
Lowe K, Allen D, Jones E, Brophy S, Moore K, James W (2007) Challenging behaviours: prevalence and topographies. J Intellect Disabil Res 51:625–636
Lowther C, Speevak M, Armour CM, Goh ES, Graham GE, Li C, Zeesman S, Nowaczyk MJ, Schultz LA, Morra A, Nicolson R, Bikangaga P, Samdup D, Zaazou M, Boyd K, Jung JH, Siu V, Rajguru M, Goobie S, Tarnopolsky MA, Prasad C, Dick PT, Hussain AS, Walinga M, Reijenga RG, Gazzellone M, Lionel AC, Marshall CR, Scherer SW, Stavropoulos DJ, Mccready E, Bassett AS (2017) Molecular characterization of NRXN1 deletions from 19,263 clinical microarray cases identifies exons important for neurodevelopmental disease expression. Genet Med 19:53–61
Ma X, Deng W, Liu X, Li M, Chen Z, He Z, Wang Y, Wang Q, Hu X, Collier DA, Li T (2011) A genome-wide association study for quantitative traits in schizophrenia in China. Genes Brain Behav 10(7):734–739
Maestrini E, Pagnamenta AT, Lamb JA, Bacchelli E, Sykes NH, Sousa I, Toma C, Barnby G, Butler H, Winchester L, Scerri TS, Minopoli F, Reichert J, Cai G, Buxbaum JD, Korvatska O, Schellenberg GD, Dawson G, De Bildt A, Minderaa RB, Mulder EJ, Morris AP, Bailey AJ, Monaco AP 2010. High-density SNP association study and copy number variation analysis of the AUTS1 and AUTS5 loci implicate the IMMP2L-DOCK4 gene region in autism susceptibility. Mol Psychiatry 15:954–968
Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, Thiruvahindrapduram B, Fiebig A, Schreiber S, Friedman J, Ketelaars CE, Vos YJ, Ficicioglu C, Kirkpatrick S, Nicolson R, Sloman L, Summers A, Gibbons CA, Teebi A, Chitayat D, Weksberg R, Thompson A, Vardy C, Crosbie V, Luscombe S, Baatjes R, Zwaigenbaum L, Roberts W, Fernandez B, Szatmari P, Scherer SW (2008) Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet 82:477–488
Mehregan H, Najmabadi H, Kahrizi K (2016) Genetic Studies in Intellectual Disability and Behavioral Impairment. Arch Iran Med 19:363–375
Moessner R, Marshall CR, Sutcliffe JS, Skaug J, Pinto D, Vincent J, Zwaigenbaum L, Fernandez B, Roberts W, Szatmari P, Scherer SW (2007) Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet 81(6):1289–1297
Moreno-De-Luca A, Myers SM, Challman TD, Moreno-De-Luca D, Evans DW, Ledbetter DH 2013. Developmental brain dysfunction: revival and expansion of old concepts based on new genetic evidence. Lancet Neurol 12:406–414
Morgan VA, Leonard H, Bourke J, Jablensky A 2008. Intellectual disability co-occurring with schizophrenia and other psychiatric illness: population-based study. Br J Psychiatry 193:364–372
Myles-Worsley M, Tiobech J, Browning SR, Korn J, Goodman S, Gentile K, Melhem N, Byerley W, Faraone SV, Middleton FA (2013) Deletion at the SLC1A1 glutamate transporter gene co-segregates with schizophrenia and bipolar schizoaffective disorder in a 5-generation family. Am J Med Genet B 162B:87–95
Nesbitt A, Bhoj EJ, McDonald Gibson K, Yu Z, Denenberg E, Sarmady M, Tischler T, Cao K, Dubbs H, Zackai EH, Santani A (2015) Exome sequencing expands the mechanism of SOX5-associated intellectual disability: a case presentation with review of sox-related disorders. Am J Med Genet A 167A(11):2548–2554
Noor A, Dupuis L, Mittal K, Lionel AC, Marshall CR, Scherer SW, Stockley T, Vincent JB, Mendoza-Londono R, Stavropoulos DJ (2015) 15q11.2 duplication encompassing only the UBE3A gene is associated with developmental delay and neuropsychiatric phenotypes. Hum Mutat 36(7):689–693
