Abstract
Vibriosis is one of the common diseases caused by gram-negative bacteria from the genus Vibrio. To treat vibriosis, vaccination has been proven to be the most effective treatment as it can avoid the risk of drugs or antibiotics resistance. Microalgae are commonly used as feed for aquatic organisms and Nannochloropsis sp. is one of the highly utilized species for fish feed. This study focused on the use of microalga, Nannochloropsis sp. as a vaccine carrier. Transgenic Nannochloropsis sp. harbouring an outer membrane protein kinase (OmpK) gene fragment of the Vibrio species namely V1, V2, CV1, CV2, CPV1 and CPV2 were utilized in this study. The stability of OmpK gene in transgenic Nannochloropsis sp. over a number of generations was evaluated. DNA and RNA from the Nannochloropsis sp. transgenic lines were extracted and subjected to PCR amplification of OmpK gene fragment. The OmpK gene fragment was successfully amplified and expressed up to the fifth generation (F5). For V1, V2, CV1 and CV2, the gene was present and expressed in fourth generation (F4) and F5 respectively but CPV1 and CPV2 the OmpK genes were present up to F4. From the results obtained, Nannochloropsis sp. is shown to be suitable as a vaccine carrier and can be utilized as a vaccine carrier to ameliorate vibriosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aquaculture is one of the industries that contribute to world’s economy where the global market for aquaculture products is about $50 billion over a year (Chen et al. 2008). This industry is known as the farming of fish, shellfish and aquatic plants where it is the fastest growing segment of the world food economy. The aquaculture industry is an efficient resource for human consumption as it has the highest protein retention compared to livestocks such as chicken, pork and beef (Gjedrem et al. 2012). Apart from that, it also has lower greenhouse gas emissions than other types of farming. However, disease is the major problem in the aquaculture industry where it causes high economic losses for the industry. It is the source of the most serious problem that can cause damage to the livelihood of farmers, loss of jobs, reduced incomes, and food insecurity. Studies showed that almost 50% of production loss was because of diseases which were more severe in developing countries (Assefa and Abunna 2018). Bacterial infection in various aquatic animal constitutes the most important source of disease problems while fungal is the second major cause of losses in aquaculture (Meyer 1991). One of the major diseases in aquaculture is vibriosis. This disease is mostly caused by a group of bacteria in the Vibrionaceae family (Allam et al. 2000). In general, the common signs of vibriosis are slow growth, lethargy and tissue necrosis, and infected fish will show skin decoloration. In addition, the presence of red lesions in the muscle and erythema at the base of fins, vent and mouth are also signs of infection (Austin and Austin 2012). To control vibriosis in aquaculture, antibiotics are widely used in various ways such as incorporation into feed as well as oral administration (Shoemaker et al. 2009). However, these approaches can cause other problems not only in aquatic animals but also in humans and the environment. The presence of antibiotics in aquaculture products will lead to allergy and toxicity in humans (Cabello et al. 2013). The frequent use of antibiotics widely in aquaculture can cause the development of antibiotics resistance. This situation can increase the health risk among organisms in aquaculture. The awareness about the usage of antibiotics is important and needs to be urgently noted. Therefore, different strategies need to be developed in order to handle this situation.
Microalgae have been used widely as biomass for animal feeds. In the aquaculture sector, microalgae are used as feedstock due to rapid growth and have complete nutrients such as balanced amino acids, vitamins, carotenoids, and polyunsaturated fatty acids (Safafar et al. 2016). In addition, microalgae are also not being used to be feedstock only but they can help to utilize carbon dioxide and have potential in water remediation. Microalgae are microorganisms that can be used as a vaccine carrier due to their various characteristics. The key feature of microalgae that makes them a good vaccine carrier candidate is that they have a rigid cell wall. This could be useful for the protection of the antigen that they harbour in order for the antigen to be delivered safely up to the gut of the fish. On top of that, microalgae are unseasonal where they can be grown throughout the year and they also can be preserved by lyophilisation. Microalgae also contain compounds that can act as vaccine adjuvants which have been known to increase vaccine’s efficiency.
