Abstract
Aquaculture is playing an increasingly important role in global food security, especially for low-income and food-deficit countries. The majority of aquaculture production occurs in freshwater earthen ponds and tilapia has quickly become one of the most widely adopted culture species in these systems. Tilapia are now farmed in over 140 countries facilitated by their ease of production, adaptability to a wide range of environmental conditions, fast growth, and high nutritional value. Typically, tilapia have been considered a hardy, disease resilient species; however, the disease is increasing with subsequent threats to the industry as their production is intensified. In this review, we discuss tilapia production, with a focus on Bangladesh as one of the top producing countries, and highlight the problems associated with disease and treatment approaches for them, including the misuse of antimicrobials. We address a key missing component in understanding health and disease processes for sustainable production in aquaculture, specifically the role played by the microbiome. Here we examine the importance of the microbiome in supporting health, focused on the symbiotic microbial community of the fish skin mucosal surface, the abiotic and biotic factors that influence the microbiome, and the shifts that are associated with diseased states. We also identify conserved taxa of skin microbiomes that may be used as indicators of health status for tilapia offering new opportunities to mitigate and manage the disease and optimize environmental growing conditions and farming practices.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The rise of aquaculture and tilapia production
Aquaculture is one of the fastest-growing food-producing sectors (Anderson et al. 2017; Barría et al. 2021) and is playing a central role in meeting the demand for nutritious and affordable food for billions of people globally (Lynch and MacMillan 2017; Tigchelaar et al. 2022). Currently, aquatic foods contribute around 17% of the global animal protein, 7% of all protein sources (FAO 2022; Obiero et al. 2019) and provide at least 20% of the per capita intake of animal protein for 3.3 billion people (FAO 2022). In many low-income and developing countries, including Bangladesh, aquatic food provides more than half of their total animal protein intake (Arthur et al. 2022; FAO 2022). Fish furthermore contain long-chained poly-unsaturated fatty acids, together with many micronutrients (e.g. calcium, iron, zinc), vitamins, and minerals, which are dietary elements especially important for nutritionally vulnerable people (Ahern et al. 2021; Arthur et al. 2022; Béné et al. 2016, 2015; HLPE 2014). Recognizing the health potential of fish, many countries, including India, Brazil, Chile, and sub-Saharan African countries, have incorporated fish into their national school-feeding programmes (Ahern et al. 2021; Béné et al. 2016; Wineman et al. 2022).
The demand for fisheries products is increasing dramatically. Illustrating this, between 1961 and 2019, global aquatic food consumption increased by 3% annually, almost twice that of the annual global population growth rate (1.6%) (FAO 2020, 2022). In line with the global demand, fisheries and aquaculture production have increased (reaching 214 million tonnes in 2020) and it is predicted that aquaculture will contribute to over half of the global fish consumption by 2030 (FAO 2020). Aquaculture now delivers 49.2% of the worldwide supply of aquatic products, most of which are used for human consumption and has a total value of US$ 281.5 billion (FAO 2022).
In Bangladesh, one of the most densely populated countries (nearly 170 million people living in an area of 148,460 km2) and poorest nations in the world (Lauria et al. 2018), fish represents about 60% of animal protein intake in the national diet (DoF 2020; Lauria et al. 2018; Shamsuzzaman et al. 2017) with an average consumption of approximately 23 kg/year per person (DoF 2020). Bangladesh now ranks third globally for inland open water capture and fifth in aquaculture production (DoF 2020; FAO 2020). More than 12% (over 20 million) of the total human population of Bangladesh depends directly or indirectly, on the fisheries sector for their livelihood (DoF 2020). In 2019–2020, aquaculture contributed 3.52% of the gross domestic product in Bangladesh and aquatic animal products (including crustaceans, fish, and molluscs) are Bangladesh’s most valuable export after textiles and footwear (OEC 2023).
Among the 87.5 million tonnes of aquatic animals produced by global aquaculture, 57.5 million tonnes of this is finfish (FAO 2022). A diverse range of fish species are cultured in different aquaculture systems across freshwater, brackish water, inland saline water, and marine waters; however, in 2020, only 28 finfish species groups made up 79% (45.4 million tonnes) of the total finfish aquaculture production (FAO 2022). Of these species, Nile tilapia (Oreochromis niloticus) is ranked third, representing 7.9% (4.5 million tonnes) of the total finfish production with other tilapias accounting for an additional 1.9% (around 1.1 million tonnes) (Fig. 1a). Only grass carp (Ctenopharyngodon idellus) and silver carp (Hypophthalmichthys molitrix) are higher produced finfish species farmed globally than tilapia (FAO 2022). Among the various species of tilapia, Nile tilapia is the most widely distributed and preferred species for aquaculture; over 90% of all commercially produced tilapia farmed outside of Africa are Nile tilapia (Prabu et al. 2019; Wang and Lu 2016). Monosex tilapia are favoured due to their fast growth and high production potential (Prabu et al. 2019), and the fact that generally they do not breed well in the natural environment, thus reducing the risk of them becoming an invasive species. Other commercially important common tilapia species include the Mozambique tilapia (O. mossambicus), blue tilapia (O. aureus), the Zanzibar tilapia (O. hornorum), and various hybrid tilapia species (Machimbirike et al. 2019; Prabu et al. 2019).
With their relative ease of farming, good market demand, and stable market price, the farming and production of tilapia have increased considerably over the last few decades (Wang and Lu 2016). They are also reasonably fast-growing, reaching around 400 g in 6/7 months (Baqui and Bhujel 2011), allowing farmers to harvest two or more crops each year depending on the harvesting size. They can also tolerate a wide range of environmental conditions and they grow well even in poor water quality conditions with low oxygen levels (Li et al. 2017a; Rebouças et al. 2015; Yu et al. 2021a). Some tilapia can also live in brackish water and can even adapt to full-strength seawater (Rahman et al. 2021; Rengmark et al. 2007), widening the areas across which they can be cultured. Tilapia are furthermore benthopelagic omnivorous fish that feeds on algae, plankton, detritus, small invertebrates, and bacterial films. This relatively low trophic need lends them well to highly sustainable forms of aquaculture practice (Tesfahun and Temesgen 2018; Wang and Lu 2016), including biofloc systems (Khanjani et al. 2022). This is reflected by the fact that 140 countries in the world have introduced at least one species of tilapia into their fish farming systems (Barroso et al. 2019; Deines et al. 2016; Fitzsimmons 2015). Tilapia farming has grown exceptionally fast compared with the aquaculture of other species, with production growing by 11% per annum for the last three decades (from 0.3 million tonnes in 1987 to 5.9 million tonnes in 2017, with a value of approximately US$11 billion (Barroso et al. 2019)). In terms of quantity, the share of tilapia in global aquaculture has increased from 1.9% in 1987 to 5.3% in 2017 and in terms of value, from 1.5% to 4.4% (Barroso et al. 2019), emphasizing its importance to global food security. Asian countries produce most of the world’s tilapia and are the major consumers of this fish species (Fig. 1b).
In Bangladesh, Mozambique tilapia (O. mossambicus) was first introduced in the 1950s but the species only got wide acceptance through the introduction of the GIFT tilapia by the Bangladesh Fisheries Research Institute in partnership with WorldFish in the 1990s (Hussain 2004; Ponzoni et al. 2010). It is generally grown in polyculture systems with carp species such as catla (Catla catla), rohu (Labeo rohita), common carp (Cyprinus carpio), silver carp (H. molitrix), and grass carp (C. idella) or catfish such as pangas (Pangasius pangasius), pabda (Ompok pabda), and shing (Gagata youssoufi), although monoculture of tilapia also occurs (Rahman et al. 2021). Bangladesh is now the third highest producer of tilapia in Asia and the fourth highest globally (DoF 2020; see Fig. 1b). Currently, in Bangladesh, tilapia is the third most farmed fish after pangas (P. pangasius) and rohu (Labeo rohita) (Fig. 2; DoF 2020). In 2019–2020, around 371,263 tonnes of tilapia were produced making up 9.69% of the annual fish production of inland water bodies (DoF 2020). The most popular method for culturing tilapia is in earthen, shallow depth (1–2 m) ponds that range from small-scale to large intensive commercial systems (DoF 2020). Almost all the produced tilapia is used for domestic consumption (Ahmed et al. 2012).
Production (tonnes) of the top ten fish species from pond aquaculture in Bangladesh for the years 2018–2019 and 2019–2020 (DoF 2020)
Diseases and their treatments in tilapia farming
To fulfil global demand, the aquaculture industry has expanded, primarily through the intensification of culture systems (Assefa and Abunna 2018). This, however, in many instances has resulted in overstocking, reduced water quality, and increased stress of the cultured fish that in turn renders the fish more susceptible to various infectious diseases. Disease is in fact now one of the major limiting factors to the growth of the aquaculture industry (Assefa and Abunna 2018). Globally, losses in the aquaculture sector due to diseases are estimated to exceed US$ 6 billion annually (Assefa and Abunna 2018; Stentiford et al. 2017; WorldBank 2014) but estimates vary, and according to Shinn et al. (2015), the global economic losses in finfish aquaculture range between US$ 1.05 to US$ 9.58 billion per year.
Tilapia are thought to be relatively disease-resistant to many common pathogens (Ferguson et al. 2014), but increasingly farmed tilapia are being shown to succumb to various disease causing organisms (Machimbirike et al. 2019; Surachetpong et al. 2020; see Table 1). Tilapia are particularly susceptible to infection by Streptococcus sp. including Streptococcus dysgalactiae, S. iniae, and S. agalactiae that are now among the most important bacterial pathogens, causing considerable economic losses throughout the world (Zamri-Saad et al. 2014). In China, for example, a large-scale outbreak of Streptococcus spp. in 2012, caused 30–80% mortality of tilapia (Chen et al. 2012; Sun et al. 2016; Zhang 2021), with a direct economic loss of approximately US$ 1.0–1.5 billion (Liu et al. 2019). Streptococcosis outbreaks can be treated with antibiotics if they are administered at the early stage of the disease, but their oral administration can be difficult as the infected fish lose appetite (Zamri-Saad et al. 2014). Barnes et al. (2022) recently reviewed the benefits of autogenous vaccination as a local solution to reduce antimicrobial resistance and protect farmed species against key pathogens. Several vaccines are available for the treatment of Streptococcosis including AQUAVAC® Strep Sa (Merck Animal Health Company, USA), AquavacTM GarvetilTM (Intervet/Schering-Plough Animal Health), and NORVAX® STREP Si (Merck Animal Health Company, USA) (Zamri-Saad et al. 2014).
Other key emerging infectious bacterial pathogens include Francisella noatunensis subsp. orientalis that causes francisellosis (Nguyen et al. 2016); Flavobacterium columnare that causes columnaris (Dong et al. 2015a; Figueiredo et al. 2005); Aeromonas hydrophila (Monir et al. 2020), A. jandaei, and A. veronii (Dong et al. 2017b) causing motile Aeromonas septicaemia (MAS); vibriosis by Vibrio spp. (Elgendy et al. 2022); and edwardsiellosis caused by Edwardsiella ictaluri and E. tarda (Dong et al. 2019; El-Yazeed and Ibrahem 2009; Nhinh et al. 2022; Park et al. 2012; Soto et al. 2012; Xu and Zhang 2014). Other opportunistic facultative fungal infections include Saprolegnia spp. (Ali et al. 2019; Zahran et al. 2017) and protozoans, including Ichthyophthirius multifiliis, Trichodina spp., and Dactylogyrus spp. (Attia et al. 2021; El-Sayed 2020; Nguyen et al. 2020; Prabu et al. 2019; Xu et al. 2015).
For the treatment of bacterial vibriosis, antibiotics are effective when supplied via medicated feed or mixed in water (El-Gohary et al. 2020). Gram-negative motile aeromonads may be treated with potassium permanganate (KMnO4) and with antibiotics in the feed (Monir et al. 2020; Prabu et al. 2019; Stratev and Odeyemi 2017). A recent study reported immersion vaccination against columnaris disease induced a strong immune response and improved survival against experimental infection of F. columnare (Kitiyodom et al. 2021). This opportunistic pathogen, in addition to fungal diseases such as white spots, can also be effectively treated via exposure to CuSO4, KMnO4, or salt/sodium chloride (El-Sayed 2020; Prabu et al. 2019). The available common treatments for treating the major disease problems caused by bacteria, parasites, fungi, etc. affecting tilapia are summarized in Table 1.
To date, eight viral diseases (five DNA and three RNA viruses) have been reported in tilapia (Table 2) some of which are causing huge losses in tilapia aquaculture (Machimbirike et al. 2019). Viral diseases like Herpes-like virus, iridoviral disease (IVD), viral nervous necrosis (VNN), tilapia lake virus (TiLV), and infectious spleen and kidney necrosis virus (ISKNV) that affect Nile tilapia and red hybrid tilapia (Dong et al. 2015b; Suebsing et al. 2016) are the most limiting viral disease-causing agents in tilapia culture. In contrast with bacterial diseases, viral diseases are much more difficult to control not only because of the lack of therapeutics but also because of the lack of knowledge of the pathogenesis of viral infections (Kibenge et al. 2012). Some commercial vaccines and selective breeding programmes have been successful in reducing the severity of some viral diseases, for most; however, there are no effective treatments or cures (Jansen et al. 2019). Viruses, therefore, tend to be considered the most important potential threats in aquaculture, including tilapia. This is well illustrated in the case of the newly emerged tilapia lake virus (TiLV) (Jansen et al. 2019).