Noor A, Lionel AC, Cohen-Woods S, Moghimi N, Rucker J, Fennell A, Thiruvahindrapuram B, Kaufman L, Degagne B, Wei J, Parikh SV, Muglia P, Forte J, Scherer SW, Kennedy JL, Xu W, McGuffin P, Farmer A, Strauss J, Vincent JB (2014) Copy number variant study of bipolar disorder in Canadian and UK populations implicates synaptic genes. Am J Med Genet B Neuropsychiatr Genet 165B(4):303–313
Ophoff RA, Escamilla MA, Service SK, Spesny M, Meshi DB, Poon W, Molina J, Fournier E, Gallegos A, Mathews C, Neylan T, Batki SL, Roche E, Ramirez M, Silva S, De Mille MC, Dong P, Leon PE, Reus VI, Sandkuijl LA et al (2002) Genomewide linkage disequilibrium mapping of severe bipolar disorder in a population isolate. Am J Hum Genet 71(3):565–574
Orellana C, Rosello M, Monfort S, Mayo S, Oltra S, Martinez F (2015) Pure duplication of 19p13.3 in three members of a family with intellectual disability and literature review. Definition of a new microduplication syndrome. Am J Med Genet A 167:1614–1620
O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, Turner EH, Stanaway IB, Vernot B, Malig M, Baker C, Reilly B, Akey JM, Borenstein E, Rieder MJ, Nickerson DA et al (2012) Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485(7397):246–250
Pagnamenta AT, Bacchelli E, De Jonge MV, Mirza G, Scerri TS, Minopoli F, Chiocchetti A, Ludwig KU, Hoffmann P, Paracchini S, Lowy E, Harold DH, Chapman JA, Klauck SM, Poustka F, Houben RH, Staal WG, Ophoff RA, O’donovan MC, Williams J, Nothen MM, Schulte-Korne G, Deloukas P, Ragoussis J, Bailey AJ, Maestrini E, Monaco AP (2010) Characterization of a family with rare deletions in CNTNAP5 and DOCK4 suggests novel risk loci for autism and dyslexia. Biol Psychiatry 68:320–328
Pantano L, Armengol L, Villatoro S, Estivill X (2008) ProSeeK: a web server for MLPA probe design. BMC Genom 9:573
Pasmant E, Sabbagh A, Hanna N, Masliah-Planchon J, Jolly E, Goussard P, Ballerini P, Cartault F, Barbarot S, Landman-Parker J, Soufir N, Parfait B, Vidaud M, Wolkenstein P, Vidaud D, France RN (2009) SPRED1 germline mutations caused a neurofibromatosis type 1 overlapping phenotype. J Med Genet 46(7):425–430
Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Almeida J, Bacchelli E, Bader GD, Bailey AJ, Baird G, Battaglia A, Berney T, Bolshakova N, Bolte S, Bolton PF, Bourgeron T, Brennan S, Brian J, Bryson SE, Carson AR, Casallo G, Casey J, Chung BH, Cochrane L, Corsello C, Crawford EL, Crossett A, Cytrynbaum C, Dawson G, de Jonge M, Delorme R, Drmic I, Duketis E, Duque F, Estes A, Farrar P, Fernandez BA, Folstein SE, Fombonne E, Freitag CM, Gilbert J, Gillberg C, Glessner JT, Goldberg J, Green A, Green J, Guter SJ, Hakonarson H, Heron EA, Hill M, Holt R, Howe JL, Hughes G, Hus V, Igliozzi R, Kim C, Klauck SM, Kolevzon A, Korvatska O, Kustanovich V, Lajonchere CM, Lamb JA, Laskawiec M, Leboyer M, Le Couteur A, Leventhal BL, Lionel AC, Liu XQ, Lord C, Lotspeich L, Lund SC, Maestrini E, Mahoney W, Mantoulan C, Marshall CR, Mcconachie H, Mcdougle CJ, Mcgrath J, Mcmahon WM, Merikangas A, Migita O, Minshew NJ, Mirza GK, Munson J, Nelson SF, Noakes C, Noor A, Nygren G, Oliveira G, Papanikolaou K, Parr JR, Parrini B, Paton T, Pickles A, Pilorge M et al (2010). Functional impact of global rare copy number variation in autism spectrum disorders. Nature, 466, 368–372