Nannochloropsis sp. is a small microalga with a diameter of 2–5 μm. This species differs from other species whereby most algae contain many kinds of chlorophyll but for Nannochloropsis species, only chlorophyll a was present within their cells. It completely lacks chlorophyll b which is tetraphyroles and chlorophyll c. Nannochloropsis species is found to have a high photosynthetic efficiency and can store lipid by converting carbon dioxide into triglyceride and polyunsaturated fatty acid and eicosapentaenoic acid (Ma et al. 2016). A study has shown that Nannochloropsis sp. had a wide potential for the development of healthier food products where they contained phenolic compounds and also antioxidant properties (Kherraf et al. 2017).
The recombinant DNA technology for microalgae is described as ‘best of both worlds’, combining the high growth rate and ease of cultivation of microorganisms with the ability to perform post-transcriptional and post-translational modifications of plants (Potvin and Zhang 2010; Rosano and Ceccarelli 2014). Most microalgae comprise a rich repertoire of species having a lot of advantages for biotechnology processes such as gene transfer technologies. So, they are allowed to act as a host for the expression of the foreign gene of interest. However, the yield of the recombinant protein expression system is still low. Nowadays, in order to overcome the problem of lower protein yield, many strategies have been performed to improve the protein expression although many obstacles need to be faced. A research was conducted where Nannochloropsis oculata was used for the production of a functional fish growth hormone. Plasmid phr-YPGHc, containing fish growth hormone (GH) cDNA, was transferred into the protoplast of these marine microalgae. As a result, after feeding the fish larvae with the transgenic microalgae, the growth of larvae is greater than those fed with wild-type microalgae. Therefore, the proposed N. oculata might be an excellent bioreactor and had the ability to produce growth hormone (Chen et al. 2008). Apart from that, one study had shown that Nannochloropsis oculata also had the ability to be used as a vaccine carrier. They utilized and modified N. oculata to express antimicrobial peptides in order to fight against bacterial pathogenic infection. To test the idea, they came out with the construction of algae-codon-optimized bovine lactoferricin (LFB) fused with a red fluorescent protein (DsRed) driven by a heat-inducible promoter, which was a heat shock protein 70A promoter combined with a ribulose-1,5-bisphosphate carboxylase or oxygenase small subunit 2′ promoter from Chlamydomonas reinhardtii. After electroporation, they examined 491 microalgal clones and generated two stable transgenic lines, each expressing a stable transgene inheritance for at least 26 months. This study was confirmed by the positive detection of the mRNA transcript and the protein of LFB-DsRed produced by the transgenic microalgae. To test the efficacy of the antimicrobial peptide LFB, medaka fish (Oryzias latipes) were adapted from freshwater to seawater and were fed with the transgenic algae by oral-in-tube delivery method. Bacterial infection with Vibrio parahaemolyticus per fish was induced 6 h after by oral-in-tube delivery as well. The fish were then fed with 1 × 108 cell transgenic algae per fish, which showed an average survival rate after a 24-h period of infection was much higher than that of the fish fed with wild-type. The result showed that the fish fed with the LFB-containing transgenic microalgae had a bactericidal defence against Vibrio parahaemolyticus infection in its digestive tract (Li and Tsai 2009).
This research focused on six transgenic lines of Nannochloropsis sp. which harbour a fragment of the OmpK gene isolated from the Vibrio species. OmpK gene was found in Vibrio parahaemolyticus and was shown to be a receptor for a broad host range vibriophage KVP40 in members of Vibrionaceae (Inoue et al. 1995). The OmpK had also been identified to act as a protective antigen against fish vibriosis caused by Vibrio alginolyticus (Qian et al. 2008). These transgenic lines were produced via homologous recombination where the nitrate reductase of the host was knocked out while integrating the OmpK gene as well as the marker gene and the stability of the transgene required validation. This study aimed to verify the presence of OmpK gene transcript in six different lines of transgenic Nannochloropsis sp. by using polymerase chain reaction (PCR) and the investigation of the expression of OmpK gene transcript in all six transgenic lines over a number of generations.