TiLV, also known as Tilapia tilapinevirus, was first reported in farmed tilapia in Israel in 2014 (Eyngor et al. 2014). It is now listed as an emerging finfish disease by the World Organization for Animal Health (OIE) with a high likelihood of being included in the list of reportable finfish diseases (Sood et al. 2021). An investigation of samples collected from Thai hatcheries from 2012 to 2017 found the presence of TiLV in the majority of tested samples (Dong et al. 2017a). Before its discovery, more than 40 countries had imported tilapia fry and fingerlings from Thailand. Recent studies have confirmed the presence of TiLV in 16 countries including some of those countries receiving tilapia from Thailand (Fig. 3). To date, TiLV has been reported from Israel (Eyngor et al. 2014), Ecuador (Bacharach et al. 2016; Ferguson et al. 2014), Colombia (Tsofack et al. 2017), Egypt (Fathi et al. 2017; Nicholson et al. 2017), Philippines (OIE 2017a), Thailand (Dong et al. 2017b; Surachetpong et al. 2017), Chinese Taipei (OIE 2017b), India (Behera et al. 2018), Malaysia (Amal et al. 2018), Indonesia (Koesharyani et al. 2018), Tanzania and Uganda (Mugimba et al. 2018), Mexico (OIE 2018), Peru (Jansen et al. 2019), the USA (Ahasan et al. 2020), and Bangladesh (Chaput et al. 2020; Debnath et al. 2020; Hossain et al. 2020) (Fig. 3). In tilapia farms affected by TiLV, mortality has been reported at rates up to 90% (Dong et al. 2017a, 2017b; Eyngor et al. 2014). In Egypt in 2015, TiLV caused a production loss of approximately 98,000 tonnes resulting in an economic loss of US$ 100 million (Fathi et al. 2017).
Global distributions of reported TiLV. The red (16), orange (32), and yellow (2) colours indicate countries with confirmed TiLV, high and lower risk of TiLV infection, respectively, through tilapia fry and/or fingerlings imported from Thailand. Algeria, Bahrain, Belgium, Burundi, Canada, China, Congo, Germany, Guatemala, Japan, Jordan, Laos, Mozambique, Myanmar, Nigeria, Pakistan, Romania, Rwanda, Saudi Arabia, Singapore, South Africa, Sri Lanka, Switzerland, Togo, Tunisia, Turkey, Turkmenistan, Ukraine, United Arab Emirate, the UK, Vietnam, and Zambia are high-risk countries; El-Salvador and Nepal are lower risk countries (Dong et al. 2017a)
The TiLV isolated from Bangladesh was found phylogenetically closely related to the several isolates of Thailand (Chaput et al. 2020; Debnath et al. 2020). In Bangladesh, TiLV has now been reported from different farms and hatcheries of at least 13 districts (Bagerhat, Barguna, Comilla, Cox's Bazar, Faridpur, Gazipur, Jessore, Khulna, Magura, Mymensingh, Narail, Satkhira, Rajbari) of the 64 districts farming tilapia (Debnath et al. 2020; Hossain et al. 2020). In Bangladesh, mortalities in the TiLV positive farms have been estimated at 50–90% in 2017 and 25–40% in 2019 (Debnath et al. 2020).
For ISKNV, reported mortalities of Nile tilapia and red hybrid tilapia (fry and juvenile) are in the range of 50–75% (Dong et al. 2015b; Figueiredo et al. 2022; Subramaniam et al. 2016; Suebsing et al. 2016). During an ISKNV disease outbreak in tilapia in Ghana (September 2018 to March 2019), morbidity and mortality rates of 60–90% were recorded, with losses of more than 10 tonnes per day in many farms (Ramírez-Paredes et al. 2021). A similar outbreak of ISKNV in the cultured Nile tilapia fish farm was also reported in Brazil in 2020 (Figueiredo et al. 2022).
VNN, first reported in tilapia larvae in France (Bigarré et al. 2009), targets the central nervous system of fish including the brain, spinal cord, and retina, and leads to neuronal necrosis and is reported to affect more than 50 fish species (Machimbirike et al. 2019). During a VNN outbreak in Thailand of farm-raised tilapia larvae, the mortality rate was between 90 and 100% with the affected fish first showing clinical signs of neurological disorder (e.g. loss of balance, corkscrew-like swimming) (Keawcharoen et al. 2015). VNN has also been reported in tilapia fry in Indonesia, but here, no mortality was recorded (Prihartini et al. 2015). In the latest report of VNN from Egypt, mass mortalities (up to 70%) occurred in the hatchery-reared Nile tilapia fry (Taha et al. 2020). This section highlights the huge impacts associated with viral diseases that are now prevalent across major expanses of the world’s most important tilapia aquaculture production sectors. Although there are many fish pathogens responsible for causing disease in aquaculture farms, there is little information available about viruses (except TiLV) causing disease in the aquaculture farms of Bangladesh. The possible reason for this is under-reporting, lack of diagnostic laboratories or support services, and training/costs of diagnostic methods, to do so.
Microbiomes of surface mucosa in tilapia
The fish skin and gills represent major pathways for pathogens to invade fish and thus act as a critical line of defence for maintaining host health by keeping microbes at bay (Glover et al. 2013; Lazado and Caipang 2014; Merrifield and Rodiles 2015). The fish skin and gill epithelial surfaces are coated in a mucus layer containing various immunogenic compounds, including antimicrobial peptides and enzymes, proteins such as immunoglobulins, defensins, lysozyme, proteases, esterase, and mucins (Gomez et al. 2013). Collectively these act as a critical component of the immune response which is active against invading microbes (Merrifield and Rodiles 2015). While it may sound like the defences of the fish’s outer mucosal surfaces represent a hostile environment for microbial growth, they in fact offer an environment of mucin-associated glycans (Barr et al. 2013) that is nutrient-rich in contrast to the relatively sparse surrounding aquatic environment. Niche-adapted microbes that can resist the immunological components are therefore bountiful in the outer mucosal surfaces of fish and form an indigenous commensal microbiota (Legrand et al. 2018; Merrifield and Rodiles 2015). In turn, these commensals act as a further line of immune defence to obstruct pathogen colonization by antagonistic activity as well as competing for adhesion sites and/or nutrients (Legrand et al. 2018; Merrifield and Rodiles 2015). Disruption of the host-microbiome symbiotic relationship may lead to major changes in microbiota community structure in a process referred to as dysbiosis (Dillon and Charnley 2002). Stressful conditions often occur in aquaculture practices and can cause shifts in the microbiota to favour the propagation of opportunistic pathogens that are often ubiquitous, naturally present in the aquatic environment and/or form part of the fish microbiome (Boutin et al. 2013; Califano et al. 2017; Rud et al. 2017). The microbiome and immune system of fish show a direct and wide-ranging connection (Kelly and Salinas 2017; Yu et al. 2021b). For instance, microbiota colonization stimulates neutrophils granulocyte in gnotobiotic zebrafish (Kanther et al. 2014). Meanwhile, in teleosts, secretory immunoglobulins (specifically IgT) are highly induced at mucosal surfaces to coat both pathobionts (potential pathological microorganisms which, under normal situations, live as non-harming symbionts) and commensals (Xu et al. 2013), preventing their intrusion into the blood. Where IgT is depleted, the microbiome becomes dysbiotic with concurrent depletion of IgT targeted microbial taxa, such as short-chain fatty acid-producing bacteria and proliferation of pathobionts associated with the disease. During disease challenges, IgT depletion also renders fish more susceptible to parasitic infection and associated mortality (Xu et al. 2020).
The advent of metabarcoding via next-generation sequencing of conserved ribosomal marker genes (16S for prokaryotes, 18S for eukaryotes, and internal transcribed spacer for fungi) has enabled the development of substantial knowledge of microbial communities comprising microbiomes in the health and disease of humans, and there is now an increasing focus on fish, albeit it much of this, research has focused on the gut microbiomes of high-value aquaculture species (i.e. Atlantic salmon and marine shrimp) due to their role in gut health, and through which feed conversion, growth, and overall productivity can be enhanced (Perry et al. 2020). Various studies however have now shown that the fish skin mucosal surface supports a unique microbial community that is discrete from communities supported by other body sites, such as the gut (Sylvain et al. 2020) and gill (Legrand et al. 2018; Minich et al. 2020b). Niche separation can be partially explained by the distinctive environment of the different mucosal surfaces. For instance, the gill supports specialist ammonia-oxidizing and denitrifying bacteria to clear these toxic metabolites excreted at the gill (van Kessel et al. 2016). The gut is typically associated with specialists such as obligate anaerobic bacteria (Sylvain et al. 2020) and those aiding digestion (Ray et al. 2012); however, the presence of niche-specialized taxa does not preclude the presence of other generalist aerobic taxa shared across body niche sites (de Bruijn et al. 2018). While the gut microbiota exists in a relatively stable and buffered environment, the outer mucosal surfaces of the skin (and gill) are in direct and continuous contact with the water of the culture environment. The microbiota of the skin niche must withstand fluctuating conditions, being largely comprised of generalist bacteria that are strongly influenced by the water microbiota and physicochemistry (Krotman et al. 2020; Sylvain et al. 2020). As such, the microbial communities of water and skin are closely interconnected, with bacterial taxa found in the surrounding water environment also forming a large composition of the skin microbiota. However, the skin microbiota is uniquely structured with specific taxa enrichment (Boutin et al. 2013; McMurtrie et al. 2022).
Environmental influences on the skin microbiome are reflected by inherent variability between fish cultured in different environments; however, conserved microbial hallmarks can be seen in skin microbiomes. Almost every published study, regardless of water salinity, has identified Proteobacteria (Pseudomonadota) as the dominant bacterial phyla of fish skin microbiomes, including tilapia (Boutin et al. 2013; Chiarello et al. 2015; Elsheshtawy et al. 2021; Krotman et al. 2020; Lokesh and Kiron 2016; McMurtrie et al. 2022; Rosado et al. 2019; Sultana et al. 2022); in particular, this comprises the Gammaproteobacteria class (Chiarello et al. 2019; Larsen et al. 2015; McMurtrie et al. 2022; Reinhart et al. 2019; Sylvain et al. 2020). Additionally, the phyla Actinobacteria (Actinomycetota) and Bacteroidetes (Bacteroidota) occur widely (Boutin et al. 2013; Chiarello et al. 2015; Lokesh and Kiron 2016; McMurtrie et al. 2022; Reinhart et al. 2019). Moving beyond coarse, taxonomic ranks can give insight into the occurrence and potential function of microbial taxa associated with specific host species. The biological significance of bacteria designated as core taxa remains unclear; however, their persistence across different environments may reflect functional importance. The two current studies of the tilapia skin microbiomes have identified core/enriched taxa including Aeromonas, Cetobacterium, Comamonadaceae, Clostridium, LD29, Acinetobacter, Vibrio, Exiguobacterium, Plesiomonas, Sphingomonas, Pseudomonas, and Paucibacter (Elsheshtawy et al. 2021; McMurtrie et al. 2022) but their functional roles/importance are largely unknown. Changes in the abundance of members of the Comamonadaceae family (many are unclassified; there are 104 known species) in the tilapia gut microbiome have been associated with aluminium exposure (Yu et al. 2019) and immunostimulatory insulin exposure in Nile tilapia (Wang et al. 2021). Comamonadaceae have also been shown to be negatively correlated with ectoparasitism in the skin microbiome of Prussian carp Carassius gibelio (Kashinskaya et al. 2021).
The genus Cetobacterium, more specifically C. somerae, remains one of the most frequently identified commensals of microbiomes in the skin (and gut and gill) of tilapia (Bereded et al. 2022; Elsaied et al. 2019; Elsheshtawy et al. 2021; McMurtrie et al. 2022). Isolates from freshwater fish guts have been shown to synthesize vitamin B12 (Sugita et al. 1991; Tsuchiya et al. 2008), which is speculated to promote fish health. Experimental monitoring studies suggest Cetobacterium is an essential component of healthy fish gut microbiomes as abundance was significantly depleted in spring viremia of carp virus (SVCV) infected common carp (Cyprinus carpio) (Meng et al. 2021) and in crucian Carp (Carassius auratus) suffering red-operculum disease (Li et al. 2017b). Additionally, the application of probiotics has been shown to increase the abundance of Cetobacterium in the skin and gill of Nile tilapia (Wang et al. 2020), while specific application of the C. somerae XMX-1 fermentation product resulted in improved gut health and disease resilience against Aeromonas veronii/hydrophila challenge (Zhou et al. 2022).
Numerous strains of Aeromonas and Pseudomonas found within tilapia skin microbiomes are widely recognized as opportunistic fish pathogens (Austin and Austin 2016; Crumlish and Austin 2020). Of these, the disease caused by Aeromonas hydrophila is particularly impactful for tilapia aquaculture (Nicholson et al. 2020). However, pathogenicity is strain-specific; therefore, microbiome surveys via metabarcoding using short hypervariable regions cannot assert pathogenic potential. Additionally, numerous Pseudomonas strains and species inhabiting fish-associated microbiomes are true commensals, protecting against invasive infections. For instance, Pseudomonas fluorescens is reported as a pathogen (Liu et al. 2015), yet selected strains have been shown to offer protection for rainbow trout (Oncorhynchus mykiss) against infection by the pathogenic oomycete Saprolegnia parasitica (González-Palacios et al. 2019) and Vibrio anguillarum (Gram et al. 1999). Similar probiotic effects of P. fluorescens have also been reported experimentally in Nile tilapia (Eissa et al. 2014). However, even for these species, the symbiotic function may change in response to host health, environmental conditions, and microbiome dysbiosis.
Given that the fish mucosal layer in the skin and gill surfaces provides the first line of defence against pathogens, characterizing shifts in the composition of these microbiomes can potentially provide insights into what changes are associated with/lead to disease outbreaks. However, the variable nature of the fish skin and gill microbiomes and the wide array of environmental and host factors that can alter them clearly complicates dissecting those key changes and shifts in the microbial assemblages that can be linked with disease onset for disease prediction.
Factors influencing microbiomes of mucosal surfaces in health and disease
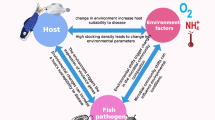
Diseases are often context-dependent and “pathogens” may exist in the culture environment biomes (water, sediment, biofilms) and host-associated microbiomes without ever causing disease. Passage to a disease state is dependent on many biotic and abiotic factors that influence the diversity and composition of fish mucosal microbiomes. Here, we review various factors known to influence the fish skin microbiome, as these factors contribute to the inherent variability of microbiome composition between individuals and may also prove influential for transitions to disease states.
