Abstract
Mitochondria dysfunction is implicated in cell death, inflammation, and autoimmunity. During viral infections, some viruses employ different strategies to disrupt mitochondria-dependent apoptosis, while others, including SARS-CoV-2, induce host cell apoptosis to facilitate replication and immune system modulation. Given mitochondrial DNAs (mtDNA) role as a pro-inflammatory damage-associated molecular pattern in inflammatory diseases, we examined its levels in the serum of COVID-19 patients and found it to be high relative to levels in healthy donors. Furthermore, comparison of serum protein profiles between healthy individuals and SARS-CoV-2-infected patients revealed unique bands in the COVID-19 patients. Using mass spectroscopy, we identified over 15 proteins, whose levels in the serum of COVID-19 patients were 4- to 780-fold higher. As mtDNA release from the mitochondria is mediated by the oligomeric form of the mitochondrial-gatekeeper—the voltage-dependent anion-selective channel 1 (VDAC1)—we investigated whether SARS-CoV-2 protein alters VDAC1 expression. Among the three selected SARS-CoV-2 proteins, small envelope (E), nucleocapsid (N), and accessory 3b proteins, the E-protein induced VDAC1 overexpression, VDAC1 oligomerization, cell death, and mtDNA release. Additionally, this protein led to mitochondrial dysfunction, as evidenced by increased mitochondrial ROS production and cytosolic Ca2+ levels. These findings suggest that SARS-CoV-2 E-protein induces mitochondrial dysfunction, apoptosis, and mtDNA release via VDAC1 modulation. mtDNA that accumulates in the blood activates the cGAS-STING pathway, triggering inflammatory cytokine and chemokine expression that contribute to the cytokine storm and tissue damage seen in cases of severe COVID-19.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Coronaviruses (CoVs) are enveloped, single-stranded RNA viruses that have a wide range of natural hosts. Among the seven CoVs known to infect humans, only three—SARS-CoV-2, MERS-CoV, and SARS-CoV—have been associated with severe acute respiratory syndrome (SARS).
SARS-CoV-2 manifests across a broad spectrum of clinical presentations that range from mild respiratory symptoms to pneumonia. In severe cases, an exaggerated inflammatory response, often termed a “cytokine storm”, ensues, characterized by elevated levels of circulating tumor necrosis factor (TNF) and interleukin-6 (IL-6) [1]. This cytokine storm attracts mononuclear cells to the lungs [2], which contributes to tissue damage and multi-organ failure, and ultimately leads to death.
The SARS-CoV genome comprises two-thirds of the RNA that is located within the first open reading frame (ORF1a/b). This translates into two polyproteins, pp1a and pp1b, that give rise to 16 non-structural proteins. The remaining portion of the viral genome encodes several structural and accessory proteins: the spike (S) protein, a transmembrane glycoprotein that is present on the viral surface, cleaved by host-cell proteases, which after anchoring enters the host cells via angiotensin receptor-2 (ACE2): the matrix (M) protein responsible for nutrient transport; the small envelope (E) protein, an integral membrane protein; and nucleocapsid (N) protein, which forms the capsid surrounding the RNA genome [2]. In addition to these proteins, SARS-CoV encodes various accessory proteins, such as 3a and 3b proteins, which play roles in virus pathology. These accessory proteins interfere with the host innate immune response, and they contribute to mitochondria dysfunction and apoptosis [3, 4].
Viruses have evolved distinct strategies to evade biochemical and immunological defenses of the host, including the regulation of apoptosis, inflammation, and immune reactions. Some viruses induce apoptosis, either by eliminating uninfected immune cells or breaking down infected cells to facilitate viral spread [5]. Viruses inhibit or induce apoptosis in various tissues and in different ways [6]—some by regulating the expression of proteins from the Bcl-2 family (such as Bid, Bcl2) or by mimicking them [5, 7], and others either directly by viral proteins [6, 8] or indirectly by activating cellular mechanism-leading to apoptosis that enhance viral infection efficiency [9] often by directly targeting the mitochondria [10]. As an example, HIV-1 induces neuronal and immunological damage through mitochondria-mediated apoptosis by at least three distinct mechanisms [11]. Viruses also modulate host cell metabolism and physiology, which results in altered cellular functions [12].
The mitochondria control cellular homeostasis, metabolism, innate immunity, apoptosis, and more [13,14,15]. They play a key function in apoptosis with VDAC1, situated on the outer mitochondrial membrane (OMM), acting as a mitochondrial gatekeeper that controls both metabolic and apoptostic functions. VDAC1 modulates apoptosis by binding anti-apoptotic proteins and mediating the release of cytochrome c and other pro-apoptotic factors [16,17,18].
Upon induction of apoptosis by various inducers, such as Ca2+ or UV irradiation, VDAC1 expression levels rise, shifting the equilibrium towards an oligomeric state with a large pore, which allows the release of mitochondrial pro-apoptotic proteins [19]. Thus, the assambly of this new oligomeric VDAC1 channel is induced by diverse initiating cascades, stressors, and apoptotic triggers [16, 18, 20,21,22]. Notably, the channel also releases mitochondrial DNA (mtDNA) [23].
mtDNA shares many similarities with immunogenic bacterial DNA and is recognized as a pro-inflammatory damage-associated molecular pattern (DAMP) that contributes to the pathogenesis of various inflammatory diseases. mtDNA was demonstrated to be an inevitable factor during cGAS-STING activation in SARS-CoV-2 infection and an inhibitor of VDAC1-oligomerization that we developed [24], VBIT-4, which suppressed mtDNA release [23] protected against cGAS-STING activation in SARS-CoV-2 infection [25].
Numerous studies have illustrated the pivotal role of the mitochondria, particularly involving VDAC1, in the pathogenic mechanisms of various viruses. These viruses employ diverse strategies, such as inducing VDAC1 overexpression or directly interacting with it, thereby modulating mitochondrial function. Additionally, silencing VDAC1 expression has been shown to significantly reduce the expression of viral proteins [19, 26]. Examples of this are: (i) in the hepatitis B virus (HBV), HBx protein forms a complex with VDAC1 that impacts mitochondrial physiology [27]; (ii) expression of the hepatitis E virus (HEV) capsid protein Orf3 increases VDAC1 levels [28]; (iii) human immunodeficiency virus type 1 (HIV-1) protein R (Vpr) interacts with VDAC1, causing cell cycle arrest and apoptosis in infected T-lymphocytes [29]; (iv) downregulation of VDAC1 expression results in reduced expression of Dengue virus (DENV) proteins NS1, NS3, NS5, and DENVE [30]; (v) infectious bursal disease virus (IBDV) protein VP5 induces apoptosis by interacting with VDAC1, and silencing VDAC1 expression decreases the expression of VP1, VP2, and VP5 [31], and (vi) coronaviruses, including SARS-CoV-2, encode both pro-apoptotic and anti-apoptotic proteins, regulating apoptosis progression [32]. A distinct population of T cells in COVID-19 patients exhibits significant upregulation of VDAC1 and VDAC1-dependent apoptosis that can be inhibited by VBIT-4 [33].
Building on these findings, the current study examined the relationship between SARS-CoV-2-derived proteins and the mitochondrial protein VDAC1, focusing on apoptosis and mtDNA release. We investigated the role of VDAC1 in the pathogenesis of SARS-CoV-2, which manifests as COVID-19. Among the array of proteins encoded by SARS-CoV, we selected four that are associated with virus pathology and are connected to mitochondria dysfunction and apoptosis [2]. The envelope (E-) protein has been shown to translocate to the cell surface, altering the host-cell membrane’s permeability to facilitate viral entry, which promotes replication, proliferation, and inflammation cascade induction by SARS-CoV [34]. The N-protein induces apoptosis via the mitochondrial apoptotic pathway under starvation of serum, increasing ROS levels, which leads to the loss of membrane potential (ΔΨm) and induces apoptosis [35]. The SARS-CoV accessory 3a protein induces extensive cell death via mitochondrial pathways [3], and the SARS-CoV accessory 3b protein, proposed to contribute to SARS pathogenesis in humans, is reported to be localized to the nucleolus and mitochondria, inducing cell growth arrest [4, 36], apoptosis, and necrosis [37]. The 3a and 3b proteins modulate the host innate immune response to SARS-CoV infection by inhibiting interferon production and signaling [38].
Our investigation reveals that the cell expression of the E-protein of SARS-CoV-2 results in VDAC1 overexpression, oligomerization, and subsequent cell death. Given that VDAC1 oligomers are implicated in the release of mtDNA [23] and considering the protective effect of a VDAC1-oligomerization inhibitor VBIT-4 against cell death [16, 18, 20,21,22] and mtDNA release [23], as well as the cytokine storm induced by SARS-CoV-2 infection [25], we analyzed the presence of mtDNA in the serum of COVID-19 patients and searched for SARS-CoV-2 protein targeting VDAC1. In this paper, we present evidence of elevated levels of mtDNA and of specific proteins in the serum of COVID-19 patients. Additionally, we show that the E-protein, by inducing VDAC1 overexpression and oligomerization, allows the release of pro-apoptotic proteins and mtDNA, leading to cell death and to inflammation. These findings strongly imply the involvement of VDAC1 in the pathology of this virus.
Materials and methods
Materials
Propidium iodide (PI), Ponseau S, acridine orange, ethidium bromide, tris (hydroxymethyl)aminomethane, Tween-20, digitonin, and protease inhibitor cocktail were obtained from Sigma (St. Louis, MO). Phosphate-buffer saline (PBS), Dulbecco’s Modified Eagle’s Medium (DMEM), supplement fetal bovine serum (FBS), and penicillin–streptomycin were obtained from Gibco (Grand Island, NY). Dimethyl sulfoxide (DMSO) was purchased from MP Biomedicals (Solon, OH). Polyvalent JetPrime transfection reagent was from Polyplus (llkirch-Graffenstaden, France) and Prime Fect was from RJH Biosciences (Edmonton, Alberta, Canada). Ethylene glycol bis(succinimidyl succinate) (EGS), MitoSOX™ red mitochondrial superoxide indicator and Fluo-4 AM cell permeant were obtained from Thermo Fisher (Waltham, MA, US). Annexin V was purchased from Enzo Life Sciences (Farmingdale, NY, US). Hank’s Balanced Salt Solution (HBSS) was from Biological Industries (Beit-Haemek, Israel). Power SYBER Green Master Mix was obtained from Applied Biosystems (Foster City, CA).
