Abstract
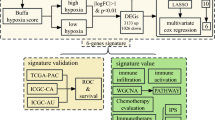
Neutrophil extracellular traps (NETs) are novel inflammatory cell death in neutrophils. Emerging studies demonstrated NETs contributed to cancer progression and metastases in multiple ways. This study intends to provide a prognostic NETs signature and therapeutic target for lung adenocarcinoma (LUAD) patients. Consensus cluster analysis performed by 38 reported NET-related genes in TCGA-LUAD cohorts. Then, WGCNA network was conducted to investigate characteristics genes in clusters. Seven machine learning algorithms were assessed for training of the model, the optimal model was picked by C-index and 1-, 3-, 5-year ROC value. Then, we constructed a NETs signature to predict the overall survival of LUAD patients. Moreover, multi-omics validation was performed based on NETs signature. Finally, we constructed stable knockdown critical gene LUAD cell lines to verify biological functions of Phospholipid Scramblase 1 (PLSCR1) in vitro and in vivo. Two NETs-related clusters were identified in LUAD patients. Among them, C2 cluster was provided as “hot” tumor phenotype and exhibited a better prognosis. Then, WGCNA network identified 643 characteristic genes in C2 cluster. Then, Coxboost algorithm proved its optimal performance and provided a prognostic NETs signature. Multi-omics revealed that NETs signature was involved in an immunosuppressive microenvironment and predicted immunotherapy efficacy. In vitro and in vivo experiments demonstrated that knockdown of PLSCR1 inhibited tumor growth and EMT ability. Besides, cocultural assay indicated that the knockdown of PLSCR1 impaired the ability of neutrophils to generate NETs. Finally, tissue microarray (TMA) for LUAD patients verified the prognostic value of PLSCR1 expression. In this study, we focus on emerging hot topic NETs in LUAD. We provide a prognostic NETs signature and identify PLSCR1 with multiple roles in LUAD. This work can contribute to risk stratification and screen novel therapeutic targets for LUAD patients.








Similar content being viewed by others
Data availability
Public datasets were analyzed in this study. TCGA-LUAD cohort was downloaded from TCGA database (http://cancergenome.nih.gov/). GSE50081, GSE72094, GSE31210, GSE81089, GSE91061, and GSE100797 were downloaded from the GEO database(http://www.ncbi.nlm.nih.gov/geo/). PRJNA482620, IMvigor210, PRJEB23709, Nathanson’s cohort, phs000452, PRJEB23709, and PRJEB25780 were obtained from Tumor Immunotherapy Gene Expression Resource databases (http://tiger.canceromics.org/).
Material availability
The original data and material were available when required.
References
Siegel RL, Miller KD, Wagle NS, Jemal A (2023) Cancer statistics, 2023. CA Cancer J Clin 73:17–48
Arbour KC, Riely GJ (2019) Systemic therapy for locally advanced and metastatic non-small cell lung cancer: a review. JAMA 322:764–774
Kargl J, Busch SE, Yang GH, Kim KH, Hanke ML, Metz HE, Hubbard JJ, Lee SM, Madtes DK, McIntosh MW, Houghton AM (2017) Neutrophils dominate the immune cell composition in non-small cell lung cancer. Nat Commun 8:14381
Németh T, Sperandio M, Mócsai A (2020) Neutrophils as emerging therapeutic targets. Nat Rev Drug Discov 19:253–275
Cui C, Chakraborty K, Tang XA, Zhou G, Schoenfelt KQ, Becker KM, Hoffman A, Chang YF, Blank A, Reardon CA et al (2021) Neutrophil elastase selectively kills cancer cells and attenuates tumorigenesis. Cell 184:3163-3177.e3121
Quail DF, Amulic B, Aziz M, Barnes BJ, Eruslanov E, Fridlender ZG, Goodridge HS, Granot Z, Hidalgo A, Huttenlocher A et al (2022) Neutrophil phenotypes and functions in cancer: a consensus statement. J Exp Med. https://doi.org/10.1084/jem.20220011
Branzk N, Lubojemska A, Hardison SE, Wang Q, Gutierrez MG, Brown GD, Papayannopoulos V (2014) Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat Immunol 15:1017–1025