Water quality
The external surface of fish is continuously exposed to water which results in a dynamic relationship between planktonic and skin surface microbial communities. Intensive aquaculture practices in ponds can lead to poor water quality, including build-up of nutrients, organic matter, and toxic products (Avnimelech and Ritvo 2003) such as nitrite (Chen et al. 2017). Furthermore, water quality parameters of aquaculture ponds follow diurnal patterns (Shoko et al. 2014). Given that planktonic pond communities can be strongly influenced by water quality (Casé et al. 2008), it is not surprising that water quality also has a significant impact on the bacterial community of fish skin. For example, in brook trout (Salvelinus fontinalis) experimentally exposed to a mixture of 11 chemical compounds, representing typical hydraulic fracturing waste, after an exposure to an antibiotic (oxytetracycline), recovery varied according to the treatment group and resulted in a shifted microbial community composition, where in particular taxa belonging to Flavobacterium were subsequently more abundant in treatment groups than in the controls (Galbraith et al. 2018). Some of the tested chemicals in that study, such as NaCl and Mg2Cl2, are routinely applied in aquaculture practices. Various water physicochemistry parameters such as water temperature, salinity, pH, and dissolved oxygen are also reported to shape fish skin microbiomes (Krotman et al. 2020). The algal content of ponds, which varies widely in tilapia earthen pond systems is also associated with changes in skin microbiomes, but given its relationship with dissolved oxygen this may be related to changes in the pond physicochemistry more generally (Kunlasak et al. 2013). Nevertheless, a lower alpha diversity of the skin microbiome has been reported in Pacific chub mackerel (Scomber japonicus), under conditions of increasing chlorophyll a concentration with a shift also in skin microbiota community structure (beta diversity) (Minich et al. 2020a).
Geographical location and habitat
Geographic location has widely been reported as one of the strongest predictors of microbial communities of fish skin; however, it is likely much of this may be related to the different environmental parameters of location (McMurtrie et al. 2022; Sylvain et al. 2020), and this, for example, can influence planktonic communities which have a close and dynamic relationship to the skin microbiome. Geographic location has been shown to be a significant and major predictor of skin microbiome structure in European catfish (Silurus glanis) (Chiarello et al. 2019); striped mullet (Mugil cephalus), red snapper (Lutjanus campechanus), spotted seatrout (Cynoscion nebulosus), sand seatrout (Cynoscion arenarius), pinfish (Lagodon rhomboides), and Atlantic croaker (Micropogonias undulatus) (Larsen et al. 2013); flag cichlid (Mesonauta festivus), pacu (Mylossoma duriventre), and black piranha (Serrasalmus rhombeus) (Sylvain et al. 2020); and redbreast tilapia (Coptodon rendalli) and shire tilapia (Oreochromis shiranus) (McMurtrie et al. 2022). Each geographical location is likely to vary in a multitude of environmental factors and habitat features that are influential to skin microbiome processes. As an extreme example of this, a comparison of Atlantic salmon (Salmo salar) from wild and hatchery-reared populations has shown that the skin microbiome of fish from an artificial environment had a lower alpha diversity and uniquely structured beta diversity and exhibited less interindividual variation (Webster et al. 2018). Among different aquaculture systems, Atlantic salmon reared in recirculating aquaculture systems were found to have a greater skin microbiota richness compared to flow through systems (Minich et al. 2020b).
Host
Host-specific factors also govern fish microbial assemblages. These are particularly important for the gastrointestinal microbiome, while environmental factors have greater relevance for the skin mucosal surface (Sylvain et al. 2020). Nevertheless, host-related factors do contribute to the skin microbiome, and in some cases, fish species have been reported as one of the most influential factors (Chiarello et al. 2018; Larsen et al. 2013). Fish skin microbiomes undoubtedly show a high degree of inter-individual variation, even amongst fish of the same species in the same environment (Berggren et al. 2022). Inter-individual variation of skin microbiomes was shown in a seminal study of artificially reared brook charr (Salvelinus fontinalis) (2nd-generation progeny following population cross) where the abundance of select commensals could be directly correlated with host genomic regions through genetic linkage analysis (Boutin et al. 2014). Genetics therefore evidently plays a complementary role, highlighting the breadth and complexity of host associated and environmental processes which govern fish microbiome composition.
Diseases/infections
In stress-free conditions, the balance of microbes in fish microbiomes works to help prevent pathogen entry and causing disease. Indigenous bacteria contained in surface mucosal layers contribute to the host defence by colonization resistance (preventing pathogen growth) (Dillon and Charnley 2002), and via the production and secretion of friction preventing polymers and complex inhibitory antimicrobials (Austin 2006; Krotman et al. 2020; Takeuchi et al. 2021; Tiralongo et al. 2020). However, a change in the host-microbiome symbiotic relationship may lead to a major change/reduction in microbial diversity. A study on the yellowtail kingfish (Seriola lalandi), for example, illustrated an overall reduction of microbial diversity in the skin and gills at disease onset, and furthermore, the emergence of a specific community (pathogenic) was reflected by a change in immune states of the host (Legrand et al. 2018). Another study on Atlantic salmon (Salmo salar L.) challenged with salmonid alphavirus reported a decrease in abundance of proteobacterial taxa and an increase of opportunistic taxa, e.g. Flavobacteriaceae, Streptococcaceae, and Tenacibaculum sp. on the skin samples (Reid et al. 2017). A recent study on striped catfish (Pangasianodon hypophthalmus) challenged with Aeromonas hydrophila similarly showed that pathogen concentration stimulates the skin immune response by altering the mucosal microbiota and increasing the prevalence of opportunistic pathogens (Chen et al. 2022). Li et al. (2017b) reported that during “red-operculum” disease in crucian carp (Carassius auratus), genera belonging to Aeromonas, Vibrio, and Shewanella were dominant while in healthy fish genera from Cetobacterium, Cyanobacterium and Clostridiaceae were more abundant. They also reported that genera present in diseased fishes were absent or rarely present in the environmental samples. An imbalance in the microbial diversity can also allow for the invasion of a secondary pathogen (de Bruijn et al. 2018; Reid et al. 2017). For example, in Atlantic salmon infected with sea lice infection, higher levels of secondary pathogens belonging to Vibrio, Flavobacterium, Tenacibaculum, and Pseudomonas genera ensue (Llewellyn et al. 2017).
While microbial alterations of the fish skin are undoubtedly associated with disease, it is nevertheless often challenging to disentangle whether microbiota shifts are a contributing factor, causal, or a direct consequence of the disease as the plethora of biotic and abiotic factors known to influence fish microbiomes undoubtedly contribute to these disease processes. This indeed may well help to explain when and why disease states arise, in some environments in the presence of a given pathogen, while in other environments, fish remain disease-free. It is furthermore worth emphasizing that there is increasing evidence that disease cannot be attributed to single aetiological agents, and disease progression is highly dependent on the collective community of microorganisms in a system (Bass et al. 2019). The one pathogen–one disease model is moving towards the pathobiome concept, where disease in the animal is now seen as the result of interactions between host-associated prokaryotes, eukaryotes, and viruses, their host, and the environment (Bass et al. 2019). Many multi-faceted diseases and syndromes are now emerging, for instance in shrimp (Kooloth Valappil et al. 2021), further emphasizing the importance of understanding the microbiomes of aquaculture species and their environment.
Effects of antibiotics on fish microbiomes
Antibiotics are used extensively to fight bacterial infections in humans or animal hosts. In aquaculture, their use is often indiscriminate, particularly in LMICs where there is very limited regulation (Thornber et al. 2020). In livestock/food-animal, an estimated 118,940 tonnes (95% confidence interval [CI] 91,455 to 172,811 tonnes) was consumed in 2013 and it is projected that around 181,650 tonnes (95% CI 136,847 to 269,465 tonnes) will be consumed by 2030 (Sriram et al. 2021; Van Boeckel et al. 2017). In 2018, the total global antibiotic consumption was 40.1 billion (95% uncertainty interval [UI] 37.2 to 43.7 billion) defined daily doses (DDD) and although the consumption rate between 2000 and 2018 remained relatively stable in high-income countries, there was a 76% increase in LMICs, South Asia (India, Pakistan, Nepal, Bangladesh, and Bhutan) where over a quarter (25·2%) of all antibiotics were consumed (Browne et al. 2021). Misuse of antibiotics has received a high profile due to the risk of antimicrobial resistance (AMR) which is estimated to result in the deaths of 0.7 million people each year (O'Neill 2016; Willyard 2017), but although the uncontrolled use of antibiotics in aquaculture (especially in LMICs) may spread AMR, this has not been well studied (Preena et al. 2020). Furthermore, beyond the long-term implications of AMR, misuse of antibiotics may have immediate implications for microbiomes associated with aquaculture as antibiotics can target commensal bacteria that support fish health, immune function, and disease resilience. Antibiotics may thus reduce the microbial community mass and/or diversity, alter community composition, and/or change the enzymatic functions (Carlson et al. 2017; Ferrer et al. 2017).
The most common ways for the administration of antibiotics in aquaculture in many countries, including Bangladesh, are by medicated feed and direct introduction into the pond water (Ali et al. 2016; Shah et al. 2014; Song et al. 2016). Antibiotics are often poorly absorbed into the body, highly water soluble, and resistant to degradation; therefore, antibiotic residues are widely detected in aquatic environments (Da Le et al. 2021; Fu et al. 2022; Hossain et al. 2017; Limbu et al. 2018; Liu et al. 2021; Sotto et al. 2017). Illustrating this, the widely used antibiotics oxytetracycline and sulfamethoxazole have been measured in rivers in China at concentrations between 174.9 ± 266.9 and 741.85 ng/L (Bai et al. 2014; Dong et al. 2016) and between 259.60 and 385.00 ng/L (Chen and Zhou 2014), respectively, and the antibiotic ciprofloxacin has been recorded in the surface water in India up to 635 ng/L (Singh et al. 2019). In a recent study of surface waters from urban and rural areas of Bangladesh, eight antibiotics were detected with the highest concentration of 1407 ng/L (ciprofloxacin), 909 ng/L (azithromycin), 728 ng/L (SMX), 205 ng/L (clarithromycin), 153 ng/L (trimethoprim), 138 ng/L (acetyl sulfamethoxazole), 77 ng/L (anhydroerythromycin), and 22 ng/L (sulfadiazine) (Angeles et al. 2020). Many aquaculture farms are located in areas adjacent to rivers that are also often used as water sources for aquaculture activities (Hossain et al. 2018). Similarly, in the surface water of the old Brahmaputra River, which is widely used for open-water-fed aquaculture, a wide range of antibiotics have been measured at a concentration of up to 13.51 ng/L for metronidazole, 17.20 ng/L for trimethoprim, 11.35 ng/L for sulfonamides, 16.68 ng/L for macrolides, and 8.80 ng/L for carbamazepine (Hossain et al. 2018). Within finfish aquaculture ponds in Bangladesh sulfamethoxazole, trimethoprim, tylosin, sulfadiazine, sulfamethazine, sulfamethizole and penicillin G have been reported at a concentration of up to 20.02 ng/L, 41.67 ng/L, 39.34 ng/L, 17.97 ng/L, 11.71 ng/L, 10.81 ng/L, 7.83 ng/L, respectively (Hossain et al. 2017). Specifically, in tilapia farms in Bangladesh, sulfadiazine, sulfamethoxazole, tylosin, and trimethoprim have been reported at concentrations of up to 17.97 ng/L, 14.46 ng/L, 4.87 ng/L, and 1.57 ng/L, respectively (Hossain et al. 2017). During monsoon periods, in many of the major freshwater finfish aquaculture areas in Asia, fish pond waters mix with river water and this may further increase the spread of antibiotic-resistance genes to and from aquaculture systems. Importantly, some antibiotics can bioaccumulate in animal tissues (Carnevali et al. 2017), exacerbating the likely development of antibiotic-resistant bacteria that can pass into the food chain (Banerjee and Ray 2017; Fu et al. 2017). It is estimated that 75% of the antibiotics administrated through feed in aquaculture enter the aquatic environment through leaching from the uneaten food and excretion of the cultured species (Hossain et al. 2017; Lalumera et al. 2004).
Total global antibiotic use in aquaculture in 2017 was estimated at 9307 tonnes (95% uncertainty interval [UI] 2869 to 40,576 tonnes) and this is predicted to rise to 11,793 tonnes (UI 3804 to 53,792 tonnes) in 2030 (Schar et al. 2020; Sriram et al. 2021). Antibiotic use in aquaculture varies depending on fish species, geographics, type of infection, and country-specific legal requirements. Suggested doses, when mixed with food for oxytetracycline and sulfamethoxazole in aquaculture, are 50–250 mg/kg of fish body weight per day for 3 to 21 days (Enis Yonar et al. 2011; Limbu et al. 2018) and 100–200 mg/kg of fish body weight per day for 5 days (Liu et al. 2017), respectively. Disease conditions, however, often cause a loss of appetite in fish (Miranda et al. 2018); therefore, much of the antibiotic dose is released directly into the aquaculture pond. Furthermore, antibiotic administration is not targeted, resulting in animals of mixed health status (diseased and non-diseased fish) being treated with antibiotics, giving rise to potential microbial dysbiosis which may render the culture organism more susceptible to disease. Experimental studies have confirmed this theory, with grass carp (Ctenopharyngodon idella) subjected to antibiotic-induced intestinal dysbiosis more susceptible to disease after challenge with Aeromonas hydrophila (Sun et al. 2022).