Subjects
Blood samples were collected from 51 hospitalized patients infected with COVID-19 and 24 healthy non-hospitalized controls without any infection, matched for age, from the COVID-19 biobank established at Shaare Zedek Medical Center (SZMC) in Jerusalem, Israel during the pandemic (March 6 to May 30, 2020). Clinical and demographic data, including age, gender, degree of oxygen support, severity of illness, and lab parameters were obtained from the computerized medical records or through interviews with study participants (see Table S1). The severity of COVID‐19 was evaluated based on the NIH guidelines (https://www-covid19treatmentguidelines-nih-gov.szmc.idm.oclc.org). Mild illness: Individuals with any of the various signs and symptoms of COVID-19 (e.g., fever, cough, sore throat, malaise, headache, muscle pain, nausea, vomiting, diarrhea, loss of taste and smell), but who do not have shortness of breath, dyspnea, or abnormal chest imaging. Moderate illness: Individuals who show evidence of lower respiratory disease during clinical assessment or imaging and who have an oxygen saturation measured by pulse oximetry (SpO2) ≥ 94% in room air at sea level. Severe illness: Individuals who have an SpO2 < 94% in room air at sea level. Critical illness: Individuals who have respiratory failure, septic shock, or multiple organ dysfunction. The study was conducted in accordance with the Helsinki guidelines and received approval from the SZMC Institutional Review Board.
Cell culture and transfection
SHSY-5Y (CRL-2266, Human neuroblastoma, epithelial), A549 (CCL-185, human lung carcinoma, epithelial-like), and PC-3 (CRL1435, human prostate, adenocarcinoma) cell lines were obtained from the American Type Culture Collection (ATCC) (Manassas, VA). They were maintained using Dulbecco’s Modified Eagle’s Medium (DMEM), supplemented with 10% inactivated fetal bovine serum (FBS), and with 100 U/mL penicillin and 100 μg/mL streptomycin. Cells were grown at 37°C in 5% CO2 in a humidified incubator. Cell lines were routinely tested for mycoplasma contamination.
Cells were transiently transfected with 1 or 2 μg DNA for 24h, 48h, or 72h. The DNA included either an empty pcDNA3.1 plasmid or the pcDNA3.1 plasmid encoding for E-, N-, or 3b protein. Transfection was performed using JetPRIME transfection reagent (Polyplus, Illkirch-Graffenstaden, France) following the manufacturer’s instructions. The N, E, and 3b genes were cloned into a pcDNA3.1 vector with BamHI (GGATCC) and EcoRI (GAATTC) cloning sites and conferred ampicillin resistance (Synbio Technologies; NJ, USA).
mtDNA isolation and analysis
Total serum DNA was extracted using a DNeasy Blood & Tissue Kit (Qiagen), according to the manufacturer’s instructions. mtDNA encoding for ND2, COX-III, D-Loop 1, and D-Loop 2 was analyzed using specific primers (Table S2) and quantified through quantitative real-time PCR (qPCR) with SYBR Green. Amplification of samples was conducted using a 7300 Real-Time PCR System (Applied Biosystems). For quantification of mtDNA released to the cytosol, cells were harvested 48h post transfection, pelleted, and re-suspended in 170 μl of buffer, containing 150 mM NaCl, 50 mM HEPES pH 7.4, and 25 µg/ml digitonin. Cells then were incubated for 10 min at room temperature onto a rotator, followed by centrifugation at 16,000 g at 4°C for 25 min. For q-RT-PCR, a 1:10 supernatant dilution was used to determine mtDNA using the specific primers listed in Table S2.
Chemical cross-linking
Following the designated treatment, cells were collected and incubated in PBS at pH 8.3 (1 mg/ml) containing the cross-linking reagent EGS (100 μM) for 15 min at 30 °C. Samples (60 µg protein) were then subjected to SDS-PAGE, followed by immunoblotting using anti-VDAC1 antibodies. Quantitative analysis of immuno-reactive VDAC1 dimer, trimer, and multimer bands was performed using Image J software.
Cell death analyses
Cell death was analyzed by propidium iodide (PI) staining (final concentration of 6.25 g/mL), followed by flow cytometry with an iCyt sy3200 Benchtop Cell Sorter/Analyzer (Sony Biotechnology Inc., San Jose, CA, USA) and analysis with EC800 software. Apoptosis was analyzed by PI and nnexin V-FITC staining, which was carried out according to the manufacturer’s instructions with minor modifications. After transfection, cells were harvested (1,500 g, 5 min), washed and re-suspended in 200 µl of binding buffer (10 mM Hepes/NaOH, pH 7.4, 140 mM NaCl, and 2.5 mM CaCl2). Annexin V-FITC/PI staining was performed, and the samples were analyzed by flow cytometry. At least 10,000 events were recorded, represented as dot plots. Cell death was also analyzed by acridine orange and ethidium bromide staining (100 g/ml) and light microscopy (LX2-KSP; Olympus).
Intracellular Ca2+ level analysis
Fluo-4-AM was used to monitor changes in cytosolic Ca2+ levels. SHSY-5Y cells were harvested 48h post transfection, collected (1,500 × g for 5 min), washed with Hank’s Balanced Salt Solution (HBSS) supplemented with 1.8 mM CaCl2 (HBSS +), and incubated with 2 μM Fluo-4 in 200 μl HBSS( +) buffer for 30 min at 37°C in a light-protected environment. After removing the excess dye by washing with HBSS( +), the cellular free Ca2+ concentration was promptly measured using an iCyt sy3200 Benchtop Cell Sorter/Analyzer (Sony Biotechnology Inc., San Jose, CA). At least 10,000 events were recorded by the FL2 detector, represented as a histogram, and analyzed by EC800 software (Sony Biotechnology Inc., San Jose, CA). Positive cells showed a shift to an enhanced level of green fluorescence (FL2).
Reactive oxygen species (ROS) level analysis
To assess mitochondrial ROS accumulation, SHSY-5Y cells were collected 48h post transfection and then treated with 5 μM MitoSOX™ Red, a mitochondrial superoxide indicator for live-cell imaging, for 10 min at 37°C. Fluorescence intensity was measured using flow cytometry (iCyt, Sony Biotechnology, San Jose, CA). Analysis was performed as in the intracellular Ca2+ level analysis above.
Gel electrophoresis and immunoblotting
Cells were lysed using lysis buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1.5 mM MgCl2, 10% glycerol, 1% Triton X-100), supplemented with a protease inhibitor cocktail for 15 min on ice. The lysates were subsequently centrifuged at 12,000xg (10 min at 4°C), and protein concentration was determined using a Lowry assay.
Protein aliquots (10–20 μg) were subjected to SDS-PAGE, and then were electro-transferred onto nitrocellulose membranes for immunostaining. The membranes were incubated with a blocking solution containing 5% non-fat dry milk and 0.1% Tween-20 in Tris-buffered saline (TBST), followed by incubation with primary antibodies. Subsequently, membranes were incubated with HRP-conjugated anti-mouse or anti-rabbit IgG as secondary antibodies. HRP activity was detected using an enhanced chemiluminescent substrate (Advantsa, San Jose, CA). Band intensity was quantified using Image J software or FUSION-FX (Vilber Lourmat, France).
Purified VDAC1 and SARS-CoV-2 proteins
Purified SARS-CoV-2 nucleocapsid N-protein (N; S014660-05) and the envelope E-protein (MBS8309649) were obtained from Synbio Technologies (NJ, USA) or from MyBioSource (San Diego, CA). VDAC1 protein was purified from rat liver mitochondria following the protocol previously described [39].
Micro-scale thermophoresis (MST) assay
MST analysis was performed using a NanoTemper Monolith NT.115 apparatus, as described previously [24, 40]. Briefly, purified VDAC1 or purified MAVS proteins were fluorescently labeled using a NanoTemper Protein labeling kit BLUE (L001, NanoTemper Technologies, GmbH). A constant protein concentration (138 nM for VDAC1 or 105 nM for MAVS) was incubated with different concentrations of the tested protein in an MST binding buffer (20mM Tris–HCl, 200mM NaCl, pH = 8) for 30 min at 37°C in the dark. Subsequently, 3–5 µl of the samples were loaded into a glass capillary (Monolith NT Capillaries), and a thermophoresis analysis was performed (LED 20%, IR laser 80%).
Liquid chromatography-high-resolution mass spectrometry (LC-HR-MS) and proteomics analysis
Protein samples were subjected to SDS-PAGE followed by Coomassie staining and protein bands of ~ 50, 19, and 14 kDa were excised and subjected to in-gel digestion using trypsin, following the manufacturer’s protocol (Promega). Peptides were then extracted from the gel and loaded onto a LC-HR-MS.
LC/MS analysis was performed using an Eksigent nano‐HPLC (model nanoLC-2D, Netherlands) connected to an LTQ Orbitrap XL ETD (Thermo Fisher Scientific, Germany & USA). Peptide separation was achieved using a reverse-phase C-18 column [Acclaim PepMap, 75 ID (µm) 15 cm long: (Thermo Fisher Scientific)]. Peptides were eluted with a 70‐min linear gradient, starting with 100% buffer A (5% acetonitrile, 0.1% formic acid) to 80% buffer B (80% acetonitrile, 0.1% formic acid) at a flow rate of 300 nl/min. Full scans were acquired at 60,000 resolutions in the Orbitrap analyzer, followed by CID MS/MS analysis of the five most abundant peaks in the data-dependent mode. Fragmentation and detection of fragments were performed in the linear ion trap, with a minimum signal trigger threshold set at 500. The maximum ion fill-time settings were 500 ms for the high-resolution full scan in the Orbitrap analyzer and 200 ms for MS/MS analysis in the ion trap. The AGC settings were 5X105 and 1X104 for the Orbitrap and linear ion trap analyzers, respectively.