Fuchs TA, Abed U, Goosmann C, Hurwitz R, Schulze I, Wahn V, Weinrauch Y, Brinkmann V, Zychlinsky A (2007) Novel cell death program leads to neutrophil extracellular traps. J Cell Biol 176:231–241
Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A (2004) Neutrophil extracellular traps kill bacteria. Science 303:1532–1535
Rayes RF, Mouhanna JG, Nicolau I, Bourdeau F, Giannias B, Rousseau S, Quail D, Walsh L, Sangwan V, Bertos N et al (2019) Primary tumors induce neutrophil extracellular traps with targetable metastasis promoting effects. JCI Insight. https://doi.org/10.1172/2Fjci.insight.128008
Jin W, Xu HX, Zhang SR, Li H, Wang WQ, Gao HL, Wu CT, Xu JZ, Qi ZH, Li S et al (2019) Tumor-infiltrating NETs predict postsurgical survival in patients with pancreatic ductal adenocarcinoma. Ann Surg Oncol 26:635–643
Masucci MT, Minopoli M, Del Vecchio S, Carriero MV (2020) The emerging role of neutrophil extracellular traps (NETs) in tumor progression and metastasis. Front Immunol 11:1749
Wu L, Saxena S, Goel P, Prajapati DR, Wang C, Singh RK (2020) Breast cancer cell-neutrophil interactions enhance neutrophil survival and pro-tumorigenic activities. Cancers (Basel) 12:2884
Park J, Wysocki RW, Amoozgar Z, Maiorino L, Fein MR, Jorns J, Schott AF, Kinugasa-Katayama Y, Lee Y, Won NH et al (2016) Cancer cells induce metastasis-supporting neutrophil extracellular DNA traps. Sci Transl Med. https://doi.org/10.1126/scitranslmed.aag1711
Surendran V, Rutledge D, Colmon R, Chandrasekaran A (2021) A novel tumor-immune microenvironment (TIME)-on-Chip mimics three dimensional neutrophil-tumor dynamics and neutrophil extracellular traps (NETs)-mediated collective tumor invasion. Biofabrication 13:035029
Kajioka H, Kagawa S, Ito A, Yoshimoto M, Sakamoto S, Kikuchi S, Kuroda S, Yoshida R, Umeda Y, Noma K et al (2021) Targeting neutrophil extracellular traps with thrombomodulin prevents pancreatic cancer metastasis. Cancer Lett 497:1–13
Zhu T, Zou X, Yang C, Li L, Wang B, Li R, Li H, Xu Z, Huang D, Wu Q (2021) Neutrophil extracellular traps promote gastric cancer metastasis by inducing epithelial-mesenchymal transition. Int J Mol Med 48:1–13
Martins-Cardoso K, Almeida VH, Bagri KM, Rossi MID, Mermelstein CS, König S, Monteiro RQ (2020) Neutrophil extracellular traps (NETs) promote pro-metastatic phenotype in human breast cancer cells through epithelial-mesenchymal transition. Cancers (Basel) 12:1542
Cools-Lartigue J, Spicer J, McDonald B, Gowing S, Chow S, Giannias B, Bourdeau F, Kubes P, Ferri L (2013) Neutrophil extracellular traps sequester circulating tumor cells and promote metastasis. J Clin Invest 123:3446–3458
Albrengues J, Shields MA, Ng D, Park CG, Ambrico A, Poindexter ME, Upadhyay P, Uyeminami DL, Pommier A, Küttner V et al (2018) Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science. https://doi.org/10.1126/science.aao4227
Mauracher LM, Posch F, Martinod K, Grilz E, Däullary T, Hell L, Brostjan C, Zielinski C, Ay C, Wagner DD et al (2018) Citrullinated histone H3, a biomarker of neutrophil extracellular trap formation, predicts the risk of venous thromboembolism in cancer patients. J Thromb Haemost 16:508–518
Teijeira A, Garasa S, Ochoa MC, Villalba M, Olivera I, Cirella A, Eguren-Santamaria I, Berraondo P, Schalper KA, de Andrea CE et al (2021) IL8, neutrophils, and NETs in a collusion against cancer immunity and immunotherapy. Clin Cancer Res 27:2383–2393
Kaltenmeier C, Yazdani H, Tohme S (2021) Neutrophil extracellular traps express PD-L1 which promotes T cell exhaustion in the tumor microenvironment. Cancer Res 81:115
Wang H, Zhang H, Wang Y, Brown ZJ, Xia Y, Huang Z, Shen C, Hu Z, Beane J, Ansa-Addo EA et al (2021) Regulatory T-cell and neutrophil extracellular trap interaction contributes to carcinogenesis in non-alcoholic steatohepatitis. J Hepatol 75:1271–1283