In fish, antibiotic exposure has also been experimentally shown to exert selective pressures on skin, gill, and gut microbiomes. Dietary doses of oxytetracycline (100 mg/kg) (Payne et al. 2021) and sulfamonomethoxine (200 and 300 mg/kg) (Ming et al. 2020) given to Nile tilapia were found to cause minimal changes to alpha and beta diversity metrics of gut microbial communities; however, these exposures were not found to cause altered abundance in taxa for genera containing reputed pathogens. Further experimental evidence, however, has shown perturbation of skin and gut microbiomes, with enrichment of potentially opportunistic pathogens, following oral exposure of yellowtail kingfish Seriola lalandi to an antibiotic cocktail of oxytetracycline (200 mg/kg), erythromycin (50 mg/kg), and metronidazole (50 mg/kg). Interestingly, this study also suggests that the skin microbiome of fish is more resilient to antibiotic exposure than the gut microbiome, with a faster return to a “normal” microbial community assemblage (Legrand et al. 2020). Long-term exposure to oxytetracycline and sulfamethoxazole has been shown to adversely affect the growth, metabolism, immunity, and intestinal morphology of Nile tilapia (Limbu et al. 2018), although it is unclear to what degree these health impacts are mediated by changes via the microbiome.
Antibiotic exposure in farmed fish also poses the risk of antimicrobial resistant gene (ARGs) selection in the resistome of fish microbiomes (Sáenz et al. 2019), fish faeces (Muziasari et al. 2017), sediment (Muziasari et al. 2016), and water (Patil et al. 2020). Controlled antibiotic exposure of Piaractus mesopotamicus to florfenicol (9.7 mg/kg) led to an increased abundance and diversity of ARGs in the fish gut microbiome, together with enrichment of Enterobacteriaceae whose genomes were associated with ARGs, including reputed nosocomial infection causing genera Citrobacter, Klebsiella, and Plesiomonas. Interestingly, antibiotic exposure also increased the co-occurrence of ARGs with mobile genetic elements, particularly transposases and phage integrases, whose abundances increased and were found to flank several ARGs (Sáenz et al. 2019). Furthermore, the sediment below the European aquaculture cage culture with a history of antibiotic use was found to be enriched in mobile genetic elements, including transposons and integrons (Muziasari et al. 2016). Together this highlights the potential for ARG spread in fish microbiomes and the wider aquaculture environment, raising the risk of AMR dissemination.
Final considerations and future research needs
Aquaculture will inevitably play a very major role in future global food security and not least for LIFD countries. Low trophic feeding finfish such as tilapia will feature strongly in the future expansion/intensification of freshwater aquaculture not least for their ability to tolerate less favourable water conditions, adaptability more generally, good nutritious qualities, and overall robustness against pathogens. However, the emergence and increasing prevalence of a wide range of diseases are limiting the future growth of the aquaculture industry with major economic consequences. This is especially the case in countries such as Bangladesh, where most production occurs in open earthen ponds in rural communities with minimal biosecurity. Research and support are urgently required to combat emerging diseases, and this requires more fundamental knowledge of disease prevalence, propagation, and pathogenesis to support the development of specific treatments and vaccines. Fundamentally it also requires the adoption of effective management and biosecurity plans, which included increasing farmer education around the management of diseases and misuse of antimicrobials.
Pertinent to these outlined action areas for aquaculture security is a need to holistically understand the functional microbial systems that interface between the pond environment and cultured fish. This in turn can help in optimizing conditions for fish growth, health, and disease avoidance. Understanding microbiomes and how they can be augmented for host protection against disease could play a very significant role in the further development of aquaculture, especially as production is intensified. Healthy skin and gill mucosal surfaces in fish are naturally colonized by pathogens (Rosado et al. 2019) that form an important part of a host’s microbiome (Califano et al. 2017; Rud et al. 2017). Generally, however, the skin microbiome in unstressed fish is dominated by taxa with probiotic and antimicrobial activity (Rosado et al. 2019), and therefore, they often do not cause disease (de Bruijn et al. 2018). In order to develop a deep and meaningful understanding of microbiomes in the mucosal surfaces of fish and how they function to prevent disease, we need to better understand the microbial assemblages of these mucosal surfaces and how they function. Next-generation sequencing has allowed for high throughput analysis of the bacterial community structure and dynamics, but less attention has been applied to assessing the functions of the microbiomes and very little attention has been directed to the viruses and bacteriophages in these mucosal communities that are likely to play a crucial rule. Given the important roles played by environmental and host-related factors in influencing mucosal microbiomes of fish, these factors too also need to be more thoroughly considered for understanding the passage of fish from healthy to diseased states.
A deeper understanding of mucosal microbiomes associated with health and disease should furthermore allow for the development and application of more effective probiotics for finfish aquaculture. Probiotics offer the opportunity to optimize health, in turn reducing the reliance on antibiotics. A combined approach to investigating the role of the microbiome during probiotic (or prebiotic) application might help improve their efficacy. Finally, a critical need in the development of microbiome research is in the identification of conserved biomarkers of the mucosal surface that can act as signals for changing health status to help in our understanding relating to disease prevention in tilapia and other fish culture practices.
Data availability
Not applicable.
References
Ahasan MS, Keleher W, Giray C, Perry B, Surachetpong W, Nicholson P, Al-Hussinee L, Subramaniam K, Waltzek Thomas B, Matthijnssens J (2020) Genomic characterization of tilapia lake virus isolates recovered from moribund Nile tilapia (Oreochromis niloticus) on a farm in the United States. Microbiol Resour Announce 9(4):e01368-e1419. https://doi.org/10.1128/MRA.01368-19
Ahern MB, Thilsted SH, Kjellevold M, Overå R, Toppe J, Doura M, Kalaluka E, Wismen B, Vargas M, Franz N (2021) Locally-procured fish is essential in school feeding programmes in sub-Saharan Africa. Foods 10(9). https://doi.org/10.3390/foods10092080
Ahmed N, Young JA, Dey MM, Muir JF (2012) From production to consumption: a case study of tilapia marketing systems in Bangladesh. Aquacult Int 20(1):51–70. https://doi.org/10.1007/s10499-011-9441-0
Ali H, Rico A, Murshed-e-Jahan K, Belton B (2016) An assessment of chemical and biological product use in aquaculture in Bangladesh. Aquaculture 454:199–209. https://doi.org/10.1016/j.aquaculture.2015.12.025
Ali SE, Gamil AAA, Skaar I, Evensen Ø, Charo-Karisa H (2019) Efficacy and safety of boric acid as a preventive treatment against Saprolegnia infection in Nile tilapia (Oreochromis niloticus). Sci Rep 9(1):18013. https://doi.org/10.1038/s41598-019-54534-y
Amal MNA, Koh CB, Nurliyana M, Suhaiba M, Nor-Amalina Z, Santha S, Diyana-Nadhirah KP, Yusof MT, Ina-Salwany MY, Zamri-Saad M (2018) A case of natural co-infection of Tilapia Lake Virus and Aeromonas veronii in a Malaysian red hybrid tilapia (Oreochromis niloticus×O. mossambicus) farm experiencing high mortality. Aquaculture 485:12–16. https://doi.org/10.1016/j.aquaculture.2017.11.019
Anderson JL, Asche F, Garlock T, Chu J (2017) Aquaculture: its role in the future of food. World agricultural resources and food security (Frontiers of Economics and Globalization), vol. 17. Emerald Publishing Limited, Bingley, pp 159–173. https://doi.org/10.1108/S1574-871520170000017011
Angeles LF, Islam S, Aldstadt J, Saqeeb KN, Alam M, Khan MA, Johura F-T, Ahmed SI, Aga DS (2020) Retrospective suspect screening reveals previously ignored antibiotics, antifungal compounds, and metabolites in Bangladesh surface waters. Sci Total Environ 712:136285. https://doi.org/10.1016/j.scitotenv.2019.136285
Ariel E, Owens L (1997) Epizootic mortalities in tilapia Oreochromis mossambicus. Dis Aquat Org 29(1):1–6. https://doi.org/10.3354/dao029001
Arthur RI, Skerritt DJ, Schuhbauer A, Ebrahim N, Friend RM, Sumaila UR (2022) Small-scale fisheries and local food systems: transformations, threats and opportunities. Fish Fish 23(1):109–124. https://doi.org/10.1111/faf.12602
Assefa A, Abunna F (2018) Maintenance of fish health in aquaculture: review of epidemiological approaches for prevention and control of infectious disease of fish. Vet Med Int 2018:5432497. https://doi.org/10.1155/2018/5432497
Attia MM, Elgendy MY, Prince A, El-Adawy MM, Abdelsalam M (2021) Morphomolecular identification of two trichodinid coinfections (Ciliophora: Trichodinidae) and their immunological impacts on farmed Nile Tilapia. Aquacult Res 52(9):4425–4433. https://doi.org/10.1111/are.15281
Austin B (2006) The bacterial microflora of fish, revised. Sci World J 6:931–945. https://doi.org/10.1100/tsw.2006.181
Austin B, Austin DA (2016) Pseudomonads. In: Austin B, Austin DA (eds) Bacterial fish pathogens: disease of farmed and wild fish. Springer International Publishing, Cham, pp 475–498
Avnimelech Y, Ritvo G (2003) Shrimp and fish pond soils: processes and management. Aquaculture 220(1):549–567. https://doi.org/10.1016/S0044-8486(02)00641-5
Bacharach E, Mishra N, Briese T, Zody MC, Kembou Tsofack JE, Zamostiano R, Berkowitz A, Ng J, Nitido A, Corvelo A, Toussaint NC, Abel Nielsen SC, Hornig M, Del Pozo J, Bloom T, Ferguson H, Eldar A, Lipkin WI (2016) Characterization of a novel orthomyxo-like virus causing mass die-offs of tilapia. mBio 7(2):e00431-16. https://doi.org/10.1128/mBio.00431-16
Bai Y, Meng W, Xu J, Zhang Y, Guo C (2014) Occurrence, distribution and bioaccumulation of antibiotics in the Liao River Basin in China. Environ Sci Process Impacts 16(3):586–593. https://doi.org/10.1039/c3em00567d
Banerjee G, Ray AK (2017) The advancement of probiotics research and its application in fish farming industries. Res Vet Sci 115:66–77. https://doi.org/10.1016/j.rvsc.2017.01.016
Baqui MA, Bhujel RC (2011) A hands-on training helped proliferation of tilapia culture in Bangladesh. In: Liping L, Fitzsimmons K (eds) Better Science, Better Fish, Better Life proceedings of the ninth international symposium on tilapia in aquaculture, Shanghai, China, 2011:311–322
Barnes AC, Silayeva O, Landos M, Dong HT, Lusiastuti A, Phuoc LH, Delamare-Deboutteville J (2022) Autogenous vaccination in aquaculture: a locally enabled solution towards reduction of the global antimicrobial resistance problem. Rev Aquac 14(2):907–918. https://doi.org/10.1111/raq.12633
Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, Stotland A, Wolkowicz R, Cutting AS, Doran KS, Salamon P, Youle M, Rohwer F (2013) Bacteriophage adhering to mucus provide a non–host-derived immunity. Proc Natl Acad Sci 110(26):10771–10776. https://doi.org/10.1073/pnas.1305923110
Barría A, Trịnh TQ, Mahmuddin M, Peñaloza C, Papadopoulou A, Gervais O, Chadag VM, Benzie JAH, Houston RD (2021) A major quantitative trait locus affecting resistance to Tilapia lake virus in farmed Nile tilapia (Oreochromis niloticus). Heredity 127(3):334–343. https://doi.org/10.1038/s41437-021-00447-4
Barroso RM, Muñoz AEP, Cai J (2019) Social and economic performance of tilapia farming in Brazil. FAO Fisheries and Aquaculture Circular No. 1181. Rome
Bass D, Stentiford GD, Wang HC, Koskella B, Tyler CR (2019) The pathobiome in animal and plant diseases. Trends Ecol Evol 34(11):996–1008. https://doi.org/10.1016/j.tree.2019.07.012