Proteins were identified and validated using SEQUEST search algorithms against the human protein sequences (NCBI and UniprotKB/SwissProt proteome database collection) operated under Proteome Discoverer 2.4 software (Thermo Fisher Scientific). Mass tolerances for precursors and fragmentations were set to 10 ppm and 0.8 Da, respectively. Only proteins containing at least two peptides of high confidence (Xcore 2 or 2.5 for doubly- or triply-charged species, respectively, or more) were selected.
An LC-HR-MS and proteomics analysis were carried out at the de Botton Institute for Protein Profiling, G-INCPM at the Weizmann Institute of Science in Rehovot, Israel, with an additional analysis performed at the Ilse Katz Institute for Nanoscale Science & Technology at Ben-Gurion University in Beer Sheva.
Machine learning and statistics
For machine-learning tasks, models were built using the Python sklearn package (version 1.4.2), with or without dimension reduction techniques such as PCA, truncatedSVD, and sparsePCA algorithms. The models were trained using the fit function of the decision tree classifier object in sklearn. Validation was performed using a leave-one-out cross-validation approach. Unless otherwise stated, a significance cutoff of α = 0.05 was applied.
The data from cells in culture are shown as the mean ± SEM of at least three independent experiments. Significance of differences was calculated by a two-tailed Student’s t-test using the T-Test function provided by Microsoft Excel. Similarly, statistical significance between healthy and COVID-12 infected individuals was determined using Student’s t-test, conducted with the T-Test function in Microsoft Excel. Statistical significance is reported at p ≤ 0.05 (*), p ≤ 0.01 (**), or p ≤ 0.001 (***).
Results
Given that certain viruses cause dysfunction of mitochondria and modulate VDAC1 expression and function [19, 26,27,28,29,30,31,32, 41], leading to cell death, inflammation, and autoimmune responses, we investigated the involvement of the mitochondria and VDAC1 in the pathology of SARS-CoV-2. Since mtDNA acts as a pro-inflammatory agent, we analyzed its levels in the serum of COVID-19 patients. In addition, we compared serum protein profiles of healthy and SARS-CoV-2-infected patients. Finally, we tested the effects of selected COVID-19 expression proteins on mitochondrial dysfunction, mtDNA release, and apoptosis induction.
Patient characterization
During the pandemic period (March 6–April 7, 2020), blood samples were collected from 51 hospitalized patients infected with COVID-19. Onset of symptoms had occurred within 10 days prior to admission in 47/51 patients. Of the patients, 23 (45%) were female, with a mean age of 62.6 ± 17.7. Twenty-four healthy subjects served as controls: 12 (50%) were female, and mean age was 57 ± 14.6. (p = 0.18).
Of the COVID-19 patients, 17 were mildly ill, 10 had moderate disease, 21 had severe disease, and three were critically ill. Age above 65 was associated with severe or critical disease (OR 4.9, 95% CI 1.45–16.5, p = 0.008), as well as mean CRP level (OR 1.2 95% CI 1.08–1.4, p = 0001). A non-significant trend towards higher ferritin and d-dimer levels was observed among severely ill patients.
Mitochondrial DNA (mtDNA) elevated in serum of COVID-19-infected patients
As SARS-CoV-2 induces COVID-19 in humans, and is linked to the activation of the cGAS-STING pathway triggered by mtDNA [25], we analyzed mtDNA levels in the blood of SARS-CoV-2-infected patients (Fig. 1). Intact mtDNA is large (16–17 kb) and anchored to the mitochondrial inner membrane (MIM) within nucleoid complexes [42]. However, as we previously demonstrated [23], short and free intra-mtDNA fragments (fmtDNA) are released from the mitochondria through the VDAC1 oligomer mega channel. The fmtDNA was isolated from heated serum samples of both healthy individuals and COVID-19 patients, and their quantities were determined using RT-qPCR specific primers (Table S2), targeting three genes and one non-coding sequence: cytochrome c oxidase subunit III (COX-III), NADH dehydrogenase 2 (ND-2), and the DNA displacement loop (D-Loop-I and D-Loop-II) (Fig. 1A). The D-Loop region, situated within the major non-coding region (NCR) of many mitochondrial genomes, is characterized by a triple-stranded structure formed by the stable integration of a third short DNA strand, known as 7S DNA. This region harbors crucial elements for transcription and replication processes [43].
High levels of mtDNA found in serum of COVID-19 patients, relative to healthy donors. A The human mtDNA genome with the selected genes for analysis are marked in the square. The qPCR expression of mtDNA, B ND-2, C D-Loop-II, D COX-III, and E D-Loop-I in healthy (n = 24) and COVID-19 (n = 51) subjects. The fold change of mtDNA in the COVID-19 patients was higher than in healthy controls. Each dot represents an individual. F Summary of mtDNA for the indicated gene present in serum of the patients presented as a fold change relative to the levels in healthy donors. Results represent the means ± SEM, *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001
High levels of mtDNA-derived fragments were found in the serum of SARS-CoV-2-infected patients compared to healthy donors (Fig. 1B–E). Specifically, while the average levels of COX-III, ND-2, and D-Loop-I were 4–sixfold higher relative to their levels in serum from healthy donors, the levels of D-Loop-II were an average of 13-fold higher (Fig. 1F). These results suggest higher production of mtDNA-D-Loop-II, indicating that the entire mtDNA molecule is not being released. This is consistent with previous findings that the sequences corresponding to a specific region within the D-Loop of the mitochondrial genome are overrepresented in the mtDNA pool [23].
We investigated whether the presence of mtDNA in sera could serve as an indicator of disease progression and/or severity. Multiple machine-learning analyses were conducted; however, the mtDNA sera levels did not emerge as a significant predictor of disease severity (Table S3).
Additionally, incorporating clinical data such as blood test results (e.g., white blood cells WBC), levels of proteins like D-dimer, ferritin, or CRP, or information about underlying conditions (e.g., hypertension) did not significantly improve prediction of disease severity.
As discussed below, the presence of plasma mtDNA exhibits a time-dependent pattern, with a peak observed at 24h post-disease onset [44]. This temporal variation may account for why our study failed to predict disease severity based on mtDNA levels, as the blood samples were collected at various time points following disease identification.
Comparison of serum protein profiles of healthy and SARS-CoV-2-infected patients
Next, we conducted a comparison of protein profiles in serum samples obtained from healthy individuals and patients infected with SARS-CoV-2 using SDS-poly acryl-amide gel electrophoresis (SDS-PAGE) (Fig. 2A). We identified three protein bands, defined according to their apparent molecular mass as p49, p17, and p14, that were present in the serum from SARS-CoV-2-infected patients, but absent in that of the healthy group. We identified three distinct protein patterns and consequently defined three patient categories based on these profiles: Profile I: p49, p17, and p14 are present; Profile II: p49 and p17 are present, while p14 is undetectable; Profile III: p49 and p14 are present, while p17 is undetectable. These are presented as the fold change (FC) relative to their levels in healthy donors (Fig. 2B).
Serum of COVID-19 patients contains high levels of selected proteins. A Serum samples (30 μg protein) from patients and healthy donors were subjected to SDS-PAGE, followed by Coomassie blue staining. The protein bands found mainly in the COVID-19 samples are indicated and labeled as p95, p52, p49, p17, and p14, according to their estimated molecular mass. B Three categories of patients were defined: Protein profiles I, II, III, presenting p49 + p17 + p14, p49 + p17, and p49 + p14, respectively, are presented as the fold change (FC) relative to their levels in healthy donors. C Quantitative analysis of p52 and p95 levels in serum samples from healthy and COVID-19 samples. The apparent reduction in p95 levels in patients seems to be due to the presence of additional protein bands in the samples, which leads to a lower relative amount of the protein
For comparison, the levels of two other proteins, p95 and p52, were analyzed and found to be similar in the sera from both COVID-19 and healthy donors (Fig. 2C).
A summary of the results of fmtDNA and differential present protein levels in serum obtained from COVID-19 patients and their diseases state is presented in Table 1.
These proteins were also subjected to multiple machine-learning analyses to test whether they could serve as an indicator of disease progression or severity. However, their predictive value, alone or in combination of mtDNA levels, was not statistically significant.
To identify these proteins, the respective bands were excised from the gel and subjected to LC–MS/MS analysis in two different proteomics centers. We selected the LC–MS/MS-identified proteins exhibiting over 18% coverage, finding that all the identified peptides were unique to the corresponding protein. These results are summarized in Table 2, showing that the FC in protein levels between patients and healthy donors ranged from a 4- to 780-fold increase, also the number of the identified peptide unique to each identified protein, the percentage of the protein sequence covered by the identified peptides. The identified peptide sequences are located within the protein sequence (Fig. S1). The functions of these proteins and their possible association with COVID-19 are presented below and extended in the Supplementary Materials.
The p49 protein band was found to consist of several proteins including:
(1) Haptoglobin, which binds to hemoglobin, forming the haptoglobin–hemoglobin complex that is subsequently cleared by the reticuloendothelial system [45]; (2) complement component C8, a beta chain that is a constituent of the membrane attack complex (MAC) involved in mediating cell lysis and, thus, affects cell membrane integrity [46]; (3) alpha-1-antichymotrypsin (AACT), a circulating serine protease inhibitor derived from the liver; (4) serum albumin, which performs various essential functions such as maintaining plasma oncotic pressure and blood pH. It also acts as a carrier protein for steroids, hemin, fatty acids, hormones, amino acids, drugs, nutrients, and metal ions [47]; and (5) CD5 antigen-like (CD5L), which is primarily secreted by macrophages in lymphoid tissues during inflammatory responses [48].