Plummer JT, Gordon AJ, Levitt P (2016) The genetic intersection of neurodevelopmental disorders and shared medical comorbidities—relations that translate from bench to bedside. Front Psychiatry 7:142
Porton B, Greenberg BD, Askland K, Serra LM, Gesmonde J, Rudnick G, Rasmussen SA, Kao HT (2013) Isoforms of the neuronal glutamate transporter gene, SLC1A1/EAAC1, negatively modulate glutamate uptake: relevance to obsessive-compulsive disorder. Transl Psychiatry 3:e259
Posey JE, Rosenfeld JA, James RA, Bainbridge M, Niu Z, Wang X, Dhar S, Wiszniewski W, Akdemir ZH, Gambin T, Xia F, Person RE, Walkiewicz M, Shaw CA, Sutton VR, Beaudet AL, Muzny D, Eng CM, Yang Y, Gibbs RA, Lupski JR, Boerwinkle E, Plon SE (2016) Molecular diagnostic experience of whole-exome sequencing in adult patients. Genet Med 18:678–685
Prasad A, Merico D, Thiruvahindrapuram B, Wei J, Lionel AC, Sato D, Rickaby J, Lu C, Szatmari P, Roberts W, Fernandez BA, Marshall CR, Hatchwell E, Eis PS, Scherer SW (2012) A discovery resource of rare copy number variations in individuals with autism spectrum disorder. G3 (Bethesda) 2(12):1665–1685
Ramos-Quiroga JA, Sanchez-Mora C, Casas M, Garcia-Martinez I, Bosch R, Nogueira M, Corrales M, Palomar G, Vidal R, Coll-Tane M, Bayes M, Cormand B, Ribases M (2014) Genome-wide copy number variation analysis in adult attention-deficit and hyperactivity disorder. J Psychiatr Res 49:60–67
Rosenfeld JA, Ballif BC, Torchia BS, Sahoo T, Ravnan JB, Schultz R, Lamb A, Bejjani BA, Shaffer LG (2010) Copy number variations associated with autism spectrum disorders contribute to a spectrum of neurodevelopmental disorders. Genet Med 12(11):694–702
Schaaf CP (2014) Nicotinic acetylcholine receptors in human genetic disease. Genet Med 16(9):649–656
Schaaf CP, Boone PM, Sampath S, Williams C, Bader PI, Mueller JM, Shchelochkov OA, Brown CW, Crawford HP, Phalen JA, Tartaglia NR, Evans P, Campbell WM, Tsai AC, Parsley L, Grayson SW, Scheuerle A, Luzzi CD, Thomas SK, Eng PA et al (2012) Phenotypic spectrum and genotype-phenotype correlations of NRXN1 exon deletions. Eur J Hum Genet 20(12):1240–1247
Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, YE K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M (2004) Large-scale copy number polymorphism in the human genome. Science 305:525–528
Spencer E, Davis J, Mikhail F, Fu C, Vijzelaar R, Zackai EH, Feret H, Meyn MS, Shugar A, Bellus G, Kocsis K, Kivirikko S, Poyhonen M, Messiaen L (2011) Identification of SPRED1 deletions using RT-PCR, multiplex ligation-dependent probe amplification and quantitative PCR. Am J Med Genet A 155A:1352–1359
Stein DJ, Lund C, Nesse RM (2013) Classification systems in psychiatry: diagnosis and global mental health in the era of DSM-5 and ICD-11. Curr Opin Psychiatry 26:493–497
Swaminathan S, Shen L, Kim S, Inlow M, West JD, Faber KM, Foroud T, Mayeux R, Saykin AJ (2012) Analysis of copy number variation in Alzheimer’s disease: the NIALOAD/NCRAD Family Study. Curr Alzheimer Res 9:801–814
Tada H, Okano HJ, Takagi H, Shibata S, Yao I, Matsumoto M, Saiga T, Nakayama KI, Kashima H, Takahashi T, Setou M, OKANO H (2010) Fbxo45, a novel ubiquitin ligase, regulates synaptic activity. J Biol Chem 285:3840–3849