Behera BK, Pradhan PK, Swaminathan TR, Sood N, Paria P, Das A, Verma DK, Kumar R, Yadav MK, Dev AK, Parida PK, Das BK, Lal KK, Jena JK (2018) Emergence of tilapia lake virus associated with mortalities of farmed Nile tilapia Oreochromis niloticus (Linnaeus 1758) in India. Aquaculture 484:168–174. https://doi.org/10.1016/j.aquaculture.2017.11.025
Béné C, Barange M, Subasinghe R, Pinstrup-Andersen P, Merino G, Hemre G-I, Williams M (2015) Feeding 9 billion by 2050 – putting fish back on the menu. Food Sec 7(2):261–274. https://doi.org/10.1007/s12571-015-0427-z
Béné C, Arthur R, Norbury H, Allison EH, Beveridge M, Bush S, Campling L, Leschen W, Little D, Squires D, Thilsted SH, Troell M, Williams M (2016) Contribution of fisheries and aquaculture to food security and poverty reduction: assessing the current evidence. World Dev 79:177–196. https://doi.org/10.1016/j.worlddev.2015.11.007
Bereded NK, Abebe GB, Fanta SW, Curto M, Waidbacher H, Meimberg H, Domig KJ (2022) The gut bacterial microbiome of Nile tilapia (Oreochromis niloticus) from lakes across an altitudinal gradient. BMC Microbiol 22(1):87. https://doi.org/10.1186/s12866-022-02496-z
Berggren H, Tibblin P, Yıldırım Y, Broman E, Larsson P, Lundin D, Forsman A (2022) Fish skin microbiomes are highly variable among individuals and populations but not within individuals. Front Microbiol 12 https://doi.org/10.3389/fmicb.2021.767770
Bigarré L, Cabon J, Baud M, Heimann M, Body A, Lieffrig F, Castric J (2009) Outbreak of betanodavirus infection in tilapia, Oreochromis niloticus (L.), in fresh water. J Fish Dis 32(8):667–673. https://doi.org/10.1111/j.1365-2761.2009.01037.x
Boutin S, Bernatchez L, Audet C, Derôme N (2013) Network analysis highlights complex interactions between pathogen, host and commensal microbiota. PLOS One 8(12):e84772. https://doi.org/10.1371/journal.pone.0084772
Boutin S, Sauvage C, Bernatchez L, Audet C, Derome N (2014) Inter individual variations of the fish skin microbiota: host genetics basis of mutualism? PLOS One 9(7):e102649. https://doi.org/10.1371/journal.pone.0102649
Browne AJ, Chipeta MG, Haines-Woodhouse G, Kumaran EPA, Hamadani BHK, Zaraa S, Henry NJ, Deshpande A, Reiner RC, Day NPJ, Lopez AD, Dunachie S, Moore CE, Stergachis A, Hay SI, Dolecek C (2021) Global antibiotic consumption and usage in humans, 2000–18: a spatial modelling study. Lancet Planet Health 5(12):e893–e904. https://doi.org/10.1016/S2542-5196(21)00280-1
de Bruijn I, Liu Y, Wiegertjes GF, Raaijmakers JM (2018) Exploring fish microbial communities to mitigate emerging diseases in aquaculture. FEMS Microbiol Ecol 94(1) https://doi.org/10.1093/femsec/fix161
Califano G, Castanho S, Soares F, Ribeiro L, Cox CJ, Mata L, Costa R (2017) Molecular taxonomic profiling of bacterial communities in a gilthead seabream (Sparus aurata) hatchery. Front Microbiol 8(204) https://doi.org/10.3389/fmicb.2017.00204
Carlson JM, Leonard AB, Hyde ER, Petrosino JF, Primm TP (2017) Microbiome disruption and recovery in the fish Gambusia affinis following exposure to broad-spectrum antibiotic. Infect Drug Resist 10:143–154. https://doi.org/10.2147/IDR.S129055
Carnevali O, Maradonna F, Gioacchini G (2017) Integrated control of fish metabolism, wellbeing and reproduction: The role of probiotic. Aquaculture 472:144–155. https://doi.org/10.1016/j.aquaculture.2016.03.037
Casé M, Leça EE, Leitão SN, Sant′Anna EE, Schwamborn R, de Moraes Junior AT (2008) Plankton community as an indicator of water quality in tropical shrimp culture ponds. Mar Pollut Bull 56(7):1343–1352. https://doi.org/10.1016/j.marpolbul.2008.02.008
Chaput DL, Bass D, Alam MM, Al Hasan N, Stentiford GD, van Aerle R, Moore K, Bignell JP, Haque MM, Tyler CR (2020) The segment matters: probable reassortment of tilapia lake virus (TiLV) complicates phylogenetic analysis and inference of geographical origin of new isolate from Bangladesh. Viruses 12(3):258. https://doi.org/10.3390/v12030258
Chen K, Zhou JL (2014) Occurrence and behavior of antibiotics in water and sediments from the Huangpu River, Shanghai, China. Chemosphere 95:604–612. https://doi.org/10.1016/j.chemosphere.2013.09.119
Chen M, Li L-P, Wang R, Liang W-W, Huang Y, Li J, Lei A-Y, Huang W-Y, Gan X (2012) PCR detection and PFGE genotype analyses of streptococcal clinical isolates from tilapia in China. Vet Microbiol 159(3):526–530. https://doi.org/10.1016/j.vetmic.2012.04.035
Chen L-H, Lin C-H, Siao R-F, Wang L-C (2022) Aeromonas hydrophila induces skin disturbance through mucosal microbiota dysbiosis in striped catfish (Pangasianodon hypophthalmus). mSphere 7(4):e00194-22. https://doi.org/10.1128/msphere.00194-22
Chen R, Deng M, He X, Hou J (2017) Enhancing Nitrate Removal from Freshwater Pond by Regulating Carbon/Nitrogen Ratio. Front Microbiol 8 https://doi.org/10.3389/fmicb.2017.01712
Chiarello M, Auguet J-C, Bettarel Y, Bouvier C, Claverie T, Graham NAJ, Rieuvilleneuve F, Sucré E, Bouvier T, Villéger S (2018) Skin microbiome of coral reef fish is highly variable and driven by host phylogeny and diet. Microbiome 6(1):147. https://doi.org/10.1186/s40168-018-0530-4
Chiarello M, Paz-Vinas I, Veyssière C, Santoul F, Loot G, Ferriol J, Boulêtreau S (2019) Environmental conditions and neutral processes shape the skin microbiome of European catfish (Silurus glanis) populations of Southwestern France. Environ Microbiol Rep 11(4):605–614. https://doi.org/10.1111/1758-2229.12774
Chiarello M, Villéger S, Bouvier C, Bettarel Y, Bouvier T (2015) High diversity of skin-associated bacterial communities of marine fishes is promoted by their high variability among body parts, individuals and species. FEMS Microbiol Ecol 91(7):fiv061 https://doi.org/10.1093/femsec/fiv061
Crumlish M, Austin B (2020) Aeromoniosis (Aeromonas salmonicida). In: Woo PTK, Leong JA, Buchmann K (eds) Climate change and infectious fish diseases. Oxfordshire Boston, Wallingford, p 211–234. https://doi.org/10.1079/9781789243277.0211
Da Le N, Hoang AQ, Hoang TTH, Nguyen TAH, Duong TT, Pham TMH, Nguyen TD, Hoang VC, Phung TXB, Le HT, Tran CS, Dang TH, Vu NT, Nguyen TN, Le TPQ (2021) Antibiotic and antiparasitic residues in surface water of urban rivers in the Red River Delta (Hanoi, Vietnam): concentrations, profiles, source estimation, and risk assessment. Environ Sci Pollut R 28(9):10622–10632. https://doi.org/10.1007/s11356-020-11329-3
Debnath PP, Delamare-Deboutteville J, Jansen MD, Phiwsaiya K, Dalia A, Hasan MA, Senapin S, Mohan CV, Dong HT, Rodkhum C (2020) Two-year surveillance of tilapia lake virus (TiLV) reveals its wide circulation in tilapia farms and hatcheries from multiple districts of Bangladesh. J Fish Dis 43(11):1381–1389. https://doi.org/10.1111/jfd.13235
Debnath PP, Dinh-Hung N, Taengphu S, Nguyen VV, Delamare-Deboutteville J, Senapin S, Vishnumurthy Mohan C, Dong HT, Rodkhum C (2022) Tilapia Lake Virus was not detected in non-tilapine species within tilapia polyculture systems of Bangladesh. J Fish Dis 45(1):77–87. https://doi.org/10.1111/jfd.13537
Declercq AM, Haesebrouck F, Van den Broeck W, Bossier P, Decostere A (2013) Columnaris disease in fish: a review with emphasis on bacterium-host interactions. Vet Res 44(1):27. https://doi.org/10.1186/1297-9716-44-27
Deines AM, Wittmann ME, Deines JM, Lodge DM (2016) Tradeoffs among ecosystem services associated with global tilapia introductions. Rev Fish Sci Aquac 24(2):178–191 https://doi.org/10.1080/23308249.2015.1115466
Dillon R, Charnley K (2002) Mutualism between the desert locust Schistocerca gregaria and its gut microbiota. Res Microbiol 153(8):503–509. https://doi.org/10.1016/S0923-2508(02)01361-X
DoF (2020) Yearbook of Fisheries Statistics of Bangladesh, 2019–20. Fisheries Resources Survey System (FRSS). vol 37. Department of Fisheries. Bangladesh: Ministry of Fisheries and Livestock, p 141
Dong HT, LaFrentz B, Pirarat N, Rodkhum C (2015) Phenotypic characterization and genetic diversity of Flavobacterium columnare isolated from red tilapia, Oreochromis sp. inThailand. J Fish Dis 38(10):901–913. https://doi.org/10.1111/jfd.12304
Dong HT, Nguyen VV, Le HD, Sangsuriya P, Jitrakorn S, Saksmerprome V, Senapin S, Rodkhum C (2015) Naturally concurrent infections of bacterial and viral pathogens in disease outbreaks in cultured Nile tilapia (Oreochromis niloticus) farms. Aquaculture 448:427–435. https://doi.org/10.1016/j.aquaculture.2015.06.027
Dong D, Zhang L, Liu S, Guo Z, Hua X (2016) Antibiotics in water and sediments from Liao River in Jilin Province, China: occurrence, distribution, and risk assessment. Environ Earth Sci 75(1202):1–10. https://doi.org/10.1007/s12665-016-6008-4
Dong HT, Ataguba GA, Khunrae P, Rattanarojpong T, Senapin S (2017) Evidence of TiLV infection in tilapia hatcheries from 2012 to 2017 reveals probable global spread of the disease. Aquaculture 479:579–583. https://doi.org/10.1016/j.aquaculture.2017.06.035
Dong HT, Siriroob S, Meemetta W, Santimanawong W, Gangnonngiw W, Pirarat N, Khunrae P, Rattanarojpong T, Vanichviriyakit R, Senapin S (2017) Emergence of tilapia lake virus in Thailand and an alternative semi-nested RT-PCR for detection. Aquaculture 476:111–118. https://doi.org/10.1016/j.aquaculture.2017.04.019
Dong HT, Senapin S, Jeamkunakorn C, Nguyen VV, Nguyen NT, Rodkhum C, Khunrae P, Rattanarojpong T (2019) Natural occurrence of edwardsiellosis caused by Edwardsiella ictaluri in farmed hybrid red tilapia (Oreochromis sp.) in Southeast Asia. Aquaculture 499:17–23. https://doi.org/10.1016/j.aquaculture.2018.09.007
Eissa N, Abou El-Gheit N, Shaheen AA (2014) Protective effect of Pseudomonas fluorescens as a probiotic in controlling fish pathogens. Am J BioSci 2(5):175–181 https://doi.org/10.11648/j.ajbio.20140205.12
Elgendy M, Moustafa M, Gaafar A, Ibrahim T (2015) Impacts of extreme cold water conditions and some bacterial infections on earthen-pond cultured Nile tilapia, Oreochromis niloticus. Res J Pharm Biol Chem Sci 6(1):136–145
Elgendy MY, Abdelsalam M, Kenawy AM, Ali SE (2022) Vibriosis outbreaks in farmed Nile tilapia (Oreochromis niloticus) caused by Vibrio mimicus and V. cholerae. Aquacult Int https://doi.org/10.1007/s10499-022-00921-8
El-Gohary MS, El Gamal AM, Atia AA, El-Dakroury MF (2020) Treatment trial of Nile tilapia (Oreochromis niloticus) experimentally infected with Vibrio alginolyticus isolated from sea bass (Dicentrarchus labrax). Pak J Biol Sci 23(12):1591–1600. https://doi.org/10.3923/pjbs.2020.1591.1600
Elsaied HE, Soliman T, Abu-Taleb HT, Goto H, Jenke-Kodam H (2019) Phylogenetic characterization of eukaryotic and prokaryotic gut flora of Nile tilapia, Oreochromis niloticus, along niches of Lake Nasser, Egypt, based on rRNA gene high-throughput sequences. Ecol Genet Genom 11:100037 https://doi.org/10.1016/j.egg.2019.100037
El-Sayed A-FM (2020) Chapter 9 - stress and diseases. In: El-Sayed A-FM (ed) Tilapia culture, 2nd edn. Academic Press, p 205–243. https://doi.org/10.1016/B978-0-12-816509-6.00009-4
Elsheshtawy A, Clokie BGJ, Albalat A, Beveridge A, Hamza A, Ibrahim A, MacKenzie S (2021) Characterization of external mucosal microbiomes of Nile tilapia and grey mullet co-cultured in semi-intensive pond systems. Front Microbiol 12 https://doi.org/10.3389/fmicb.2021.773860
El-Yazeed HA, Ibrahem MD (2009) Studies on Edwardsiella tarda infection in catfish and Tilapia nilotica. J Vet Med Res 19(1):44–50 https://doi.org/10.21608/jvmr.2009.77808
Enis Yonar M, Mişe Yonar S, Silici S (2011) Protective effect of propolis against oxidative stress and immunosuppression induced by oxytetracycline in rainbow trout (Oncorhynchus mykiss, W.). Fish Shellfish Immunol 31(2):318–325 https://doi.org/10.1016/j.fsi.2011.05.019
Eyngor M, Zamostiano R, Tsofack JEK, Berkowitz A, Bercovier H, Tinman S, Lev M, Hurvitz A, Galeotti M, Bacharach E, Eldar A (2014) Identification of a novel RNA virus lethal to tilapia. J Clin Microbiol 52(12):4137–4146. https://doi.org/10.1128/JCM.00827-14
FAO (2020) The State of World Fisheries and Aquaculture. Sustainability in action. The State of the World, Rome. https://doi.org/10.4060/ca9229en