Within the p17 protein band, several proteins were identified, including:
(1) C-reactive protein (CRP), known for its rapid elevation in response to inflammation and infection [49, 50]; (2) apolipoprotein A-1 (ApoA1), a primary protein constituent of high-density lipoprotein (HDL) particles; (3) alpha-1-anti-trypsin (AAT), a serine protease inhibitor that protects the lungs from neutrophil elastase which can damage lung tissue [51]; (4) immunoglobulin Joining chain (J chain), which is essential for the formation and stabilization of polymeric Ig structures [52]; (5) retinoic acid receptor responder 2 (CRABP-II), a soluble cytosolic protein belonging to the family of intracellular lipid-binding proteins. It transports retinoic acid (RA), thereby enhancing its transcriptional activity [53]; and (6) retinol-binding protein 4 (RBP4), a transporter protein responsible for carrying retinol from liver stores to peripheral tissues [54]. Upon association with transthyretin (TTR), the retinol/RBP4/TTR complex is released into the bloodstream to deliver retinol to tissues [54].
The p14 protein band was found to contain several proteins:
(1) Transthyretin (TTR or TBPA), a tetrameric transporter of the thyroid hormone thyroxine and the retinol-binding protein (RBP) bound to retinol [55]; (2) serum amyloid A4 (SAA4), an apolipoprotein found in high-density lipoproteins. Different isoforms of SAA are expressed constitutively at different levels or in response to inflammatory stimuli [56]; (3) apolipoprotein C-IV (APOC4), a lipid-binding protein with a role in lipoprotein metabolism [57]; and (4) immunoglobulin kappa constant (IGKC), representing one of the immunoglobulin (Ig) isotypes.
Finally, among the proteins exhibiting increased expression levels are those associated with immune response and inflammation. Notably, three of the identified proteins—CRABP-II, RBP4, and transthyretin—are involved in retinoid signaling, thus, influencing the regulation of Type I interferon synthesis and potentially contributing to excessive inflammation.
Effects of the expression of the three SARS-CoV-2 encoded proteins on mitochondria-mediated apoptosis, VDAC1 expression levels, and VDAC1 oligomeric state
Next, we selected SARS-CoV-2 proteins known to potentially induce mitochondrial dysfunction and apoptosis [3, 4]—the small envelope E- and nucleocapsid N-, and 3a- and 3b- proteins (Fig. 3A)—and tested their cell death induction and VDACI involvement. While the plasmid encoding 3a could not be successfully produced in bacteria, plasmids encoding for the E- and N- and 3b- proteins were successfully generated. Cells were transfected with E, N, 3b, or empty plasmid and, subsequently, assessed for apoptosis induction, VDAC1 expression levels, and VDAC1 oligomerization (Figs. 3B–D and 4). E-protein expression, but not N- and 3b, led to an increase in VDAC1 expression levels (Fig. 3B).
SHSY-5Y cell expression of SARS-CoV-2 E-protein induces VDAC1 overexpression and oligomerization. A (The SARS-CoV-2 genome contains the replicase gene, which encodes two open reading frames: ORF1a and ORF1b (two-thirds). The other genome encodes several ORFs including the spike glycoprotein (S), membrane (M), envelope (E), and nucleocapsid (N). ORFs encode several accessory proteins with their functions less well defined. ORFs 1a and 1b translated to produce the replicase pp1a and pp1ab polyproteins b [58]. B SHSY-5Y cells were transfected with 2μg pcDNA3.1 plasmid (empty) or encoding for proteins E, N, or 3b, and 48h post transfection, cells were harvested, lysed, and subjected to SDS-PAGE and immunoblotting using anti-VDAC1 antibodies. VDAC1 expression levels are presented in relative units (RUs), relative to their levels in the empty plasmid-transfected cells and are shown below the immunoblot. Coomassie blue stained of the blot is shown as a loading control. C, D SHSY-5Y cells were transfected with 1or 2 μg of the indicated plasmid (empty, E, N, or 3b) and 48h post transfection, cells were harvested and subjected to cross-linking using EGS (100 μM, 1 mg protein/ml) to monitor VDAC1 oligomerization as revealed by immunoblotting (C), and VDAC1 dimers were quantified using ImageJ software (D)
SHSY-5Y cell expression of SARS-CoV-2 E-protein induces cell death. A, B SHSY-5Y cells were transfected with 2 μg of pcDNA3.1 plasmid, empty, or encoding for E-, N-, or 3b proteins. 24h, 48h, or 72h post transfection, cells were harvested and analyzed for cell death using PI staining and flow cytometry. Quantitative analysis (A) and representative FACS histograms for the expressed viral proteins (B) are shown. Results represent the means ± SEM (n = 3) *p ≤ 0.05; **p ≤ 0.01; **p ≤ 0.001; NS non-specific. C, D Cells were transfected with 2 μg of pcDNA3.1 (empty) plasmid or encoding for E-, N-, or 3b proteins, and 48h post transfection, they were analyzed for cell death by staining with acridine orange and ethidium bromide, visualized with a microscope (LX2-KSP; Olympus). Representative images, captured by a CCD camera are shown, with the labeled squares pointing to the areas enlarged below (C), and quantification of ethidium bromide staining intensity (D). E, F SHSY-5Y cells were transfected with 2 μg of pcDNA3.1 plasmid (empty) or encoding for E-protein, and 48h post transfection, cells were subjected to PI/annexin V-FITC staining and flow cytometry analysis. Green and red boxes present live cells and dead cells, respectively. F Quantitative analysis of three independent experiments as shown in E. Results represent the means ± SEM (n = 3), **p ≤ 0.01; ****p ≤ 0.0001; NS = non-specific
Next, VDAC1 oligomerization was assessed through chemical cross-linking with EGS and Western blotting using anti-VDAC1 antibodies. Several anti-VDAC1 antibody-labeled protein bands corresponding to VDAC1 dimers, trimers, tetramers, and higher-order multimers were detected in cells transfected to express E-, but not N-, or 3b protein (Fig. 3C, D).
Subsequently, we examined the impact of cells transfected with plasmids encoding E-, N-, and 3b proteins, and an empty plasmid on the induction of apoptotic cell death, as analyzed using several methods. In SHSY-5Y cells, only the E-protein induced apoptotic cell death, observed at 48 and 72h post transfection, as analyzed by propidium iodide (PI) and FACS analysis (Fig. 4A, B), aligning with its highest induction of VDAC1 expression and oligomerization (Fig. 3B–D).
To mitigate cell death caused by the empty plasmid, various transfection agents such as Prime Fect (RJH Biosciences; Edmonton, Alberta, Canada) and Jet Prime (Polyplus; Illkirch-Graffenstaden, France) were tested. However, cell transfection with the empty plasmid still resulted in some degree of cell death with all the transfection reagents used (data not shown).
Apoptosis induced by the expression of the three plasmids encoding for E-, but not N- or 3b protein, was also analyzed by acridine orange and ethidium bromide staining (Fig. 4C, D), showing that the expression of E-protein, but no other proteins, induced apoptotic cell death, supporting the results obtained by PI staining (Fig. 4A, B).
Apoptosis induced by the expression of the E-protein was further confirmed using PI/annexin V-FITC staining (Fig. 4E, F). The FACS plots (Fig. 4E) show that the E-protein induced apoptotic, but not necrotic cell death.
Combining E-, N-, or 3b proteins expression led to levels of cell death induction comparable to those observed with E-protein alone (data not shown).
To verify the expression of these proteins, we used available antibodies against the viral proteins N and E. Only those targeting the N-protein found to be highly specific, as demonstrated in the immunoblot (Fig. S2A). Using PCR with specific primers, we confirmed the expression of both N- and E-proteins, but not 3b (Fig. S2B, C). Thus, the lack of effect of the 3b protein may be due to it not being expressed for unknown reasons.
The induction of cell death by the expressed E-protein exhibited cell type dependency as demonstrated using lung cancer A549, prostate cancer PC-3, and neuroblastoma SHSY-5Y cells. The highest levels of apoptosis were observed in SHSY-5Y and PC-3 cells, with lesser induction observed in A549 cells (Fig. 5A, B).
SARS-CoV-2 E-protein induces VDAC1 overexpression, and oligomerization is cell type-dependent. A, B A549, SHSY-5Y and PC-3 cells were transfected with 2 μg of pcDNA3.1 plasmid (empty) or encoding for proteins E, N, or 3b, and 48h post transfection, cells were analyzed for cell death using PI staining and flow cytometry (A). Representative FACS histograms are shown (B). C–E Transfected cell lines A549, SHSY-5Y, and PC-3 were analyzed for VDAC1 expression levels by immunoblotting (C) and quantification (D) and subjected to cross-linking using EGS (1mg protein/ml,100μM) (E). The positions of VDAC1 monomers, dimers, trimers, and multimers and of the molecular weight standards are indicated. VDAC1 dimer levels are presented relative to their levels in the empty plasmid-transfected cells (RUs) and are shown below the immunoblot. Results represent the means ± SEM (n = 3) *p ≤ 0.05; **p ≤ 0.01; ****p ≤ 0.0001; NS non-specific
Analysis of E-protein-induced VDAC1 overexpression, oligomerization, and apoptosis revealed that SHSY-5Y and PC-3 cells exhibited the highest levels of VDAC1, whereas A549 cells showed lower levels (Fig. 5C–E). Similar results were obtained for E-protein-induced VDAC1 oligomerization (Fig. 5E). Thus, SHSY-5Y was selected for most of the performed studies.
The effects of E-protein expression on the three different cell lines (Fig. 5) support the association between VDAC between VDAC1 expression levels, oligomerization, and apoptosis.
Expression of E-protein reduced ROS production and induced an increase in cytosolic Ca2+ and fmtDNA release
Since apoptosis is often accompanied by mitochondria dysfunction, we examined the impact of the expressed E-protein on cellular Ca2+ levels and mitochondrial superoxide using MitoSOX (Fig. 6A–D). Cell transfected to express E-protein exhibited elevated cytosolic Ca2+ ([Ca2+]i) levels, as monitored using Fluo-4-AM (Fig. 6A, C). Similarly, ROS levels were also increased (Fig. 6B, D).