Materials and methods
Culture preparation, maintenance and conditions
Transgenic lines of Nannochloropsis sp. used in this study were obtained from the Plant-Algae Biochemistry and Biotechnology Laboratory. The following abbreviations will be used throughout this article which are WT (wild type), V1 (transformed using VCP_OMPK_ZA1 vector), V2 (transformed using VCP_OMPK_ZA2 vector), CV1 (transformed using CRISPR_NR and VCP_OMPK_ZA1 vector), CV2 (transformed using CRISPR_NR and VCP_OMPK_ZA2 vector), CPV1 (transformed using CRISPR_NR and PCR product of VCP_OMPK_ZA1 vector) and CPV2 (transformed using CRISPR_NR and PCR product of VCP_OMPK_ZA2 vector). The vectors construct used in developing the transgenic lines are as shown in Fig. 1 and also patented under the patent file no. PI 2020000621. For the cultivation of wild-type Nannochloropsis sp., Guillard F/2 medium was used with sodium nitrate as the nitrogen source while for the cultivation of transgenic lines, the Guillard F/2 medium was used with ammonium chloride as a nitrogen sources. Cultures were continuously shaken at 130 rpm on an orbital shaker at 24 °C with 12 h of light illumination and 12 h in dark.
Identification of microalgal species via the amplification of 18S rRNA gene fragment
Genomic DNA of Nannochloropsis sp. was extracted using the DNeasy Plant mini kit (QIAGEN). The extracted DNA was quantified by using a NanoDrop Spectrophotometer (Genova Nano, UK). The amplification of the 18S rRNA gene was carried out using universal primers with the following conditions: one cycle of initial denaturation (95 °C, 4 min 30 s), 34 cycles of annealing (50–63 °C, 1 min 30 s), 34 cycles of extension (72 °C, 10 min) and a cycle of final extension (4 °C, infinity) using the Bio-Rad, T100, USA, thermal cycler. PCR products were then cloned and sent for sequencing to verify the amplified fragment’s sequence using the 1st Base Sequencing Service (1st Base, Singapore). The result obtained was then analysed through Basic Local Alignment tools (BLAST) (http://blast.ncbi.nlm.nih.gov) to validate the sequence of the fragment of interest.
Expression of heterologous gene in transgenic Nannochloropsis sp.
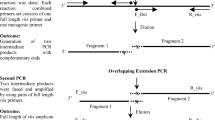
Each transgenic line obtained from the transformation procedure was maintained accordingly based on the number of subculturing from the original transformation plate (Fig. 2). The extraction of RNA from the Nannochloropsis sp. transgenic lines was carried out as described by Fern et al. (2017). The extracted RNA was stored at ‑ 80 °C prior to further analysis. Reverse transcription PCR was performed using Tetro™ cDNA Synthesis Kit (Bioline, Canada). The synthesized complementary DNAs (cDNAs) were stored at ‑ 20 °C for polymerase chain reaction (PCR).
Amplification of OmpK gene from genomic DNA and cDNA
Transgenic lines were first determined by the integration of the expression cassette with the host gDNA via amplification of nitrate reductase using NR_980_F (TCAATGGACGAACTGGTCGC) and NR_980_R (CTTGCCATCGTCGAGGGAG) primers with an expected size of 980 bp (wild type), 3703 to 4208 bp (transgenic lines) at F4. On the other hand, the OmpK gene fragment was amplified from the gDNA and cDNA using OmpK_F (ATGCGTAAATCACTTTTAGCTCTTAGCC) and OmpK_R (TTAGAACTTGTAAGTTACTGC) primers with an expected band size of 750 bp. The housekeeping gene (GAPDH) used in this study was amplified from cDNA of wild-type as well as Nannochloropsis sp. transgenic lines using GAPDH_F (CCTGTGCGTCCACGGGCGC) and GAPDH_R (GAGGGTGGTCTCGGAAGCC) primers with an expected size of 502 bp. Amplification of the gene fragment from gDNA and cDNA was carried out using exTEN 2X PCR Master Mix (Apical Scientific, Singapore) from gDNA and cDNA of wild-type and transgenic Nannochloropsis sp. Amplification of OmpK gene fragment from gDNA indicates the stability of the foreign gene fragment in the Nannochloropsis sp. gDNA, while amplification of OmpK gene transcript from cDNA indicates the positive expression of the transcript in the transgenic line.