FAO (2022) The State of World Fisheries and Aquaculture 2022. Towards Blue Transformation. The State of World Fisheries and Aquaculture (SOFIA). FAO, Rome, Italy. p 266 https://doi.org/10.4060/cc0461en
Fathi M, Dickson C, Dickson M, Leschen W, Baily J, Muir F, Ulrich K, Weidmann M (2017) Identification of Tilapia Lake Virus in Egypt in Nile tilapia affected by ‘summer mortality’ syndrome. Aquaculture 473:430–432. https://doi.org/10.1016/j.aquaculture.2017.03.014
Ferguson HW, Kabuusu R, Beltran S, Reyes E, Lince JA, del Pozo J (2014) Syncytial hepatitis of farmed tilapia, Oreochromis niloticus (L.): a case report. J Fish Dis 37(6):583–589 https://doi.org/10.1111/jfd.12142
Ferrer M, Méndez-García C, Rojo D, Barbas C, Moya A (2017) Antibiotic use and microbiome function. Biochem Pharmacol 134:114–126. https://doi.org/10.1016/j.bcp.2016.09.007
Figueiredo HCP, Klesius PH, Arias CR, Evans J, Shoemaker CA, Pereira DJ Jr, Peixoto MTD (2005) Isolation and characterization of strains of Flavobacterium columnare from Brazil. J Fish Dis 28(4):199–204. https://doi.org/10.1111/j.1365-2761.2005.00616.x
Figueiredo HCP, Tavares GC, Dorella FA, Rosa JCC, Marcelino SAC, Pierezan F, Pereira FL (2022) First report of infectious spleen and kidney necrosis virus in Nile tilapia in Brazil. Transbound Emerg Dis 69(5):3008–3015. https://doi.org/10.1111/tbed.14217
Fitzsimmons K (2015) Market stability: Why Tilapia supply and demand have avoided the boom and busts of other commodities. In: 4th International Trade and Technical Conference and position on Tilapia. Kuala Lumpur, Malaysia
Fu J, Yang D, Jin M, Liu W, Zhao X, Li C, Zhao T, Wang J, Gao Z, Shen Z, Qiu Z, Li J-W (2017) Aquatic animals promote antibiotic resistance gene dissemination in water via conjugation: role of different regions within the zebra fish intestinal tract, and impact on fish intestinal microbiota. Mol Ecol 26(19):5318–5333. https://doi.org/10.1111/mec.14255
Fu C, Xu B, Chen H, Zhao X, Li G, Zheng Y, Qiu W, Zheng C, Duan L, Wang W (2022) Occurrence and distribution of antibiotics in groundwater, surface water, and sediment in Xiong’an New Area, China, and their relationship with antibiotic resistance genes. Sci Total Environ 807:151011. https://doi.org/10.1016/j.scitotenv.2021.151011
Galbraith H, Iwanowicz D, Spooner D, Iwanowicz L, Keller D, Zelanko P, Adams C (2018) Exposure to synthetic hydraulic fracturing waste influences the mucosal bacterial community structure of the brook trout (Salvelinus fontinalis) epidermis. AIMS Microbiology 4(3):413. https://doi.org/10.3934/microbiol.2018.3.413
Glover CN, Bucking C, Wood CM (2013) The skin of fish as a transport epithelium: a review. J Comp Physiol B 183(7):877–891. https://doi.org/10.1007/s00360-013-0761-4
Gomez D, Sunyer JO, Salinas I (2013) The mucosal immune system of fish: the evolution of tolerating commensals while fighting pathogens. Fish Shellfish Immunol 35(6):1729–1739. https://doi.org/10.1016/j.fsi.2013.09.032
González-Palacios C, Fregeneda-Grandes J-M, Aller-Gancedo J-M (2019) Biocontrol of saprolegniosis in rainbow trout (Oncorhynchus mykiss Walbaum) using two bacterial isolates (LE89 and LE141) of Pseudomonas fluorescens. J Fish Dis 42(2):269–275. https://doi.org/10.1111/jfd.12928
Gram L, Melchiorsen J, Spanggaard B, Huber I, Nielsen Torben F (1999) Inhibition of Vibrio anguillarum by Pseudomonas fluorescens AH2, a possible probiotic treatment of fish. Appl Environ Microbiol 65(3):969–973. https://doi.org/10.1128/AEM.65.3.969-973.1999
Hedrick RP, Fryer JL, Chen SN, Kou GH (1983) Characteristics of four birnaviruses isolated from fish in Taiwan. Fish Pathol 18(2):91–97. https://doi.org/10.3147/jsfp.18.91
HLPE (2014) Sustainable fisheries and aquaculture for food security and nutrition. A report by the High Level Panel of Experts on Food Security and Nutrition of the Committee on World Food Security. Rome
Hossain A, Nakamichi S, Habibullah-Al-Mamun M, Tani K, Masunaga S, Matsuda H (2017) Occurrence, distribution, ecological and resistance risks of antibiotics in surface water of finfish and shellfish aquaculture in Bangladesh. Chemosphere 188:329–336. https://doi.org/10.1016/j.chemosphere.2017.08.152
Hossain A, Nakamichi S, Habibullah-Al-Mamun M, Tani K, Masunaga S, Matsuda H (2018) Occurrence and ecological risk of pharmaceuticals in river surface water of Bangladesh. Environ Res 165:258–266. https://doi.org/10.1016/j.envres.2018.04.030
Hossain MMM, Uddin MI, Hossain MM, Islam H, Al-Amin FN, Afroz R (2020) Molecular detection of tilapia lake virus (TiLV) in farmed mono-sex Nile tilapia (Tilapia niloticus) in Bangladesh. Asian J Sci Res 13(1):67–78. https://doi.org/10.3923/ajsr.2020.67.78
Hussain M (2004) Farming of tilapia: breeding plans, mass seed production and aquaculture techniques. Mymensingh, Bangladesh
Jansen MD, Dong HT, Mohan CV (2019) Tilapia lake virus: a threat to the global tilapia industry? Rev Aquac 11(3):725–739. https://doi.org/10.1111/raq.12254
Kanther M, Tomkovich S, Xiaolun S, Grosser MR, Koo J, Flynn EJ III, Jobin C, Rawls JF (2014) Commensal microbiota stimulate systemic neutrophil migration through induction of Serum amyloid A. Cell Microbiol 16(7):1053–1067. https://doi.org/10.1111/cmi.12257
Kashinskaya EN, Simonov EP, Andree KB, Vlasenko PG, Polenogova OV, Kiriukhin BA, Solovyev MM (2021) Microbial community structure in a host–parasite system: the case of Prussian carp and its parasitic crustaceans. J Appl Microbiol 131(4):1722–1741. https://doi.org/10.1111/jam.15071
Keawcharoen J, Techangamsuwan S, Ponpornpisit A, Lombardini ED, Patchimasiri T, Pirarat N (2015) Genetic characterization of a betanodavirus isolated from a clinical disease outbreak in farm-raised tilapia Oreochromis niloticus (L.) in Thailand. J Fish Dis 38(1):49–54 https://doi.org/10.1111/jfd.12200
Kelly C, Salinas I (2017) Under pressure: interactions between commensal microbiota and the teleost immune system. Front Immunol 8:559–559. https://doi.org/10.3389/fimmu.2017.00559
Khanjani MH, Sharifinia M, Hajirezaee S (2022) Recent progress towards the application of biofloc technology for tilapia farming. Aquaculture 552:738021 https://doi.org/10.1016/j.aquaculture.2022.738021
Kibenge FSB, Godoy MG, Fast M, Workenhe S, Kibenge MJT (2012) Countermeasures against viral diseases of farmed fish. Antiviral Res 95(3):257–281. https://doi.org/10.1016/j.antiviral.2012.06.003
Kitiyodom S, Yata T, Thompson KD, Costa J, Elumalai P, Katagiri T, Temisak S, Namdee K, Rodkhum C, Pirarat N (2021) Immersion vaccination by a biomimetic-mucoadhesive nanovaccine induces humoral immune response of red tilapia (Oreochromis sp.) against Flavobacterium columnare challenge. Vaccines 9(11) https://doi.org/10.3390/vaccines9111253
Koesharyani I, Gardenia L, Widowati Z, Khumaira K, Rustianti D (2018) Studi kasus infeksi tilapia lake virus (TiLV) pada ikan nila (Oreochromis niloticus). Jurnal Riset Akuakultur 13(1):85–92 https://doi.org/10.15578/jra.13.1.2018.85-92
Kooloth Valappil R, Stentiford GD, Bass D (2021) The rise of the syndrome – sub-optimal growth disorders in farmed shrimp. Rev Aquac 13(4):1888–1906. https://doi.org/10.1111/raq.12550
Krotman Y, Yergaliyev TM, Alexander Shani R, Avrahami Y, Szitenberg A (2020) Dissecting the factors shaping fish skin microbiomes in a heterogeneous inland water system. Microbiome 8(1):9. https://doi.org/10.1186/s40168-020-0784-5
Kunlasak K, Chitmanat C, Whangchai N, Promya J, Lebel L (2013) Relationships of dissolved oxygen with chlorophyll-a and phytoplankton composition in tilapia ponds. Int J Geosci 4(05):46. https://doi.org/10.4236/ijg.2013.45B008
Lalumera GM, Calamari D, Galli P, Castiglioni S, Crosa G, Fanelli R (2004) Preliminary investigation on the environmental occurrence and effects of antibiotics used in aquaculture in Italy. Chemosphere 54(5):661–668. https://doi.org/10.1016/j.chemosphere.2003.08.001
Larsen A, Tao Z, Bullard SA, Arias CR (2013) Diversity of the skin microbiota of fishes: evidence for host species specificity. FEMS Microbiol Ecol 85(3):483–494. https://doi.org/10.1111/1574-6941.12136
Larsen AM, Bullard SA, Womble M, Arias CR (2015) Community structure of skin microbiome of Gulf killifish, Fundulus grandis, is driven by seasonality and not exposure to oiled sediments in a Louisiana salt marsh. Microb Ecol 70(2):534–544. https://doi.org/10.1007/s00248-015-0578-7
Lauria V, Das I, Hazra S, Cazcarro I, Arto I, Kay S, Ofori-Danson P, Ahmed M, Hossain MAR, Barange M, Fernandes JA (2018) Importance of fisheries for food security across three climate change vulnerable deltas. Sci Total Environ 640–641:1566–1577. https://doi.org/10.1016/j.scitotenv.2018.06.011
Lazado CC, Caipang CMA (2014) Mucosal immunity and probiotics in fish. Fish Shellfish Immunol 39(1):78–89. https://doi.org/10.1016/j.fsi.2014.04.015
Legrand TPRA, Catalano SR, Wos-Oxley ML, Wynne JW, Weyrich LS, Oxley APA (2020) Antibiotic-induced alterations and repopulation dynamics of yellowtail kingfish microbiota. Anim Microbiome 2(1):26. https://doi.org/10.1186/s42523-020-00046-4
Legrand TPRA, Catalano SR, Wos-Oxley ML, Stephens F, Landos M, Bansemer MS, Stone DAJ, Qin JG, Oxley APA (2018) The inner workings of the outer surface: skin and gill microbiota as indicators of changing gut health in yellowtail kingfish. Front Microbiol 8(2664) https://doi.org/10.3389/fmicb.2017.02664
Li HL, Gu XH, Li BJ, Chen CH, Lin HR, Xia JH (2017) Genome-wide QTL analysis identified significant associations between hypoxia tolerance and mutations in the GPR132 and ABCG4 genes in Nile tilapia. Mar Biotechnol 19(5):441–453. https://doi.org/10.1007/s10126-017-9762-8
Li T, Li H, Gatesoupe F-J, She R, Lin Q, Yan X, Li J, Li X (2017) Bacterial signatures of “red-operculum” disease in the gut of crucian carp (Carassius auratus). Microb Ecol 74(3):510–521. https://doi.org/10.1007/s00248-017-0967-1
Limbu SM, Zhou L, Sun SX, Zhang ML, Du ZY (2018) Chronic exposure to low environmental concentrations and legal aquaculture doses of antibiotics cause systemic adverse effects in Nile tilapia and provoke differential human health risk. Environ Int 115:205–219. https://doi.org/10.1016/j.envint.2018.03.034
Liu L, Chi H, Sun L (2015) Pseudomonas fluorescens: identification of Fur-regulated proteins and evaluation of their contribution to pathogenesis. Dis Aquat Org 115(1):67–80. https://doi.org/10.3354/dao02874
Liu X, Steele JC, Meng XZ (2017) Usage, residue, and human health risk of antibiotics in Chinese aquaculture: a review. Environ Pollut 223:161–169. https://doi.org/10.1016/j.envpol.2017.01.003
Liu L, Lu D-Q, Xu J, Luo H-L, Li A-X (2019) Development of attenuated erythromycin-resistant Streptococcus agalactiae vaccine for tilapia (Oreochromis niloticus) culture. J Fish Dis 42(5):693–701. https://doi.org/10.1111/jfd.12977
Liu C, Tan L, Zhang L, Tian W, Ma L (2021) A review of the distribution of antibiotics in water in different regions of china and current antibiotic degradation pathways. Front Environ Sci 9 https://doi.org/10.3389/fenvs.2021.692298
Llewellyn MS, Leadbeater S, Garcia C, Sylvain FE, Custodio M, Ang KP, Powell F, Carvalho GR, Creer S, Elliot J, Derome N (2017) Parasitism perturbs the mucosal microbiome of Atlantic Salmon. Sci Rep 7(1):43465. https://doi.org/10.1038/srep43465
Lokesh J, Kiron V (2016) Transition from freshwater to seawater reshapes the skin-associated microbiota of Atlantic salmon. Sci Rep 6(1):19707. https://doi.org/10.1038/srep19707
Lynch AJ, MacMillan JR (2017) The role of fish in a globally changing food system. In: Hatfield JL, Sivakumar MVK, Prueger JH (eds) Agroclimatology: Linking Agriculture to Climate. 60:579–593
Machimbirike VI, Jansen MD, Senapin S, Khunrae P, Rattanarojpong T, Dong HT (2019) Viral infections in tilapines: More than just tilapia lake virus. Aquaculture 503:508–518. https://doi.org/10.1016/j.aquaculture.2019.01.036
McGrogan D, Ostland V, Byrne P, Ferguson H (1998) Systemic disease involving an iridovirus-like agent in cultured tilapia, Oreochromis niloticus L. - a case report. J Fish Dis 21(2):149–152 https://doi.org/10.1046/j.1365-2761.1998.00082.x