E-protein increases intracellular Ca2+ levels, ROS production and mtDNA release (A–D) SHSY-5Y cells were transfected with 2 μg of pcDNA3.1 plasmid (empty) or encoding for protein E, and 48h post transfection, cells were subjected to analysis of the levels of intracellular Ca2+ using Fluo-4 (A, C), mitochondrial ROS with MitoSOX-Red (B, D), followed by flow cytometry. Representative FACS histograms (A, B) and quantitative analysis (C, D) are shown. E Cytosolic fmtDNA levels were analyzed as indicated in the Methods section. Cells were transfected with 1.5 μg of pcDNA3.1 plasmid (empty) or encoding for E-protein, and 48h post-transfection, analyzed for fmtDNA using qPCR and primers specific to the D-Loop-II. Cells were also subjected to cell death analysis using PI staining and flow cytometry. Results represent the means ± SEM (n = 3) *p ≤ 0.05; **p ≤ 0.01
Recently, we showed that fmtDNA is released from the mitochondria via oligomeric VDAC1, and that VBIT-4 inhibits fmtDNA release, type-Ι interferon signaling, and disease severity in a mouse model of lupus [23]. Given that the E-protein led to VDAC1 overexpression and oligomerization, we investigated whether it also triggers fmtDNA release into the cytosol (Fig. 6E). The results reveal that the E-protein does indeed induce this release.
Interaction of the recombinant viral E- and N-proteins with purified VDAC1 and MAVS
The direct interaction of purified VDAC1 and purified MAVS with the purified viral N- and E-proteins (Fig. 7A) was analyzed using Microscale thermophoresis (MST) [24] (Fig. 7B–D). Both proteins interacted with VDAC1 with a half maximal binding (C50) of 220 ± 26 nM and 1100 ± 140 nM for the N- and E-proteins, respectively. This suggests relatively high-affinity binding, with the N-protein displaying higher binding affinity than the E-protein (Fig. 7B, C).
Interaction of SARS-CoV-2 purified N- and E-proteins with purified VDAC1 and MAVS. A SDS-PAGE of purified VDAC1, MAVS, and SARS-CoV-2 E- and N-proteins stained with Coomassie blue. B–D The interactions of VDAC1 with purified E- or N-protein (B, C) and of MAVS with the N-protein (D) were analyzed using MST, as described in the Methods section. The concentration of E- or N-protein giving 50% of the maximal binding (C50) is indicated
The mitochondrial antiviral protein MAVS is anchored to the OMM and plays an important role in innate immunity. It also contributes to the formation of an antiviral and signaling immune system by activating pro-inflammatory cytokines to combat virus infection within the cell [59, 60].
Given that the MAVS–VDAC1 interaction may underline MAVS’s action as an antiviral protein [61] and its significance in innate immunity, we investigated MAVS binding to the N-protein. The findings demonstrate that MAVS indeed interacts with the N-protein, exhibiting high affinity with a C50 of 195 ± 21 nM (Fig. 7D).
Discussion
SARS-CoV-2 invades the mitochondria of infected cells, resulting in altered mitochondrial signaling, modifies cellular energy metabolism, induces apoptosis, mitophagy, and abnormal levels of mitochondrial proteins [62]. The link between VDAC1 and SARS-CoV-2 was demonstrated, by the overexpression of VDAC1 in a population of T cells, suppression of mtDNA release, cGAS-STING pathway activation and improved survival in the T cells of COVID-19 patients when treated with the VDAC1-interacting compound VBIT-4 [33]. mtDNA plays a critical role in cGAS-STING activation during SARS-CoV-2 infection, and VBIT-4 has been shown to protect against this activation [25]. Moreover, elevated levels of circulating fmtDNA are associated with an increased probability of intensive care unit admission higher mortality risk and disease severity, influencing the immune response in patients [23, 44, 63,64,65,66].
Additionally, VDAC1 levels were found to be decreased in extracellular vesicle isolated from the blood of acutely infected patients but not in those with long COVID-19 [67].
Elevated fmtDNA in serum of COVID-19-infected patients
Alterations in mtDNA content have been shown to disrupt cell metabolism and impair innate immune responses, contributing to inflammatory pathology, including the upregulation of interferon-stimulated genes [68]. Numerous reports have highlighted the association between the presence of mtDNA outside the mitochondria and various human disorders that include neuromuscular and neurodegenerative diseases, cancer, diabetes, cardiovascular disorders, and systemic lupus erythematosus [69].
Viruses can manipulate host-cell mitochondria and damage mtDNA to exert control over cellular functions. For example, the Zta protein encoded by the Ebola virus impacts mtDNA replication [70]. Hepatitis C virus (HCV) infection results in mtDNA damage, and the herpes simplex virus produces UL12.5 protein, which leads to mtDNA degradation [71]. Moreover, HIV and HCV infections cause mtDNA depletion in co-infected patients [72].
Previous studies have demonstrated that in COVID-19 patients elevated levels of circulating cell-free fmtDNA are associated with an increased probability of intensive care unit admission and heightened risk of death [64,65,66]. Furthermore, fmtDNA was found in the plasma of COVID-19 patients, with high mtDNA levels correlated with disease severity, and influencing the immune response in these patients [23, 44, 63]. These findings underscore the potential use of fmtDNA levels as a prognostic marker for COVID-19 severity and outcomes.
Here, we analyzed serum fmtDNA levels in both severe and non-severe COVID-19 patients and found a significant increase in fmtDNA levels in their serum compared to healthy donors (Fig. 1). Specifically, the levels of the fmtDNA displacement loop (D-loop)-II, a non-coding region, showed a 24-fold increase, while COX-III and ND-2 exhibited increases of 4- and sevenfold, respectively.
However, we did not observe a clear relationship between COVID-19 serum fmtDNA levels and disease severity. Critically ill COVID-19 patients displayed an early increase in plasma levels of ND1 fmtDNA, with a peak at 24h [44]. This may explain why in our study no clear correlation was found between disease severity and fmtDNA levels, as the blood samples were taken from patients at different time points following disease onset.
The mechanism underlying fmtDNA release in response to SARS-CoV-2 remains unclear. Based on recent findings demonstrating that fmtDNA is released from the mitochondria via oligomeric VDAC1 [23], we suggest that a similar mechanism would be applied in COVID-19 (Fig. 8). We showed that the VDAC1 oligomerization inhibitor, VBIT-4, effectively inhibits fmtDNA release, type-I interferon signaling, and disease severity in a mouse model of lupus [23]. Similarly, VBIT-4 promotes survival in the T cells of COVID-19 patients [33]. The roles of VDAC1 in fmtDNA release, promoting cell death and inflammation and involvement in the pathogenesis of COVID-19 are discussed below.
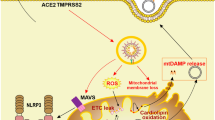
Proposed model for SARS-CoV-2 E-protein inducing VDAC1 overexpression and oligomerization, cell death, and inflammation activation. A Epithelial cell expressing the SARS-CoV-2 E-protein results in VDAC1 overexpression and VDAC1 oligomerization, forming a large channel mediating the release of apoptogenic proteins such as cytochrome c (Cyto c) and apoptosis-inducing factor (AIF), leading to cell death and of fmtDNA (a). Released fmtDNA activates the cyclic GMP-AMP synthase (cGAS)-STING pathway, leading to induction of expression of a type 1 interferon and inflammatory response (b). The pathways in a and b during SARS-CoV-2 infection play a role in epithelial cell death and pulmonary inflammation, with fmtDNA acting to induce an inflammatory response [25]. B. When macrophages engulf the dead cell debris, including the mtDNA released to the cytosol (c), cGAS-STING signaling is triggered in these cells, causing the macrophages to produce a large amount of IFN that results in a robust inflammatory response. C Extracellular mtDNA can also activate neutrophil extracellular traps (NETs). Thus, SARS-CoV-2 infection causes mitochondrial damage in epithelial cells, resulting in the release of fmtDNA to the cytosol, which activates the cGAS-STING pathway and causes excessive inflammation, affecting lung pathology in the late stage of SARS-CoV-2 infection. mitochondria/VDAC1
Different serum protein profile of COVID-19-infected patients compared to healthy people
A number of biomarkers have been identified for COVID-19 [73, 74]. These include several proinflammatory cytokines, neuron-specific enolase, lactate dehydrogenase (LDH), aspartate transaminase (AST), neutrophil count, troponins, creatine kinase, myoglobin, D-dimer, and brain natriuretic peptide, among others [75]. Monitoring these molecular markers can provide valuable insights into diagnosis, prognosis, and severity, and outcomes of the disease in COVID-19 patients, aiding in clinical management and decision making.
A recent proteomic analysis of extracellular vesicles found in the saliva of COVID-19 patients uncovered a protein signature associated with immune response processes, oxygen transport, and antioxidant mechanisms [76].
Our SDS-PAGE analysis of sera from COVID-19 patients and healthy donors revealed three major protein bands with molecular masses of 49, 17, and 14 kDa, defined as unique to patients, where their serum includes either two or all three of the proteins (Fig. 2, Table 2). Subsequent proteomic analysis using LC–MS/MS, has identified over 20 proteins within these bands, each distinguished by several unique peptides. From these, we selected 15 proteins with the highest sequence coverage and scores, all displaying very high levels compared to those in healthy patients (Table 2).