Papayannopoulos V, Metzler KD, Hakkim A, Zychlinsky A (2010) Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol 191:677–691
Lewis HD, Liddle J, Coote JE, Atkinson SJ, Barker MD, Bax BD, Bicker KL, Bingham RP, Campbell M, Chen YH et al (2015) Inhibition of PAD4 activity is sufficient to disrupt mouse and human NET formation. Nat Chem Biol 11:189–191
Tang F, Tie Y, Tu C, Wei X (2020) Surgical trauma-induced immunosuppression in cancer: recent advances and the potential therapies. Clin Transl Med 10:199–223
Papayannopoulos V (2018) Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol 18:134–147
Hu J, Yu A, Othmane B, Qiu D, Li H, Li C, Liu P, Ren W, Chen M, Gong G et al (2021) Siglec15 shapes a non-inflamed tumor microenvironment and predicts the molecular subtype in bladder cancer. Theranostics 11:3089–3108
Xu L, Deng C, Pang B, Zhang X, Liu W, Liao G, Yuan H, Cheng P, Li F, Long Z et al (2018) TIP: a web server for resolving tumor immunophenotype profiling. Cancer Res 78:6575–6580
Avila Cobos F, Alquicira-Hernandez J, Powell JE, Mestdagh P, De Preter K (2020) Benchmarking of cell type deconvolution pipelines for transcriptomics data. Nat Commun 11:5650
Cai MC, Zhao X, Cao M, Ma P, Chen M, Wu J, Jia C, He C, Fu Y, Tan L et al (2020) T-cell exhaustion interrelates with immune cytolytic activity to shape the inflamed tumor microenvironment. J Pathol 251:147–159
Cabrita R, Lauss M, Sanna A, Donia M, Skaarup Larsen M, Mitra S, Johansson I, Phung B, Harbst K, Vallon-Christersson J et al (2020) Author correction: tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 580:E1
Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, Albright A, Cheng JD, Kang SP, Shankaran V et al (2017) IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 127:2930–2940
Galon J, Bruni D (2019) Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat Rev Drug Discov 18:197–218
Too NSH, Ho NCW, Adine C, Iyer NG, Fong ELS (2021) Hot or cold: bioengineering immune contextures into in vitro patient-derived tumor models. Adv Drug Deliv Rev 175:113791
Zhang J, Huang D, Saw PE, Song E (2022) Turning cold tumors hot: from molecular mechanisms to clinical applications. Trends Immunol 43:523–545
Gerard CL, Delyon J, Wicky A, Homicsko K, Cuendet MA, Michielin O (2021) Turning tumors from cold to inflamed to improve immunotherapy response. Cancer Treat Rev 101:102227
Wang Y, Zhao Q, Zhao B, Zheng Y, Zhuang Q, Liao N, Wang P, Cai Z, Zhang D, Zeng Y, Liu X (2022) Remodeling tumor-associated neutrophils to enhance dendritic cell-based HCC neoantigen nano-vaccine efficiency. Adv Sci (Weinh) 9:e2105631
Raftopoulou S, Valadez-Cosmes P, Mihalic ZN, Schicho R, Kargl J (2022) Tumor-mediated neutrophil polarization and therapeutic implications. Int J Mol Sci 23:3218
Singel KL, Emmons TR, Khan ANH, Mayor PC, Shen S, Wong JT, Morrell K, Eng KH, Mark J, Bankert RB et al (2019) Mature neutrophils suppress T cell immunity in ovarian cancer microenvironment. JCI Insight. https://doi.org/10.1172/jci.insight.122311
Rogers T, DeBerardinis RJ (2021) Metabolic plasticity of neutrophils: relevance to pathogen responses and cancer. Trends Cancer 7:700–713
Kron A, Alidousty C, Scheffler M, Merkelbach-Bruse S, Seidel D, Riedel R, Ihle MA, Michels S, Nogova L, Fassunke J et al (2018) Impact of TP53 mutation status on systemic treatment outcome in ALK-rearranged non-small-cell lung cancer. Ann Oncol 29:2068–2075
Kodigepalli KM, Bowers K, Sharp A, Nanjundan M (2015) Roles and regulation of phospholipid scramblases. FEBS Lett 589:3–14
Bassé F, Stout JG, Sims PJ, Wiedmer T (1996) Isolation of an erythrocyte membrane protein that mediates Ca2+-dependent transbilayer movement of phospholipid. J Biol Chem 271:17205–17210
Dal Col J, Lamberti MJ, Nigro A, Casolaro V, Fratta E, Steffan A, Montico B (2022) Phospholipid scramblase 1: a protein with multiple functions via multiple molecular interactors. Cell Commun Signal 20:78