McMurtrie J, Alathari S, Chaput DL, Bass D, Ghambi C, Nagoli J, Delamare-Deboutteville J, Mohan CV, Cable J, Temperton B, Tyler CR (2022) Relationships between pond water and tilapia skin microbiomes in aquaculture ponds in Malawi. Aquaculture 558:738367. https://doi.org/10.1016/j.aquaculture.2022.738367
Meng K-F, Ding L-G, Wu S, Wu Z-B, Cheng G-F, Zhai X, Sun R-H, Xu Z (2021) Interactions between commensal microbiota and mucosal immunity in teleost fish during viral infection with SVCV. Front Immunol 12 https://doi.org/10.3389/fimmu.2021.654758
Merrifield DL, Rodiles A (2015) The fish microbiome and its interactions with mucosal tissues. In: Beck BH, Peatman E (eds) Mucosal Health in Aquaculture. Academic Press, San Diego, pp 273–295
Ming J, Fu Z, Ma Z, Zhou L, Zhang Z, Song C, Yuan X, Wu Q (2020) The effect of sulfamonomethoxine treatment on the gut microbiota of Nile tilapia (Oreochromis niloticus). MicrobiologyOpen 9(11):e1116 https://doi.org/10.1002/mbo3.1116
Minich JJ, Petrus S, Michael JD, Michael TP, Knight R, Allen EE (2020) Temporal, environmental, and biological drivers of the mucosal microbiome in a wild marine fish. Scomber Japonicus Msphere 5(3):e00401-e420. https://doi.org/10.1128/mSphere.00401-20
Minich JJ, Poore GD, Jantawongsri K, Johnston C, Bowie K, Bowman J, Knight R, Nowak B, Allen EE, Liu S-J (2020) Microbial ecology of Atlantic salmon (Salmo salar) hatcheries: impacts of the built environment on fish mucosal microbiota. Appl Environ Microbiol 86(12):e00411-e420. https://doi.org/10.1128/AEM.00411-20
Miranda CD, Godoy FA, Lee MR (2018) Current status of the use of antibiotics and the antimicrobial resistance in the Chilean salmon farms. Front Microbiol 9(1284) https://doi.org/10.3389/fmicb.2018.01284
Monir SM, Yusoff SM, Mohamad A, Ina-Salwany MY (2020) Vaccination of tilapia against motile Aeromonas septicemia: a review. J Aquat Anim Health 32(2):65–76. https://doi.org/10.1002/aah.10099
Mugimba KK, Chengula AA, Wamala S, Mwega ED, Kasanga CJ, Byarugaba DK, Mdegela RH, Tal S, Bornstein B, Dishon A, Mutoloki S, David L, Evensen Ø, Munang’andu HM (2018) Detection of tilapia lake virus (TiLV) infection by PCR in farmed and wild Nile tilapia (Oreochromis niloticus) from Lake Victoria. J Fish Dis 41(8):1181–1189. https://doi.org/10.1111/jfd.12790
Mulei IR, Nyaga PN, Mbuthia PG, Waruiru RM, Njagi LW, Mwihia EW, Gamil AAA, Evensen Ø, Mutoloki S (2018) Infectious pancreatic necrosis virus isolated from farmed rainbow trout and tilapia in Kenya is identical to European isolates. J Fish Dis 41(8):1191–1200. https://doi.org/10.1111/jfd.12807
Muziasari WI, Pärnänen K, Johnson TA, Lyra C, Karkman A, Stedtfeld RD, Tamminen M, Tiedje JM, Virta M (2016) Aquaculture changes the profile of antibiotic resistance and mobile genetic element associated genes in Baltic Sea sediments. FEMS Microbiol Ecol 92(4):fiw052 https://doi.org/10.1093/femsec/fiw052
Muziasari WI, Pitkänen LK, Sørum H, Stedtfeld RD, Tiedje JM, Virta M (2017) The resistome of farmed fish feces contributes to the enrichment of antibiotic resistance genes in sediments below Baltic Sea fish farms. Front Microbiol 7 https://doi.org/10.3389/fmicb.2016.02137
Nguyen VV, Dong HT, Senapin S, Pirarat N, Rodkhum C (2016) Francisella noatunensis subsp. orientalis, an emerging bacterial pathogen affecting cultured red tilapia (Oreochromis sp.) in Thailand. Aquacult Res 47(11):3697–3702 https://doi.org/10.1111/are.12802
Nguyen VV, Dong HT, Senapin S, Kayansamruaj P, Pirarat N, Rung-ruangkijkrai T, Tiawsirisup S, Rodkhum C (2020) Synergistic infection of Ichthyophthirius multifiliis and Francisella noatunensis subsp. orientalis in hybrid red tilapia (Oreochromis sp.). Microb Pathog 147:104369 https://doi.org/10.1016/j.micpath.2020.104369
Nhinh DT, Giang NTH, Van Van K, Dang LT, Dong HT, Hoai TD (2022) Widespread presence of a highly virulent Edwardsiella ictaluri strain in farmed tilapia, Oreochromis spp. Transbound Emerg Dis 69(5):e2276–e2290. https://doi.org/10.1111/tbed.14568
Nicholson P, Fathi MA, Fischer A, Mohan C, Schieck E, Mishra N, Heinimann A, Frey J, Wieland B, Jores J (2017) Detection of Tilapia Lake Virus in Egyptian fish farms experiencing high mortalities in 2015. J Fish Dis 40(12):1925–1928. https://doi.org/10.1111/jfd.12650
Nicholson P, Mon-on N, Jaemwimol P, Tattiyapong P, Surachetpong W (2020) Coinfection of tilapia lake virus and Aeromonas hydrophila synergistically increased mortality and worsened the disease severity in tilapia (Oreochromis spp.). Aquaculture 520:734746 https://doi.org/10.1016/j.aquaculture.2019.734746
Obiero K, Meulenbroek P, Drexler S, Dagne A, Akoll P, Odong R, Kaunda-Arara B, Waidbacher H (2019) The contribution of fish to food and nutrition security in Eastern Africa: emerging trends and future outlooks. Sustainability 11(6):1636. https://doi.org/10.3390/su11061636
Observatory of Economic Complexity (2020) Products exports/imports. https://oec.world/en/profile/country/bgd. Accessed 16 Mar 2023
OIE (2017a) Immediate notifications and follow-ups. Tilapia lake virus, the Philippines. Retrieved from https://www.woah.org/wahis_2/public/wahid.php/Revie%20wrepo%20rt/Revie%20w?page_refer%20=MapFu%20llEve%20ntReport&repor%20tid=25278
OIE (2017b) Tilapia lake virus disease, Chinese Taipei. Retrieved from http://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?reportid=24033.
OIE (2018) Immediate notification. Tilapia lake virus. Mexico. Retrieved from https://www.woah.org/wahis_2/publi%20c/wahid.php/Revie%20wreport/Revie%20w?page_refer%20=MapFu%20llEve%20ntRep%20ort&repor%20tid=27650.
O'Neill J (2016) Tackling drug-resistant infections globally: final report and recommendations. Government of the United Kingdom
Paperna I (1973) Lymphocystis in fish from east African lakes. J Wildl Dis 9(4):331–335. https://doi.org/10.7589/0090-3558-9.4.331
Park SB, Aoki T, Jung TS (2012) Pathogenesis of and strategies for preventing Edwardsiella tarda infection in fish. Vet Res 43(1):67. https://doi.org/10.1186/1297-9716-43-67
Patil HJ, Gatica J, Zolti A, Benet-Perelberg A, Naor A, Dror B, Al Ashhab A, Marman S, Hasan NA, Colwell RR, Sher D, Minz D, Cytryn E (2020) Temporal resistome and microbial community dynamics in an intensive aquaculture facility with prophylactic antimicrobial treatment. Microorganisms 8(12) https://doi.org/10.3390/microorganisms8121984
Payne CJ, Turnbull JF, MacKenzie S, Crumlish M (2021) Investigating the effect of an oxytetracycline treatment on the gut microbiome and antimicrobial resistance gene dynamics in Nile tilapia (Oreochromis niloticus). Antibiotics 10(10):1213. https://doi.org/10.3390/antibiotics10101213
Perry WB, Lindsay E, Payne CJ, Brodie C, Kazlauskaite R (2020) The role of the gut microbiome in sustainable teleost aquaculture. ProcRSocB 287(1926):20200184. https://doi.org/10.1098/rspb.2020.0184
Ponzoni RW, Khaw HL, Yee HY (2010) GIFT: the story since leaving ICLARM (now known as the WorldFish Center): socioeconomic, access and benefit sharing and dissemination aspects. Penang, Malaysia
Prabu E, Rajagopalsamy C, Ahilan B, Jeevagan IJMA, Renuhadevi M (2019) Tilapia–an excellent candidate species for world aquaculture: a review. Annu Res Rev Biol 31(3):1–14. https://doi.org/10.9734/arrb/2019/v31i330052
Preena PG, Swaminathan TR, Rejish Kumar VJ, Bright Singh IS (2020) Unravelling the menace: detection of antimicrobial resistance in aquaculture. Lett Appl Microbiol 71(1):26–38. https://doi.org/10.1111/lam.13292
Prihartini NC, Yanuhar U, Maftuch (2015) Betanodavirus infections in tilapia seed (Oreochromis sp.), in Indonesia. J Life Sci Biomed 5(4):106–109
Rahman ML, Shahjahan M, Ahmed N (2021) Tilapia farming in Bangladesh: adaptation to climate change. Sustainability 13(14):7657. https://doi.org/10.3390/su13147657
Ramírez-Paredes JG, Paley RK, Hunt W, Feist SW, Stone DM, Field TR, Haydon DJ, Ziddah PA, Nkansa M, Guilder J, Gray J, Duodu S, Pecku EK, Awuni JA, Wallis TS, Verner-Jeffreys DW (2021) First detection of infectious spleen and kidney necrosis virus (ISKNV) associated with massive mortalities in farmed tilapia in Africa. Transbound Emerg Dis 68(3):1550–1563. https://doi.org/10.1111/tbed.13825
Ray AK, Ghosh K, Ringø E (2012) Enzyme-producing bacteria isolated from fish gut: a review. Aquacult Nutr 18(5):465–492. https://doi.org/10.1111/j.1365-2095.2012.00943.x
Rebouças VT, dos Santos Lima FR, de Holanda Cavalcantedo Carmo e Sá DMV (2015) Tolerance of Nile tilapia juveniles to highly acidic rearing water. Acta Sci 37(3):227–233. https://doi.org/10.4025/actascianimsci.v37i3.27031
Reid KM, Patel S, Robinson AJ, Bu L, Jarungsriapisit J, Moore LJ, Salinas I (2017) Salmonid alphavirus infection causes skin dysbiosis in Atlantic salmon (Salmo salar L.) post-smolts. PLoS One 12(3):e0172856 https://doi.org/10.1371/journal.pone.0172856
Reinhart EM, Korry BJ, Rowan-Nash AD, Belenky P (2019) Defining the distinct skin and gut microbiomes of the northern pike (Esox lucius). Front Microbiol 10(2118) https://doi.org/10.3389/fmicb.2019.02118
Rengmark AH, Slettan A, Lee WJ, Lie Ø, Lingaas F (2007) Identification and mapping of genes associated with salt tolerance in tilapia. J Fish Biol 71(sc):409–422 https://doi.org/10.1111/j.1095-8649.2007.01664.x
Rizkiantino R, Pasaribu FH, Soejoedono RD, Purnama S, Wibowo DB, Wibawan IWT (2021) Experimental infection of Enterococcus faecalis in red tilapia (Oreochromis hybrid) revealed low pathogenicity to cause streptococcosis. Open Vet J 11(2):309–318. https://doi.org/10.5455/OVJ.2021.v11.i2.16
Rosado D, Jo C, Pr-L M, Raquel X, Ricardo S (2019) Characterization of the skin and gill microbiomes of the farmed seabass (Dicentrarchus labrax) and seabream (Sparus aurata). Aquaculture 500:57–64. https://doi.org/10.1016/j.aquaculture.2018.09.063
Rud I, Kolarevic J, Holan AB, Berget I, Calabrese S, Terjesen BF (2017) Deep-sequencing of the bacterial microbiota in commercial-scale recirculating and semi-closed aquaculture systems for Atlantic salmon post-smolt production. Aquacult Eng 78:50–62. https://doi.org/10.1016/j.aquaeng.2016.10.003
Sáenz JS, Marques TV, Barone RSC, Cyrino JEP, Kublik S, Nesme J, Schloter M, Rath S, Vestergaard G (2019) Oral administration of antibiotics increased the potential mobility of bacterial resistance genes in the gut of the fish Piaractus mesopotamicus. Microbiome 7(1):24. https://doi.org/10.1186/s40168-019-0632-7
Schar D, Klein EY, Laxminarayan R, Gilbert M, Van Boeckel TP (2020) Global trends in antimicrobial use in aquaculture. Sci Rep 10(1):21878. https://doi.org/10.1038/s41598-020-78849-3
Shah SQA, Cabello FC, L’Abée-Lund TM, Tomova A, Godfrey HP, Buschmann AH, Sørum H (2014) Antimicrobial resistance and antimicrobial resistance genes in marine bacteria from salmon aquaculture and non-aquaculture sites. Environ Microbiol 16(5):1310–1320. https://doi.org/10.1111/1462-2920.12421
Shamsuzzaman MM, Islam MM, Tania NJ, Abdullah Al-Mamun M, Barman PP, Xu X (2017) Fisheries resources of Bangladesh: present status and future direction. Aquac Fish 2(4):145–156. https://doi.org/10.1016/j.aaf.2017.03.006
Shinn A, Pratoomyot J, Bron J, Paladini G, Brooker E, Brooker A (2015) Economic impacts of aquatic parasites on global finfish production. Global Aquac Advocate 2015:58–61
Shlapobersky M, Sinyakov MS, Katzenellenbogen M, Sarid R, Don J, Avtalion RR (2010) Viral encephalitis of tilapia larvae: primary characterization of a novel herpes-like virus. Virology 399(2):239–247. https://doi.org/10.1016/j.virol.2010.01.001
Shoko AP, Limbu SM, Mrosso HDJ, Mgaya YD (2014) A comparison of diurnal dynamics of water quality parameters in Nile tilapia (Oreochromis niloticus, Linnaeus, 1758) monoculture and polyculture with African sharp tooth catfish (Clarias gariepinus, Burchell, 1822) in earthen ponds. Int Aquat Res 6(1):56. https://doi.org/10.1007/s40071-014-0056-8
Singh RR, Angeles LF, Butryn DM, Metch JW, Garner E, Vikesland PJ, Aga DS (2019) Towards a harmonized method for the global reconnaissance of multi-class antimicrobials and other pharmaceuticals in wastewater and receiving surface waters. Environ Int 124:361–369. https://doi.org/10.1016/j.envint.2019.01.025