Taylor MR, Jirikowic J, Wells C, Springer M, Mcgavran L, Lunt B, Swisshelm K (2010) High prevalence of array comparative genomic hybridization abnormalities in adults with unexplained intellectual disability. Genet Med 12:32–38
Tebartz van Elst, L, Maier S, Fangmeier T, Endres D, Mueller GT, Nickel K, Ebert D, Lange T, Hennig J, Biscaldi M, Riedel A, Perlov E (2014) Disturbed cingulate glutamate metabolism in adults with high-functioning autism spectrum disorder: evidence in support of the excitatory/inhibitory imbalance hypothesis. Mol Psychiatry 19:1314–1325
Vinas-Jornet M, Esteba-Castillo S, Gabau E, Ribas-Vidal N, Baena N, San J, Ruiz A, Coll MD, Novell R, Guitart M (2014) A common cognitive, psychiatric, and dysmorphic phenotype in carriers of NRXN1 deletion. Mol Genet Genomic Med 2:512–521
Waga C, Okamoto N, Ondo Y, Fukumura-Kato R, Goto Y, Kohsaka S, Uchino S (2011) Novel variants of the SHANK3 gene in Japanese autistic patients with severe delayed speech development. Psychiatr Genet 21(4):208–211
Walss-Bass C, Soto-Bernardini MC, Johnson-Pais T, Leach RJ, Ontiveros A, Nicolini H, Mendoza R, Jerez A, Dassori A, Chavarria-Siles I, Escamilla MA, Raventos H (2009) Methionine sulfoxide reductase: a novel schizophrenia candidate gene. Am J Med Genet B Neuropsychiatr Genet 150B(2):219–225
Wang C, Koide T, Kimura H, Kunimoto S, Yoshimi A, Nakamura Y, Kushima I, Banno M, Kawano N, Takasaki Y, Xing J, Noda Y, Mouri A, Aleksic B, Ikeda M, Okada T, Iidaka T, Inada T, Iwata N, Ozaki N (2014) Novel rare variants in F-box protein 45 (FBXO45) in schizophrenia. Schizophr Res 157:149–156
Wang K, Zhang H, Ma D, Bucan M, Glessner JT, Abrahams BS, Salyakina D, Imielinski M, Bradfield JP, Sleiman PM, Kim CE, Hou C, Frackelton E, Chiavacci R, Takahashi N, Sakurai T, Rappaport E, Lajonchere CM, Munson J, Estes A et al (2009) Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature 459(7246):528–533
Weiss LA, Arking DE, Daly MJ, Chakravarti A (2009) A genome-wide linkage and association scan reveals novel loci for autism. Nature 461(7265):802–808
Willatt L, Cox J, Barber J, Cabanas ED, Collins A, Donnai D, Fitzpatrick DR, Maher E, Martin H, Parnau J, Pindar L, Ramsay J, Shaw-Smith C, Sistermans EA, Tettenborn M, Trump D, de Vries BB, Walker K, Raymond FL (2005) 3q29 microdeletion syndrome: clinical and molecular characterization of a new syndrome. Am J Hum Genet 77:154–160
Wisniowiecka-Kowalnik B, Nesteruk M, Peters SU, Xia Z, Cooper ML, Savage S, Amato RS, Bader P, Browning MF, Haun CL, Duda AW 3rd, Cheung SW, Stankiewicz P (2010) Intragenic rearrangements in NRXN1 in three families with autism spectrum disorder, developmental delay, and speech delay. Am J Med Genet B Neuropsychiatr Genet 153B(5):983–993
Wolfe K, Strydom A, Morrogh D, Carter J, Cutajar P, Eyeoyibo M, Hassiotis A, Mccarthy J, Mukherjee R, Paschos D, Perumal N, Read S, Shankar R, Sharif S, Thirulokachandran S, Thygesen JH, Patch C, Ogilvie C, Flinter F, Mcquillin A, BASS N (2016) Chromosomal microarray testing in adults with intellectual disability presenting with comorbid psychiatric disorders. Eur J Hum Genet 25:66–72
Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M (2008) Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet 40:880–885