Among these proteins were: haptoglobin, that forms a complex haptoglobin–hemoglobin that is subsequently removed by the reticuloendothelial system [45]. It is associated with inflammation and numerous diseases [77]; CD5 antigen-like (CD5L), secreted primarily from macrophages during an inflammatory response [48]. It is associated with autophagy regulation, cell polarization, apoptosis inhibition, and lipid metabolism regulation, and is implicated in various diseases resulting from acute or chronic inflammation, including infectious, metabolic, and autoimmune conditions [78]; apolipoprotein A-1 (ApoA1), a major protein constituent of high-density lipoprotein (HDL) particles linked to the pathogenesis of cardiometabolic diseases, atherosclerosis, thrombosis, diabetes, cancer, and neurological disorders [79]; transthyretin (TTR), which transports thyroxine and retinol-binding protein (RBP) bound to retinol [55]. Its misfolding and aggregation can lead to amyloid transthyretin (ATTR) formation. Patients with ATTR amyloidosis are particularly vulnerable to COVID-19 [80]; retinoic acid receptor responder 2 (CRABP-II), involved in transporting retinoic acid (RA) from the cytosol to the nucleus, enhancing cell transcriptional activity [53]; retinol-binding protein 4 (RBP4), which transports retinol from liver stores to peripheral tissues [54]. When associated with TTR, the retinol/RBP4/TTR complex is released into the bloodstream to deliver retinol to tissues [54]. We found an increase in both RBP4 and TTR in the serum of COVID-19 patients (Table 2), consistent with the secretion of this complex into circulation [54]; apolipoprotein C-IV (APOC4), a lipid-binding protein critical for activating lipoprotein lipase [57]; and immunoglobulin kappa constant (IGKC), an immunoglobulin isotype involved in immune response pathways such as the Fc epsilon RI pathway and NFAT in immune response.
Other detected proteins have previously been reported as elevated in COVID-19 patients’ serum. Among these is C-reactive protein (CRP), with an increase of over 290-fold (Table 2). This is consistent with previous findings of elevated CRP levels in the serum of COVID-19 patients [81, 82] being correlated with COVID-19 severity [83], suggests it as a potential early marker for predicting disease severity [84, 85]. CRP functions by activating the classic complement pathway through binding to phosphocholine expressed on bacterial cell surfaces, such as pneumococcus bacteria [86].
Another protein with markedly increased levels (274-fold) in COVID-19 patients is the complement component C8-beta chain, a crucial constituent of the membrane attack complex (MAC) responsible for mediating cell lysis and affecting cell membrane integrity [46], playing a role in both innate and adaptive immune responses.
Levels of alpha-1-antichymotrypsin (AACT) protein were found to be increased 23-fold. AACT functions as a serine protease inhibitor and acts in bacterial killing by neutrophils and is induced during inflammation [87].
Similarly, levels of the serine protease inhibitor alpha-1-anti-trypsin (AAT), which protects the lungs from neutrophil elastase damage [51] were increased by 26-fold, possibly reflecting the body’s response to protect the lungs. Levels of immunoglobulin Joining chain (J chain) showed an increase of 21-fold. This protein is essential for the formation and stabilization of polymeric Ig structure [52], indicating its potential role in the immune response in COVID-19 patients.
These findings indicate the presence in the serum of COVID-19 patients of proteins associated with both innate and adaptive immune responses, and inflammatory processes. This aligns with the pathophysiological mechanisms of COVID-19, which heavily involve the immune system. [88].
COVID-19 proteins induced apoptosis, inflammation, and mitochondria dysfunction via induction of VDAC1 overexpression and its oligomerization
Numerous viruses exert their pathogenic effects by disrupting mitochondrial activities, either directly targeted by viral proteins or influenced by alterations in their cellular milieu, deregulated Ca2+ homeostasis, ER stress, and/or apoptosis [89]. The connection between mitochondrial dysfunction and SARS-CoV-2 infection, including hijacking of host mitochondria [90], altering mitochondrial metabolism [91], and inducing mtDNA release [25] has been demonstrated.
Here, as mitochondria dysfunction by viruses often involves modulation of VDAC1 expression levels or its activity via the interaction of their specific proteins with VDAC1 [26,27,28,29,30, 92, 93], we aimed to elucidate the role of VDAC1 in COVID-19 pathology. We selected four SARS-CoV-2-encoded proteins and proposed to mediate their pathology via inducing mitochondria dysfunction and apoptosis. We illustrated their effects on VDAC1 expression levels, promoting its oligomerization, and inducing apoptosis.
Of the three SARS-CoV-2 proteins E, 3b, and N expressed in cells, the E-protein induced VDAC1 overexpression and its oligomerization, increased ROS production, and elevated intracellular Ca2+ levels, apoptosis, and the release of fmtDNA (Figs. 3–6). The E-protein, known for altering the permeability of host-cell membranes and being implicated in the inflammatory cascade triggered by SARS-CoV [34], exhibited the most significant activity.
The proposed pathways activated by SARS-CoV-2 E-protein leading to activation of apoptosis and an inflammatory response are presented in Fig. 8. E-protein induces VDAC1 overexpression and its oligomerization, resulting in the formation of a large channel that mediates the release of pro-apoptotic proteins [16, 18, 20, 22, 24] and mtDNA [23] from the mitochondria to the cytosol, leading to apoptosis activation and an inflammatory response (Fig. 8A).
Lymphocytes from COVID-A patients exhibit mitochondrial dysfunction, a distinct metabolic profile, and are susceptible to apoptosis due to elevated VDAC1 levels [33]. Additionally, a specific T-cell subset observed in COVID-A patients displays overexpression of both VDAC1 and H3K27me3, an epigenetic marker. The survival of these cells was rescued by VBIT-4, an inhibitor of VDAC1 oligomerization [33]. These findings align with VBIT-4’s ability to suppress VDAC1 oligomerization resulting from its overexpression, thereby preventing cell death [19]. Since VDAC1 oligomers mediate mtDNA released from the mitochondria [23], as expected, the E-protein stimulated fmtDNA release (Fig. 6E).
VDAC1 overexpression, its oligomerization, and apoptosis were triggered by various apoptosis-inducing conditions including chemotherapy drugs and UV irradiation [16,17,18, 20, 94] in several disorders such as Alzheimer’s disease [95], type 2 diabetes [96], and autoimmune diseases such as lupus [23] and inflammatory bowel diseases [97]. Also, several viral proteins elevate VDAC1 expression [28, 33, 92]. Hence, similar to chemotherapy drugs, stress conditions, various diseases, and viruses, SARS-CoV-2 E-protein induces VDAC1 overexpression.
The increase in VDAC1 levels upon E-protein expression may result from increased intracellular Ca2+ levels (Fig. 6C), which are required for VDAC1 overexpression induced by various chemicals and conditions [20, 98]. Additionally, since purified protein E directly interacts with VDAC1 (Fig. 7C), it may stabilize VDAC1 and prevent its degradation.
Direct interaction of purified E-and N-proteins with VDAC1 and MAVS
Here, we showed that both the purified E- and N-proteins interact directly with purified VDAC1 (Fig. 7). However, the precise function of these interactions remains unclear. It is possible that the E-protein may directly stimulate VDAC1 oligomerization to form a larger channel, as we found with mtDNA [23] and amyloid beta (Aβ) [99]. On the other hand, the interaction of the N-protein with VDAC1 may modulate VDAC1 activity, potentially involving functions unrelated to cell death, but associated with other multifunctional roles of VDAC1.
We also showed interaction of the N-protein with the mitochondrial protein MAVS, which plays a crucial role in antiviral responses, and signals the immune system by activating pro-inflammatory cytokines to combat viral infections within cells [59]. MAVS exerts its activity only when associated with the mitochondria [60]. Numerous viruses were shown to displace MAVS from the mitochondria, thereby impeding the IFN-I response [100, 101] and escape from host defenses [60]. Recently [61], we demonstrated that MAVS interacts with VDAC1 and suggested that this interaction lies at the base of MAVS’ antiviral action. The N-protein, by binding to VDAC1 or/and MAVS potentially hinders MAVS’ signaling to the immune system through MAVS’ detachment from the mitochondria/VDAC1.
Cytosolic and extracellular mtDNA in patients with COVID-19 and dysregulation of the immune and inflammatory systems
In this study, we demonstrated elevated levels of fmtDNA in the serum of COVID-19 patients and in the cytosol of cells expressing E-protein. The release of mtDNA reflects the formation of a large channel associated with VDAC1 oligomerization that mediates the mtDNA release [23] and of pro-apoptotic proteins [16, 18, 20, 22, 24], resulting in cell death and inflammation (Fig. 8).
We propose that VDAC1 overexpression and the formation of a mega channel through its oligomerization lead to fmtDNA release and activate the cGAS–STING pathway that triggers type I interferon responses (Fig. 8A). This is supported by findings that the VDAC1 oligomerization inhibitor, VBIT-4, protected against cGAS-STING activation in the cytokine storm derived from SARS-CoV-2 infection [25].
When mtDNA is released from damaged or dying cells into the cytoplasm or circulation, it can impact immunity [102], as shown by mtDNA’s influence on the immune response in COVID-19 [44, 63,64,65,66]. Extracellular mtDNA can activate macrophages when they engulf the dead cell debris into the cytosol. This triggers cGAS-STING signaling in these cells, causing the macrophages to produce a large amount of IFN, resulting in an inflammatory response (Fig. 8B). Extracellular fmtDNA can also activate neutrophils leading to the release of web-like structures—neutrophil extracellular traps (NETs) (Fig. 8C). High levels of NETs were found in both the peripheral blood and lung tissues of COVID-19 patients and contribute to disease pathogenesis [103,104,105,106]. Activated neutrophils also release lytic enzymes, ROS, cytokines, histones, mtDNA, and nucleus (nDNA) [107]. mtDNA within NETs contribute to the function of these extracellular traps in host defense mechanisms [108] (Fig. 8C).
Consequently, the release of mtDNA into the cytoplasm and extracellular environment triggers various pattern recognition receptors that contribute to the immune response, promoting an inflammatory cascade in patients with COVID-19 [109].
While this study did not directly show that SARS-CoV-2 infection or the E-protein induces mtDNA release and activates the cGAS-STING pathway and a cytokine storm, the connection between mtDNA release and cGAS-STING activation is well established [110,111,112]. Additionally, it has been demonstrated that VBIT-4, which inhibits mtDNA release and cGAS-STING pathway activation, as well as interferon-stimulating genes [23], protects against cGAS-STING activation in SARS-CoV-2 infection [25].