Results and discussion
Molecular identification of Nannochloropsis sp.
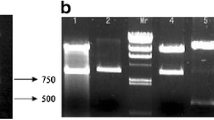
The amplification of 18S rRNA gene fragments was done using the universal 18S primer. The amplification of 18S rRNA gene fragments was carried out with the following forward and reverse primers, 5′-CAGAGGTGAAATTCTTGGA-3′ and 5′-TCACCTACGGAAACCTTGTTACGAC-3′. Amplification of a band ranging from 800 to 900 bp indicates successful amplification and the sequencing results obtained showed significant sequence similarity with Nannochloropsis sp. (Fig. 3).
Molecular identification of Nannochloropsis sp. via 18S gene fragment amplification and sequencing. a Amplification of 18S rRNA gene fragment from gDNA of Nannochloropsis sp. Lane M represents 100-bp DNA ladder (HyperLadder™ 100-bp ladder, NEB) and lane 1 represents the amplified gene fragment from Nannochloropsis sp. genomic DNA template. b Sequencing results obtained from BLAST tool which showed high sequence similarity of up to 82% with Nannochloropsis sp.
Integration of expression cassette site in transgenic Nannochloropsis sp. gDNA
DNAs extracted from the transgenic lines were analysed using PCR to amplify the homologous region where hypothetically is the site of integration. Analysis using PCR showed that survived transgenic lines of Nannochloropsis sp. transformants contain the expression cassette which is indicated by the single band between 10 and 1.5 kb (Fig. 4). The expected band size for transgenic lines transfected with VCP_OMPK_ZA1 and its derivatives is 3703 bp whereas for transgenic lines transfected withVCP_OMPK_ZA2 and its derivatives, it is at 4208 bp. If no integration of the vector into the Nannochloropsis sp. genome had occurred, a band of a size of 980 bp should be present similar to WT (wild type). All transgenic lines showed the presence of an expression cassette (Fig. 5).
Amplification of nitrate reductase to determine the integration of the expression cassette into the host genome. M1 and M2 represent 100-bp DNA ladder (Apical Scientific, Singapore) and 1-kbp DNA ladder (Apical Scientific, Singapore) respectively followed by six transgenic lines (V1, V2, CV1, CV2, CPV1 and CPV2) and wild type (WT)
Amplification of OmpK gene fragment in fourth generation (a), fifth generation (b) and sixth generation (c) of all six Nannochloropsis sp. transgenic line gDNA. M represents 100-bp DNA ladder (Apical Scientific, Singapore) followed by wild-type (WT), and six transgenic lines (V1, V2, CV1, CV2, CPV1 and CPV2)
Presence and stability of heterologous gene in transgenic Nannochloropsis sp. gDNA
Polymerase chain reaction (PCR) was performed to detect the presence of OmpK gene fragment with an amplicon size of 800–900 bp. In F4 and F5, successful amplification of the OmpK gene fragment for transgenic lines V1, V2, CV1 and CV2 is shown. However, for CPV1 and CPV2, the OmpK gene fragment was amplified in F4 only. As shown in Fig. 3, no amplification can be detected for both of these transgenic lines in F5. Unfortunately, in the sixth generation (F6), all six transgenic lines showed no amplification of the OmpK gene fragment where no single band appeared at 800–900 bp size. Therefore, the presence of the OmpK gene fragment can be considered to be stable to up to F5 in some transgenic Nannochlorosis sp. lines.