Acknowledgements
Financial support was received from the “Instituto de Salud Carlos III” (FIS, grant no. PI080778), “Fundació Parc Taulí–Institut d’Investigació i Innovació Parc Taulí I3PT” (Grant Nos. CIR2009/33, CIR2010/034) and “Fundació Barnola-Vallribera 2011”. We acknowledge the following individuals for their help with this study: MC Mata, MC Dominguez, A Veraguas, M Garcia, O Villa and C Hernando. Finally, we especially thank the patients and families who participated in this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Marina Viñas-Jornet, Susanna Esteba-Castillo, Neus Baena, Núria Ribas-Vidal, Anna Ruiz, David Torrents-Rodas, Elisabeth Gabau, Elisabet Vilella, Lourdes Martorell, Lluís Armengol, Ramon Novell and Míriam Guitart declare that they have no conflict of interest.
Ethical Approval
All procedures performed in this study were in accordance with the ethical standards of the institutional ethics committee (CEIC 2009/582) of the Parc Taulí Hospital Universitari and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Animal Rights
This article does not contain any studies with animals performed by any of the authors.
Informed Consent
Informed written consent was obtained from all individual participants included in the study.
Additional information
Edited by Michael Lyons.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10519_2018_9902_MOESM1_ESM.xlsx
Additional file 1 Rare CNVs identified in the adult population affected by intellectual disability and co-morbid psychiatric/behavioural disorders. (Pat Id) Patient identification; (CNV, ISCN 2016) Copy number variant according to the International System for Human Cytogenetic Nomenclature edited in 2016; (bp) Base pair; (MAT) Maternal; (PAT) Paternal; (MAT and PAT) Inherited from both progenitors; (CGHa) Comparative genomic hybridization array; (BAC FISH) Bacterial artificial chromosome clones for fluorescent in situ hybridization; (MLPA) Multiplex ligation-dependent probe amplification; (MLPA autism) MRC-Holland Probemix P343; (MLPA subtel) MRC-Holland Probemix P036 and P070; (MS-MLPA) Methylation-specific MLPA with MRC-Holland Probemix ME028; (NA) Not analysed; (*) Polymorphic variant reported here due to its putative clinical relevance; (NP) Not present (XLSX 64 KB)
10519_2018_9902_MOESM2_ESM.xlsx
Additional file 2 Pathogenic CNVs and variants of unknown significance (VOUS) likely pathogenic (pVOUS): summary of data from literature (Coe et al., 2014 and DDD 2017) as well as public databases. (Pat Id) Patient identification; (chr) chromosome; (bp) base pair; (ISCN 2016) International System for Human Cytogenetic Nomenclature edited in 2016; (CNV) Copy number variant; (DD) Developmental delay (XLSX 34 KB)
10519_2018_9902_MOESM3_ESM.xlsx
Additional file 3 7q31 copy number variants reported in Database of Genomic Variants. (DGV) Database of Genomic Variants; (CNVR) Copy number variant region; (bp) base pair (XLSX 12 KB)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Viñas-Jornet, M., Esteba-Castillo, S., Baena, N. et al. High Incidence of Copy Number Variants in Adults with Intellectual Disability and Co-morbid Psychiatric Disorders. Behav Genet 48, 323–336 (2018). https://doi.org/10.1007/s10519-018-9902-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10519-018-9902-6