In summary, our study revealed elevated levels of mtDNA in the serum of COVID-19 patients. Additionally, we demonstrated that the E-protein of COVID-19 enhances the overexpression and oligomerization of VDAC1, which leads to cell death. Furthermore, VDAC1 oligomers mediate the release of mtDNA into the cytosol, where it triggers the activation of toll-like receptors and the cGAS-STING signaling axis. This activation prompts the expression of various inflammatory cytokines and chemokines in a cytokine storm, which is associated with severe tissue damage in infectious or non-infectious conditions, particularly in viral respiratory infections such as H5N1 influenza, SARS-CoV-1, and SARS-CoV-2 [113, 114].
The findings presented here suggest that COVID-19 pathology is associated with the induction of VDAC1 overexpression and oligomerization, leading to cell death and the release of fmtDNA, which is associated with the cytokine storm that directly influences SARS-CoV-2 severity and the COVID-19 illness.
Data availability
No datasets were generated or analysed during the current study.
References
Mehta P et al (2020) COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet 395(10229):1033–1034
Cui J, Li F, Shi ZL (2019) Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 17(3):181–192
Padhan K et al (2008) Severe acute respiratory syndrome coronavirus 3a protein activates the mitochondrial death pathway through p38 MAP kinase activation. J Gen Virol 89(Pt 8):1960–1969
Yuan X et al (2006) Mitochondrial location of severe acute respiratory syndrome coronavirus 3b protein. Mol Cells 21(2):186–191
Galluzzi L et al (2008) Viral control of mitochondrial apoptosis. PLoS Pathog 4(5):e1000018
Benedict CA, Norris PS, Ware CF (2002) To kill or be killed: viral evasion of apoptosis. Nat Immunol 3(11):1013–1018
Chen GG et al (2001) Decreased expression of Bid in human hepatocellular carcinoma is related to hepatitis B virus X protein. Eur J Cancer 37(13):1695–1702
Mori I et al (2004) Virus-induced neuronal apoptosis as pathological and protective responses of the host. Rev Med Virol 14(4):209–216
Amara A, Mercer J (2015) Viral apoptotic mimicry. Nat Rev Microbiol 13(8):461–469
Saxena R et al (2023) Modulation of mitochondria by viral proteins. Life Sci 313:121271
Perfettini JL et al (2005) Mechanisms of apoptosis induction by the HIV-1 envelope. Cell Death Differ 12(Suppl 1):916–923
Akira S, Uematsu S, Takeuchi O (2006) Pathogen recognition and innate immunity. Cell 124(4):783–801
McBride HM, Neuspiel M, Wasiak S (2006) Mitochondria: more than just a powerhouse. Curr Biol 16(14):R551–R560
Angajala A et al (2018) Diverse roles of mitochondria in immune responses: novel insights into immuno-metabolism. Front Immunol 9:1605
Breda CNS et al (2019) Mitochondria as central hub of the immune system. Redox Biol 26:101255
Shoshan-Barmatz V, Mizrachi D, Keinan N (2013) Oligomerization of the mitochondrial protein VDAC1: from structure to function and cancer therapy. Prog Mol Biol Transl Sci 117:303–334
Shoshan-Barmatz V et al (2010) VDAC, a multi-functional mitochondrial protein regulating cell life and death. Mol Aspects Med 31(3):227–285
Shoshan-Barmatz V, Maldonado EN, Krelin Y (2017) VDAC1 at the crossroads of cell metabolism, apoptosis and cell stress. Cell Stress 1(1):11–36
Shoshan-Barmatz V, Shteinfer-Kuzmine A, Verma A (2020) VDAC1 at the intersection of cell metabolism, apoptosis, and diseases. Biomolecules 10(11):1485
Keinan N et al (2013) The role of calcium in VDAC1 oligomerization and mitochondria-mediated apoptosis. Biochim Biophys Acta 1833(7):1745–1754
Ben-Hail D, Shoshan-Barmatz V (2016) VDAC1-interacting anion transport inhibitors inhibit VDAC1 oligomerization and apoptosis. Biochim Biophys Acta 1863(7):1612–1623
Keinan N, Tyomkin D, Shoshan-Barmatz V (2010) Oligomerization of the mitochondrial protein voltage-dependent anion channel is coupled to the induction of apoptosis. Mol Cell Biol 30(24):5698–5709
Kim J et al (2019) VDAC oligomers form mitochondrial pores to release mtDNA fragments and promote lupus-like disease. Science 366(6472):1531–1536
Ben-Hail D et al (2016) Novel compounds targeting the mitochondrial protein VDAC1 inhibit apoptosis and protect against mitochondrial dysfunction. J Biol Chem 291(48):24986–25003
Di Domizio J et al (2022) The cGAS-STING pathway drives type I IFN immunopathology in COVID-19. Nature 603(7899):145
Zamarin D et al (2005) Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog 1(1):e4
Rahmani Z et al (2000) Hepatitis B virus X protein colocalizes to mitochondria with a human voltage-dependent anion channel, HVDAC3, and alters its transmembrane potential. J Virol 74(6):2840–2846
Moin SM, Panteva M, Jameel S (2007) The hepatitis E virus Orf3 protein protects cells from mitochondrial depolarization and death. J Biol Chem 282(29):21124–21133
Qiao H, McMillan JR (2007) Gelsolin segment 5 inhibits HIV-induced T-cell apoptosis via Vpr-binding to VDAC. FEBS Lett 581(3):535–540
Jitobaom K, Tongluan N, Smith DR (2016) Involvement of voltage-dependent anion channel (VDAC) in dengue infection. Sci Rep 6:35753
Lin W et al (2015) The association of receptor of activated protein kinase C 1(RACK1) with infectious bursal disease virus viral protein VP5 and voltage-dependent anion channel 2 (VDAC2) inhibits apoptosis and enhances viral replication. J Biol Chem 290(13):8500–8510
Ye Z et al (2008) A SARS-CoV protein, ORF-6, induces caspase-3 mediated, ER stress and JNK-dependent apoptosis. Biochim Biophys Acta 1780(12):1383–1387
Thompson EA et al (2021) Metabolic programs define dysfunctional immune responses in severe COVID-19 patients. Cell Rep 34(11):108863
Schoeman D, Fielding BC (2019) Coronavirus envelope protein: current knowledge. Virol J 16(1):69
Zhang L et al (2007) SARS-CoV nucleocapsid protein induced apoptosis of COS-1 mediated by the mitochondrial pathway. Artif Cells Blood Substit Immobil Biotechnol 35(2):237–253
Yuan X et al (2005) Nucleolar localization of non-structural protein 3b, a protein specifically encoded by the severe acute respiratory syndrome coronavirus. Virus Res 114(1–2):70–79
Khan S et al (2006) Over-expression of severe acute respiratory syndrome coronavirus 3b protein induces both apoptosis and necrosis in Vero E6 cells. Virus Res 122(1–2):20–27
Kopecky-Bromberg SA et al (2007) Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol 81(2):548–557
Ben-Hail D, Shoshan-Barmatz V (2014) Purification of VDAC1 from rat liver mitochondria. Cold Spring Harb Protoc 2014(1):94–99
Wienken CJ et al (2010) Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun 1:100
Deniaud A, Brenner C, Kroemer G (2004) Mitochondrial membrane permeabilization by HIV-1 Vpr. Mitochondrion 4(2–3):223–233
Iborra FJ, Kimura H, Cook PR (2004) The functional organization of mitochondrial genomes in human cells. BMC Biol 2:9
Nicholls TJ, Minczuk M (2014) In D-loop: 40 years of mitochondrial 7S DNA. Exp Gerontol 56:175–181
Edinger F et al (2022) Peak plasma levels of mtDNA serve as a predictive biomarker for COVID-19 in-hospital mortality. J Clin Med 11(23):7161
Naryzny SN, Legina OK (2021) Haptoglobin as a biomarker. Biochem Moscow-Suppl Series B-Biomed Chem 15(3):184–198
Brannen CL, Sodetz JM (2007) Incorporation of human complement C8 into the membrane attack complex is mediated by a binding site located within the C8beta MACPF domain. Mol Immunol 44(5):960–965
Mishra V, Heath RJ (2021) Structural and biochemical features of human serum albumin essential for eukaryotic cell culture. Int J Mol Sci 22(16):8411
Sanjurjo L et al (2015) AIM/CD5L: a key protein in the control of immune homeostasis and inflammatory disease. J Leukoc Biol 98(2):173–184
Marnell L, Mold C, Du Clos TW (2005) C-reactive protein: ligands, receptors and role in inflammation. Clin Immunol 117(2):104–111
McFadyen JD et al (2020) C-reactive protein and its structural isoforms: an evolutionary conserved marker and central player in inflammatory diseases and beyond. Subcell Biochem 94:499–520
Hunt JM, Tuder R (2012) Alpha 1 anti-trypsin: one protein, many functions. Curr Mol Med 12(7):827–835
Castro CD, Flajnik MF (2014) Putting J chain back on the map: how might its expression define plasma cell development? J Immunol 193(7):3248–3255
Budhu AS, Noy N (2002) Direct channeling of retinoic acid between cellular retinoic acid-binding protein II and retinoic acid receptor sensitizes mammary carcinoma cells to retinoic acid-induced growth arrest. Mol Cell Biol 22(8):2632–2641
Steinhoff JS, Lass A, Schupp M (2021) Biological functions of RBP4 and its relevance for human diseases. Front Physiol 12:659977
Ueda M (2022) Transthyretin: its function and amyloid formation. Neurochem Int 155:105313
Sun L, Ye RD (2016) Serum amyloid A1: structure, function and gene polymorphism. Gene 583(1):48–57
Wolska A, Reimund M, Remaley AT (2020) Apolipoprotein C-II: the re-emergence of a forgotten factor. Curr Opin Lipidol 31(3):147–153
Wu CR et al (2022) Structure genomics of SARS-CoV-2 and its omicron variant: drug design templates for COVID-19. Acta Pharmacol Sin 43(12):3021–3033
Seth RB et al (2005) Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122(5):669–682