Sinyakov MS, Belotsky S, Shlapobersky M, Avtalion RR (2011) Vertical and horizontal transmission of tilapia larvae encephalitis virus: the bad and the ugly. Virology 410(1):228–233. https://doi.org/10.1016/j.virol.2010.11.004
Song C, Zhang C, Fan L, Qiu L, Wu W, Meng S, Hu G, Kamira B, Chen J (2016) Occurrence of antibiotics and their impacts to primary productivity in fishponds around Tai Lake, China. Chemosphere 161:127–135. https://doi.org/10.1016/j.chemosphere.2016.07.009
Sood N, Verma DK, Paria A, Yadav SC, Yadav MK, Bedekar MK, Kumar S, Swaminathan TR, Mohan CV, Rajendran KV, Pradhan PK (2021) Transcriptome analysis of liver elucidates key immune-related pathways in Nile tilapia Oreochromis niloticus following infection with tilapia lake virus. Fish Shellfish Immunol 111:208–219. https://doi.org/10.1016/j.fsi.2021.02.005
Soto E, Griffin M, Arauz M, Riofrio A, Martinez A, Cabrejos ME (2012) Edwardsiella ictaluri as the causative agent of mortality in cultured Nile tilapia. J Aquat Anim Health 24(2):81–90 https://doi.org/10.1080/08997659.2012.675931
Sotto RB, Medriano CD, Cho Y, Kim H, Chung IY, Seok KS, Song KG, Hong SW, Park Y, Kim S (2017) Sub-lethal pharmaceutical hazard tracking in adult zebrafish using untargeted LC-MS environmental metabolomics. J Hazard Mater 339:63–72. https://doi.org/10.1016/j.jhazmat.2017.06.009
Sriram A, Kalanxhi E, Kapoor G, Craig J, Balasubramanian R, Brar S, Criscuolo N, Hamilton A, Klein E, Tseng K (2021) State of the world’s antibiotics 2021: a global analysis of antimicrobial resistance and its drivers. Center for Disease Dynamics, Economics & Policy, Washington, DC, USA
Stentiford GD, Sritunyalucksana K, Flegel TW, Williams BAP, Withyachumnarnkul B, Itsathitphaisarn O, Bass D (2017) New paradigms to help solve the global aquaculture disease crisis. PLoS Path 13(2):e1006160. https://doi.org/10.1371/journal.ppat.1006160
Stratev D, Odeyemi OA (2017) An overview of motile Aeromonas septicaemia management. Aquacult Int 25(3):1095–1105. https://doi.org/10.1007/s10499-016-0100-3
Subramaniam K, Gotesman M, Smith CE, Steckler NK, Kelley KL, Groff JM, Waltzek TB (2016) Megalocytivirus infection in cultured Nile tilapia Oreochromis niloticus. Dis Aquat Org 119(3):253–258. https://doi.org/10.3354/dao02985
Suebsing R, Pradeep PJ, Jitrakorn S, Sirithammajak S, Kampeera J, Turner WA, Saksmerprome V, Withyachumnarnkul B, Kiatpathomchai W (2016) Detection of natural infection of infectious spleen and kidney necrosis virus in farmed tilapia by hydroxynapthol blue-loop-mediated isothermal amplification assay. J Appl Microbiol 121(1):55–67. https://doi.org/10.1111/jam.13165
Sugita H, Miyajima C, Deguchi Y (1991) The vitamin B12-producing ability of the intestinal microflora of freshwater fish. Aquaculture 92:267–276. https://doi.org/10.1016/0044-8486(91)90028-6
Sultana S, Khan MN, Hossain MS, Dai J, Rahman MS, Salimullah M (2022) Community structure and functional annotations of the skin microbiome in healthy and diseased catfish, Heteropneustes fossilis. Front Microbiol 13 https://doi.org/10.3389/fmicb.2022.856014
Sun J, Fang W, Ke B, He D, Liang Y, Ning D, Tan H, Peng H, Wang Y, Ma Y, Ke C, Deng X (2016) Inapparent Streptococcus agalactiae infection in adult/commercial tilapia. Sci Rep 6(1):26319. https://doi.org/10.1038/srep26319
Sun B-Y, He W, Yang H-X, Tian D-Y, Jian P-Y, Wu K, Yang C-G, Song X-H (2022) Increased susceptibility to Aeromonas hydrophila infection in grass carp with antibiotic-induced intestinal dysbiosis. Aquaculture 552:737969 https://doi.org/10.1016/j.aquaculture.2022.737969
Surachetpong W, Janetanakit T, Nonthabenjawan N, Tattiyapong P, Sirikanchana K, Amonsin A (2017) Outbreaks of tilapia lake virus infection, Thailand, 2015–2016. Emerging Infect Dis 23(6):1031. https://doi.org/10.3201/eid2306.161278
Surachetpong W, Roy SRK, Nicholson P (2020) Tilapia lake virus: the story so far. J Fish Dis 43(10):1115–1132. https://doi.org/10.1111/jfd.13237
Sylvain F-É, Holland A, Bouslama S, Audet-Gilbert É, Lavoie C, Val AL, Derome N (2020) Fish skin and gut microbiomes show contrasting signatures of host species and habitat. Appl Environ Microbiol 86(16):e00789-e820. https://doi.org/10.1128/AEM.00789-20
Taha E, Shawky M, Ahmed B, Moustafa M, Yousif A, Abdelaziz M (2020) Emergence of viral nervous necrosis is associated with mass mortality in hatchery-reared tilapia (Oreochromis niloticus) in Egypt. Aquacult Int 28(5):1811–1823. https://doi.org/10.1007/s10499-020-00559-4
Takeuchi M, Fujiwara-Nagata E, Katayama T, Suetake H (2021) Skin bacteria of rainbow trout antagonistic to the fish pathogen Flavobacterium psychrophilum. Sci Rep 11(1):7518. https://doi.org/10.1038/s41598-021-87167-1
Tesfahun A, Temesgen M (2018) Food and feeding habits of Nile tilapia Oreochromis niloticus (L.) in Ethiopian water bodies: a review. Int J Fish Aquat Stud 6(1):43–47
Thornber K, Verner-Jeffreys D, Hinchliffe S, Rahman MM, Bass D, Tyler CR (2020) Evaluating antimicrobial resistance in the global shrimp industry. Rev Aquac 12(2):966–986. https://doi.org/10.1111/raq.12367
Tigchelaar M, Leape J, Micheli F, Allison EH, Basurto X, Bennett A, Bush SR, Cao L, Cheung WWL, Crona B, DeClerck F, Fanzo J, Gelcich S, Gephart JA, Golden CD, Halpern BS, Hicks CC, Jonell M, Kishore A, Koehn JZ, Little DC, Naylor RL, Phillips MJ, Selig ER, Short RE, Sumaila UR, Thilsted SH, Troell M, Wabnitz CCC (2022) The vital roles of blue foods in the global food system. Glob Food Sec 33:100637 https://doi.org/10.1016/j.gfs.2022.100637
Tiralongo F, Messina G, Lombardo BM, Longhitano L, Li Volti G, Tibullo D (2020) Skin mucus of marine fish as a source for the development of antimicrobial agents. Front Mar Sci 7 https://doi.org/10.3389/fmars.2020.541853
Tsofack JEK, Zamostiano R, Watted S, Berkowitz A, Rosenbluth E, Mishra N, Briese T, Lipkin WI, Kabuusu RM, Ferguson H, del Pozo J, Eldar A, Bacharach E (2017) Detection of tilapia lake virus in clinical samples by culturing and nested reverse transcription-PCR. J Clin Microbiol 55(3):759–767. https://doi.org/10.1128/JCM.01808-16
Tsuchiya C, Sakata T, Sugita H (2008) Novel ecological niche of Cetobacterium somerae, an anaerobic bacterium in the intestinal tracts of freshwater fish. Lett Appl Microbiol 46(1):43–48. https://doi.org/10.1111/j.1472-765X.2007.02258.x
Van Boeckel TP, Glennon EE, Chen D, Gilbert M, Robinson TP, Grenfell BT, Levin SA, Bonhoeffer S, Laxminarayan R (2017) Reducing antimicrobial use in food animals. Science 357(6358):1350–1352. https://doi.org/10.1126/science.aao1495
van Kessel MAHJ, Mesman RJ, Arshad A, Metz JR, Spanings FAT, van Dalen SCM, van Niftrik L, Flik G, Wendelaar Bonga SE, Jetten MSM, Klaren PHM, Op den Camp HJM (2016) Branchial nitrogen cycle symbionts can remove ammonia in fish gills. Environ Microbiol Rep 8(5):590–594. https://doi.org/10.1111/1758-2229.12407
Wang M, Lu M (2016) Tilapia polyculture: a global review. Aquacult Res 47(8):2363–2374. https://doi.org/10.1111/are.12708
Wang M, Yi M, Lu M, Gao F, Liu Z, Huang Q, Li Q, Zhu D (2020) Effects of probiotics Bacillus cereus NY5 and Alcaligenes faecalis Y311 used as water additives on the microbiota and immune enzyme activities in three mucosal tissues in Nile tilapia Oreochromis niloticus reared in outdoor tanks. Aquac Rep 17:100309. https://doi.org/10.1016/j.aqrep.2020.100309
Wang T, Zhang N, Yu X-B, Qiao F, Chen L-Q, Du Z-Y, Zhang M-L (2021) Inulin alleviates adverse metabolic syndrome and regulates intestinal microbiota composition in Nile tilapia (Oreochromis niloticus) fed with high-carbohydrate diet. Br J Nutr 126(2):161–171. https://doi.org/10.1017/S000711452000402X
Webster TMU, Consuegra S, Hitchings M, de Leaniz CG (2018) Interpopulation variation in the Atlantic salmon microbiome reflects environmental and genetic diversity. Appl Environ Microbiol 84(16):e00691-e718. https://doi.org/10.1128/aem.00691-18
Willyard C (2017) The drug-resistant bacteria that pose the greatest health threats. Nature 543(7643):15–15. https://doi.org/10.1038/nature.2017.21550
Wineman A, Ekwueme MC, Bigayimpunzi L, Martin-Daihirou A, de Gois V. N. Rodrigues EL, Etuge P, Warner Y, Kessler H, Mitchell A (2022) School meal programs in Africa: regional results from the 2019 Global Survey of School Meal Programs. Front Public Health 10 https://doi.org/10.3389/fpubh.2022.871866
WorldBank (2014) Reducing disease risk in aquaculture. World Bank Report Number 88257-GLB
Xu T, Zhang X-H (2014) Edwardsiella tarda: an intriguing problem in aquaculture. Aquaculture 431:129–135. https://doi.org/10.1016/j.aquaculture.2013.12.001
Xu Z, Parra D, Gómez D, Salinas I, Zhang Y-A, LvG J, Heinecke RD, Buchmann K, LaPatra S, Sunyer JO (2013) Teleost skin, an ancient mucosal surface that elicits gut-like immune responses. PNAS 110(32):13097–13102. https://doi.org/10.1073/pnas.1304319110
Xu D-H, Shoemaker CA, Zhang D (2015) Treatment of Trichodina sp reduced load of Flavobacterium columnare and improved survival of hybrid tilapia. Aquac Rep 2:126–131. https://doi.org/10.1016/j.aqrep.2015.09.007
Xu Z, Akizawa F, Casadei E, Shibasaki Y, Ding Y, Sauters TJC, Yu Y, Salinas I, Sunyer JO (2020) Specialization of mucosal immunoglobulins in pathogen control and microbiota homeostasis occurred early in vertebrate evolution. Sci Immunol 5(44):eaay3254 https://doi.org/10.1126/sciimmunol.aay3254
Yu X, Megens H-J, Mengistu SB, Bastiaansen JWM, Mulder HA, Benzie JAH, Groenen MAM, Komen H (2021) Genome-wide association analysis of adaptation to oxygen stress in Nile tilapia (Oreochromis niloticus). BMC Genomics 22(1):426. https://doi.org/10.1186/s12864-021-07486-5
Yu Y-Y, Ding L-G, Huang Z-Y, Xu H-Y, Xu Z (2021) Commensal bacteria-immunity crosstalk shapes mucosal homeostasis in teleost fish. Rev Aquac 13(4):2322–2343. https://doi.org/10.1111/raq.12570
Yu L, Qiao N, Li T, Yu R, Zhai Q, Tian F, Zhao J, Zhang H, Chen W (2019) Dietary supplementation with probiotics regulates gut microbiota structure and function in Nile tilapia exposed to aluminum. PeerJ 7:e6963 https://doi.org/10.7717/peerj.6963
Zahran E, Hafez EE, Hossain FMA, Elhadidy M, Shaheen AA (2017) Saprolegniosis in Nile tilapia: identification, molecular characterization, and phylogenetic analysis of two novel pathogenic Saprolegnia strains. J Aquat Anim Health 29(1):43–49 https://doi.org/10.1080/08997659.2016.1259691
Zamri-Saad M, Amal M, Siti-Zahrah A, Zulkafli AR (2014) Control and prevention of streptococcosis in cultured tilapia in Malaysia: a review. Pertanika J Trop Agric Sci 37(4):389–410
Zhang Z (2021) Research advances on tilapia streptococcosis. Pathogens 10(5) https://doi.org/10.3390/pathogens10050558
Zhou W, Xie M, Xie Y, Liang H, Li M, Ran C, Zhou Z (2022) Effect of dietary supplementation of Cetobacterium somerae XMX-1 fermentation product on gut and liver health and resistance against bacterial infection of the genetically improved farmed tilapia (GIFT, Oreochromis niloticus). Fish Shellfish Immunol 124:332–342. https://doi.org/10.1016/j.fsi.2022.04.019
Funding
SCD was supported by The University of Exeter, WorldFish, and Cefas; JM was supported by the BBSRC/South West Biosciences Doctoral Training Partnership (BB/M009122/1) with CASE partners WorldFish and Cefas. This work was further supported by the CGIAR Research Program on Fish Agri-Food Systems (FISH) led by WorldFish.
Author information
Authors and Affiliations
Contributions
Conceptualization: SCD and CRT; funding acquisition: CRT, BT, and CVM; methodology: SCD; supervision: CRT and BT; writing: SCD, JM, and CRT; editing: SCD, JM, CVM, JD, BT, and CRT.
Corresponding authors
Ethics declarations
Ethical approval
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Handling Editor: Amany Abbass
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Debnath, S.C., McMurtrie, J., Temperton, B. et al. Tilapia aquaculture, emerging diseases, and the roles of the skin microbiomes in health and disease. Aquacult Int 31, 2945–2976 (2023). https://doi.org/10.1007/s10499-023-01117-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10499-023-01117-4