Expression of heterologous gene in transgenic Nannnochloropsis sp.
RNA Extraction was performed on the wild-type Nannochloropsis sp. and six lines of transgenic Nannochloropsis sp. namely as V1, V2, CV1, CV2, CPV1 and CPV2. Two generations of transgenic Nannochloropsis sp. namely the fourth generation (F4) and fifth generation (F5) were analysed. The expression of the housekeeping gene chosen in this study is as shown in Fig. 6a for generations 4 and 5 of the transgenic lines. The GAPDH is one of the most common housekeeping genes used in the comparison of gene expression data and it showed stable expression throughout despite the generation and line. In comparison, the expression of OmpK gene transcript for all transgenic lines in both generations 4 and 5 was evaluated (Fig. 6b. Based on the results obtained, the expression of the OmpK gene transcript in F4 can be detected in all six transgenic lines except in CPV2. It is postulated that the expression of the OmpK gene transcript may be too low for detection via RT-PCR in CPV2. For F5, the expression of OmpK gene transcript can be detected only in V1, V2, CV1 and CV2. Wild-type Nannochloropsis sp. acted as a control for both generations. The results correlate with the presence of the OmpK gene fragment where it could still be detected up to the 5th generation.
The expression of GAPDH (a) and OmpK (b) gene transcripts in fourth generation (F4) and fifth generation (F5) of wild-type and six transgenic lines of Nannochloropsis sp. obtained via RT-PCR. M represents 100-bp DNA ladder (Apical Scientific, Singapore) followed by wild-type (WT), and six transgenic lines (V1, V2, CV1, CV2, CPV1 and CPV2)
Overall, this study showed that there was a correlation between the presence of OmpK gene fragment in the transgenic lines and the expression of the OmpK gene. The detection and expression of the heterologous gene introduced into the cells could only be detected up to the fifth generation. For future work, expression analysis should be done using a quantitative approach like quantification polymerase chain reaction (qPCR) where this variation of PCR can detect the lower amount of expression effectively than the RT-PCR. Further optimization of the vector or transformation method in delivering the heterologous gene into the Nannochloropsis sp. to increase its stability in the genome is highly recommended and needed to produce a better vaccine carrier. Protein expression should also be considered in identifying a similar protein coding or structure of the original antigen.
Conclusion
This study described the presence, stability and expression of the OmpK gene fragment introduced into Nannochloropsis sp. as it could be a potential vaccine delivery system. Six transgenic lines namely V1, V2, CV1, CV2, CPV1 and CPV2 were subjected to PCR analysis for the detection of presence and stability and RT-PCR analysis for the expression of the gene transcript. From the results obtained, the presence of the transgene can be detected and shown to be stable in transgenic lines V1, V2, CV1 and CV2 up to the F5. The expression of the OmpK gene transcript can also be detected up to the F5. Overall, transgenic lines V1, V2, CV1 and CV2 showed presence, stability and expression of the OmpK gene up to the F5. From the results obtained, Nannochloropsis sp. can be considered a suitable vaccine carrier due to the stability of the transgene introduced. It is hoped that this study will contribute to the prevention of vibriosis disease which will then keep our aquaculture industry sustainable.