Sun Q et al (2006) The specific and essential role of MAVS in antiviral innate immune responses. Immunity 24(5):633–642
Trishna S et al (2023) Overexpression of the mitochondrial anti-viral signaling protein, MAVS, in cancers is associated with cell survival and inflammation. Mol Ther Nucl Acids 33:713–732
Rurek M (2024) Mitochondria in COVID-19: from cellular and molecular perspective. Front Physiol 15:1406635
Leonard AJ et al (2022) Circulating, cell-free mitochondrial DNA marks disease severity in COVID-19. Am J Respir Crit Care Med 205:A2927
Fernández-Ayala DJM, Navas P, López-Lluch G (2020) Age-related mitochondrial dysfunction as a key factor in COVID-19 disease. Exp Gerontol 142:111147
Scozzi D et al (2021) Circulating mitochondrial DNA is an early indicator of severe illness and mortality from COVID-19. JCI Insight. https://doi.org/10.1172/jci.insight.143299
Valdes-Aguayo JJ et al (2021) Peripheral blood mitochondrial DNA levels were modulated by SARS-CoV-2 infection severity and its lessening was associated with mortality among hospitalized patients with COVID-19. Front Cell Infect Microbiol 11:754708
Goetzl EJ, Yao PJ, Kapogiannis D (2023) Prediction of post-acute-sequelae of COVID-19 by cargo protein biomarkers of blood total extracellular vesicles in acute COVID-19. Am J Med 136(8):824–829
West AP, Shadel GS (2017) Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat Rev Immunol 17(6):363–375
Perez-Trevino P, Velasquez M, Garcia N (2020) Mechanisms of mitochondrial DNA escape and its relationship with different metabolic diseases. Biochim Biophys Acta Mol Basis Dis 1866(6):165761
Wiedmer A et al (2008) Epstein-Barr virus immediate-early protein Zta co-opts mitochondrial single-stranded DNA binding protein to promote viral and inhibit mitochondrial DNA replication. J Virol 82(9):4647–4655
Martinez R et al (1996) The product of a 1.9-kb mRNA which overlaps the HSV-1 alkaline nuclease gene (UL12) cannot relieve the growth defects of a null mutant. Virology 215(2):152–164
de Mendoza C et al (2007) Mitochondrial DNA depletion in HIV-infected patients with chronic hepatitis C and effect of pegylated interferon plus ribavirin therapy. AIDS 21(5):583–588
Andrianto MJ et al (2021) Biomarkers of endothelial dysfunction and outcomes in coronavirus disease 2019 (COVID-19) patients: a systematic review and meta-analysis. Microvasc Res 138:104224
Kermali M et al (2020) The role of biomarkers in diagnosis of COVID-19-a systematic review. Life Sci 254:117788
Battaglini D et al (2022) Laboratory biomarkers for diagnosis and prognosis in COVID-19. Front Immunol 13:857573
Weber L et al (2024) Proteomic analysis of salivary extracellular vesicles from COVID-19 patients reveals a specific anti-COVID-19 response protein signature. Int J Mol Sci 25(7):3704
Quaye IK (2008) Haptoglobin, inflammation and disease. Trans R Soc Trop Med Hyg 102(8):735–742
Sanchez-Moral L et al (2021) Multifaceted roles of CD5L in infectious and sterile inflammation. Int J Mol Sci 22(8):4076
Cochran BJ et al (2021) APOA1: a protein with multiple therapeutic functions. Curr Atheroscler Rep 23(3):11
Brannagan TH 3rd et al (2021) ATTR amyloidosis during the COVID-19 pandemic: insights from a global medical roundtable. Orphanet J Rare Dis 16(1):204
Wang D et al (2020) Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 323(11):1061–1069
Chen N et al (2020) Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395(10223):507–513
Nazemi P et al (2023) Serum C-reactive protein greater than 75 mg/dL as an early available laboratory predictor of severe COVID-19: a systematic review. Immun Inflamm Dis 11(12):e1130
Ali N (2020) Elevated level of C-reactive protein may be an early marker to predict risk for severity of COVID-19. J Med Virol 92(11):2409–2411
Potempa LA et al (2020) Insights into the use of c-reactive protein as a diagnostic index of disease severity in COVID-19 Infections. Am J Trop Med Hyg 103(2):561–563
Sproston NR, Ashworth JJ (2018) Role of C-reactive protein at sites of inflammation and infection. Front Immunol. https://doi.org/10.3389/fimmu.2018.00754
Janciauskiene S et al (2018) The multifaceted effects of alpha1-antitrypsin on neutrophil functions. Front Pharmacol 9:341
Shen J et al (2023) Innate and adaptive immunity to SARS-CoV-2 and predisposing factors. Front Immunol 14:1159326
Moreno-Altamirano MMB, Kolstoe SE, Sanchez-Garcia FJ (2019) Virus control of cell metabolism for replication and evasion of host immune responses. Front Cell Infect Microbiol 9:95
Singh KK et al (2020) Decoding SARS-CoV-2 hijacking of host mitochondria in COVID-19 pathogenesis. Am J Physiol Cell Physiol 319(2):C258–C267
Ajaz S et al (2021) Mitochondrial metabolic manipulation by SARS-CoV-2 in peripheral blood mononuclear cells of patients with COVID-19. Am J Physiol Cell Physiol 320(1):C57–C65
Han C et al (2017) Voltage-dependent anion channel 1 interacts with ribonucleoprotein complexes to enhance infectious bursal disease virus polymerase activity. J Virol 91(16):e00584
Fongsaran C et al (2014) Voltage dependent anion channel is redistributed during Japanese encephalitis virus infection of insect cells. Sci World J 2014:976015
Shoshan-Barmatz V et al (2015) The mitochondrial voltage-dependent anion channel 1 in tumor cells. Biochim Biophys Acta 1848(10):2547–2575
Verma A et al (2022) Targeting the overexpressed mitochondrial protein VDAC1 in a mouse model of Alzheimer’s disease protects against mitochondrial dysfunction and mitigates brain pathology. Transl Neurodegener 11(1):58
Zhang E et al (2019) Preserving insulin secretion in diabetes by inhibiting VDAC1 overexpression and surface translocation in beta cells. Cell Metab 8(29):64–77
Verma A et al (2022) The role of the mitochondrial protein VDAC1 in inflammatory bowel disease: a potential therapeutic target. Mol Ther 30(2):726–744
Weisthal S et al (2014) Ca(2+)-mediated regulation of VDAC1 expression levels is associated with cell death induction. Biochim Biophys Acta 1843(10):2270–2281
Smilansky A et al (2015) The voltage-dependent anion channel 1 mediates amyloid beta toxicity and represents a potential target for Alzheimer disease therapy. J Biol Chem 290(52):30670–30683
Li XD et al (2005) Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci USA 102(49):17717–17722
Mukherjee A et al (2011) The coxsackievirus B 3C protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling. PLoS Pathog 7(3):e1001311
Piantadosi CA (2020) Mitochondrial DNA, oxidants, and innate immunity. Free Radic Biol Med 152:455–461
Thierry AR, Roch B (2020) Neutrophil extracellular traps and by-products play a key role in COVID-19: pathogenesis, risk factors, and therapy. J Clin Med 9(9):2942
Janiuk K, Jablonska E, Garley M (2021) Significance of NETs formation in COVID-19. Cells 10(1):151
Joshi MB et al (2022) Modulation of neutrophil (dys)function by Ayurvedic herbs and its potential influence on SARS-CoV-2 infection. J Ayurveda Integr Med 13(1):100424
Cavalcante-Silva LHA et al (2021) Neutrophils and COVID-19: the road so far. Int Immunopharmacol 90:107233
Papayannopoulos V (2018) Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol 18(2):134–147
Lood C et al (2016) Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat Med 22(2):146–153
Riley JS, Tait SW (2020) Mitochondrial DNA in inflammation and immunity. EMBO Rep 21(4):e49799
Hu H et al (2022) Mitochondrial VDAC1: a potential therapeutic target of inflammation-related diseases and clinical opportunities. Cells 11(19):3174
Zheng W et al (2023) How the innate immune DNA sensing cGAS-STING pathway is involved in apoptosis. Int J Mol Sci 24(3):3029
Tan HY et al (2022) cGAS and DDX41-STING mediated intrinsic immunity spreads intercellularly to promote neuroinflammation in SOD1 ALS model. Iscience 25(6):104404
Liu Q, Zhou YH, Yang ZQ (2016) The cytokine storm of severe influenza and development of immunomodulatory therapy. Cell Mol Immunol 13(1):3–10
Soy M, Keser G, Atagunduz P (2021) Pathogenesis and treatment of cytokine storm in COVID-19. Turk J Biol 45(4):372–389
Acknowledgements
We thank the Krieger Foundation for funding this study.
Funding
Open access funding provided by Ben-Gurion University. This work was supported by the Krieger Foundation.
Author information
Authors and Affiliations
Contributions
A.S-K., A.V., and H.P. carried out the experiments, data analysis, and figure preparation. R.B. and E.R. carried out the bioinformatic analysis. A.B-Y., E.B-S. and E.S. provided COVID-19 and healthy serum samples and the relevant information, and Y.M. offered discussion of the results. V.S-B. provided experimental oversight, interpreted the results, arranged the figure presentation, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This study was approved by the Shaare Zedek Medical Center Institutional Review Board.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shteinfer-Kuzmine, A., Verma, A., Bornshten, R. et al. Elevated serum mtDNA in COVID-19 patients is linked to SARS-CoV-2 envelope protein targeting mitochondrial VDAC1, inducing apoptosis and mtDNA release. Apoptosis (2024). https://doi.org/10.1007/s10495-024-02025-5
Accepted:
Published:
DOI: https://doi.org/10.1007/s10495-024-02025-5