Liao WC, Lin TJ, Liu YC, Wei YS, Chen GY, Feng HP, Chang YF, Chang HT, Wang CL, Chi HC et al (2022) Nuclear accumulation of KPNA2 impacts radioresistance through positive regulation of the PLSCR1-STAT1 loop in lung adenocarcinoma. Cancer Sci 113:205–220
Huang P, Liao R, Chen X, Wu X, Li X, Wang Y, Cao Q, Dong C (2020) Nuclear translocation of PLSCR1 activates STAT1 signaling in basal-like breast cancer. Theranostics 10:4644–4658
Andreasson J, Bodén E, Fakhro M, von Wachter C, Olm F, Malmsjö M, Hallgren O, Lindstedt S (2022) Exhaled phospholipid transfer protein and hepatocyte growth factor receptor in lung adenocarcinoma. Respir Res 23:369
Talukder AH, Bao M, Kim TW, Facchinetti V, Hanabuchi S, Bover L, Zal T, Liu YJ (2012) Phospholipid scramblase 1 regulates Toll-like receptor 9-mediated type I interferon production in plasmacytoid dendritic cells. Cell Res 22:1129–1139
Lizak M, Yarovinsky TO (2012) Phospholipid scramblase 1 mediates type i interferon-induced protection against staphylococcal α-toxin. Cell Host Microbe 11:70–80
Sadanari H, Takemoto M, Ishida T, Otagiri H, Daikoku T, Murayama T, Kusano S (2022) The interferon-inducible human PLSCR1 protein is a restriction factor of human cytomegalovirus. Microbiol Spectr 10:e0134221
Martinelli S, Urosevic M, Daryadel A, Oberholzer PA, Baumann C, Fey MF, Dummer R, Simon HU, Yousefi S (2004) Induction of genes mediating interferon-dependent extracellular trap formation during neutrophil differentiation. J Biol Chem 279:44123–44132
Kantari C, Pederzoli-Ribeil M, Amir-Moazami O, Gausson-Dorey V, Moura IC, Lecomte MC, Benhamou M, Witko-Sarsat V (2007) Proteinase 3, the Wegener autoantigen, is externalized during neutrophil apoptosis: evidence for a functional association with phospholipid scramblase 1 and interference with macrophage phagocytosis. Blood 110:4086–4095
Martin KR, Kantari-Mimoun C, Yin M, Pederzoli-Ribeil M, Angelot-Delettre F, Ceroi A, Grauffel C, Benhamou M, Reuter N, Saas P et al (2016) Proteinase 3 is a phosphatidylserine-binding protein that affects the production and function of microvesicles. J Biol Chem 291:10476–10489
Hidalgo A, Chilvers ER, Summers C, Koenderman L (2019) The neutrophil life cycle. Trends Immunol 40:584–597
Acknowledgements
We thank all the healthy donors who took part in this study. We thank nurses in the Department of Oncology at PLA General Hospital for their assistance in sample collection.
Funding
This research fund by the National Natural Science Foundation of China under Grant No. 81902912, the Military Health Special Research Project under grant 20BJZ37, and the key project of the National Health Commission under Grant No. GWJJ2021100304.
Author information
Authors and Affiliations
Contributions
Xuan Han: data curation, writing–original draft. Yaping Long: methodology, editing, validation. Yao Li: visualization, software. Qi Xiong: editing. Jinfeng Li: IHC assistant. Liangliang Wu: cell cultural guidance. Qiaowei Liu: conceptualization. Bo Yang: conceptualization, writing review and editing. Yi Hu: check and approve.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests in this paper.
Ethics approval and consent to participate
This study was approved by the ethics committee of the Chinese PLA General Hospital [No. IACUC-DWZX-2023–524]. This study’s healthy donors signed informed consent before blood sample collection. The blood sample used in this study followed the principles of the Helsinki Declaration.
Consent for publication
All the authors and patients signed the consent for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Han, A.X., Long, B.Y., Li, C.Y. et al. Machine learning framework develops neutrophil extracellular traps model for clinical outcome and immunotherapy response in lung adenocarcinoma. Apoptosis (2024). https://doi.org/10.1007/s10495-024-01947-4
Accepted:
Published:
DOI: https://doi.org/10.1007/s10495-024-01947-4