References
Allam B, Paillard C, Michel A (2000) Alterations in hemolymph and extrapallial fluid parameters in the Manila clam, Ruditapes philippinarum, challenged with the pathogen Vibrio tapetis. J Invertebr Pathol 76(1):63–69. https://doi.org/10.1006/jipa.2000.4940
Assefa A, Abunna F (2018) Maintenance of fish health in aquaculture: review of epidemiological approaches for prevention and control of infectious disease of fish. Vet Med Int 5432497:1–10. https://doi.org/10.1155/2018/5432497
Austin B, Austin DA (2012) Vibrionaceae representatives. Springer, In Bacterial fish pathogens. Dordrecht, pp 357–411. https://doi.org/10.1007/978-94-007-4884-2 ISBN: 978-94-007-4884-2
Cabello FC, Godfrey HP, Tomova A, Ivanova L, Dölz H, Millanao A, Buschmann AH (2013) Antimicrobial use in aquaculture re-examined: its relevance to antimicrobial resistance and to animal and human health. Environ Microbiol 5(7):1917–1942. https://doi.org/10.1111/1462-2920.12134
Chen HL, Li SS, Huang R, Tsai HJ (2008) Conditional production of a functional fish growth hormone in the transgenic line of Nannochloropsis Oculata (Eustigmatophyceae). J Phycol 44(3):768–776. https://doi.org/10.1111/j.1529-8817.2008.00508.x
Fern LL, Abidin AAZ, Yusof ZNB (2017) Upregulation of thiamine (vitamin B1) biosynthesis gene upon stress application in Anabaena sp. and Nannochloropsis oculata. J Plant Biotechnol 44:462–471. https://doi.org/10.5010/JPB.2017.44.4.462
Gjedrem T, Robinson N, Rye M (2012) The importance of selective breeding in aquaculture to meet future demands for animal protein: a review. Aquaculture 350-351:117–129. https://doi.org/10.1016/j.aquaculture.2012.04.008
Inoue T, Matsuzaki T, Tanaka S (1995) A 26-kDa outer membrane protein, OmpK, common to Vibrio species is the receptor for a broad-host-range vibriophage, KVP40. FEMS Microbiol Lett 125(1):101–105. https://doi.org/10.1111/j.1574-6968.1995.tb07342.x
Kherraf A, Wafaa T, Tehami W, Boufeldja W, Yahla I, Dra GA, Mansour IFZ, BENALI M (2017) Determination of the nutritional and functional metabolites of Nannochloropsis gaditana produced in Algeria and of its antioxidant activity. Der Pharma Chem 9(14):8–13 https://www.derpharmachemica.com/
Li SS, Tsai HJ (2009) Transgenic microalgae as a non-antibiotic bactericide producer to defend against bacterial pathogen infection in the fish digestive tract. Fish Shellfish Immun 26(2):316–325. https://doi.org/10.1016/j.fsi.2008.07.004
Meyer FP (1991) Aquaculture disease and health management. J Anim Sci 69(10):4201–4208. https://doi.org/10.2527/1991.69104201x
Potvin G, Zhang Z (2010) Strategies for high-level recombinant protein expression in transgenic microalgae: a review. Biotechnol Adv 28(6):910–918. https://doi.org/10.1016/j.biotechadv.2010.08.006
Qian RH, Xiao ZH, Zhang CW, Chu WY, Wang LS, Zhou HH, Wei YW, Yu L (2008) A conserved outer membrane protein as an effective vaccine candidate from Vibrio alginolyticus. Aquaculture 278(1–4):5–9. https://doi.org/10.1016/j.aquaculture.2008.03.010
Rosano GL, Ceccarelli E (2014) A. Recombinant protein expression in microbial systems. Front Microbiol 5(341):1–2. https://doi.org/10.3389/fmicb.2014.00341
Safafar H, Hass MZ, Møller P, Holdt SL, Jacobsen C (2016) High-EPA biomass from Nannochloropsis salina cultivated in a flat-panel PhotoBioreactor on a process water-enriched growth medium. Mar Drugs 14(8):1–19. https://doi.org/10.3390/md14080144
Shoemaker CA, Klesius PH, Evans JJ, Arias CR (2009) Use of modified live vaccines in aquaculture. J World Aquacult Soc 40(5):573–585. https://doi.org/10.1111/j.1749-7345.2009.00279
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Abidin, A.A.Z., Othman, N.A., Yusoff, F.M. et al. Determination of transgene stability in Nannochloropsis sp. transformed with immunogenic peptide for oral vaccination against vibriosis. Aquacult Int 29, 477–486 (2021). https://doi.org/10.1007/s10499-020-00634-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10499-020-00634-w