Abstract
DNA fragmentation factor is a heterodimer complex of the nuclease CAD and its specific inhibitor ICAD, which can be activated during apoptosis to induce DNA fragmentation. Although ICAD expression levels have been quantified in a variety of human cancer cells, the mechanism of ICAD gene regulation remains unknown. In this study, we identified a 106-bp TATA-less region upstream of the transcription start site as a basal promoter of the human ICAD gene. An E-Box motif, which binds transcription factors of the basic helix-loop-helix/leucine zipper family, is responsible for transcriptional activity, as demonstrated using mutated promoter-reporters. A chromatin immunoprecipitation assay further demonstrated that Myc binds to an endogenous ICAD promoter. The functional importance of Myc in the regulation of ICAD transcription was also demonstrated by knock-down of c-Myc and N-Myc gene expression, as well as their ectopic expression. Structural analysis of the human ICAD promoter and identification of factors which regulate its activity might further our understanding of the biological role of ICAD with respect to regulation of apoptosis and cancer development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Apoptosis or programmed cell death ensures the elimination of unwanted cells during normal development and homeostasis [1]. This process is progressively inactivated during malignant development and loss of the capacity for apoptosis is a hallmark of malignant cells. Chromatin condensation and internucleasomal DNA fragmentation are typical nuclear features and well-recognized events in apoptosis. DNA fragmentation factor (DFF) is a heterodimer protein composed of a 40-kDa caspase-activated DNase (CAD) otherwise known as DFF40, and its cognate 45-kDa inhibitor (inhibitor of CAD: ICAD or DFF45 [2–8]). Both human genes map to 1p36 [9, 10]. CAD is thought to be responsible for the majority of nuclear activity resulting in chromosomal DNA fragmentation. When apoptosis is activated, ICAD is cleaved by executor caspases, mainly caspase-3, into three fragments, after which it dissociates from CAD, resulting in CAD activation.
Thus, the DFF complex may play a role in malignant transformation, and up- or down-regulation of ICAD/CAD expression might correlate with cancer aggression. Expression levels of ICAD have been examined in a variety of human cancers. For example, ICAD expression is down-regulated during the exponential growth phase of human colon carcinoma cells [11]. In some neuroblastomas, preferential ICAD expression is observed in low-stage, but not in their high grade [12]. Other research suggests that down-regulation of ICAD may contribute to tumor growth and lymph node metastasis in esophageal carcinoma [13], and that ICAD expression might serve as a marker of aggressive tumor behavior with an associated poor prognosis in ovarian cancer [14]. We have previously demonstrated that the hepatitis C virus core protein, which not only encodes the viral nucleocapsid but has a number of properties enabling persistent viral infection, induces gene expression of ICAD, thereby increasing steady-state levels of the ICAD protein [15]. To date, the mechanism of transcriptional regulation of the ICAD gene is not well understood. Analysis of the gene structure of mouse ICAD showed that a 118-bp flanking region of the ICAD gene is required for promoter activity [16]. In this study, we identified a functional promoter of the human ICAD gene and investigated the role of c-Myc and N-Myc in regulation of the ICAD promoter.
Materials and methods
Cell cultures
Human hepatoblastoma cells (Huh-7), human oral squamous carcinoma cells (HSC-2, HSC-3 and Ca22-9), mouse neuroblastoma cells (IMR-32 and GOTO) and mouse 3T3 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 100 units/ml of penicillin, and 100 mg/ml of streptomycin and kept at 37°C in a 5% CO2 incubator.
Cloning of the 5′-upstream region of the human ICAD gene
Human genomic DNA was isolated from Huh-7 cell lines using the QIAamp DNA Mini kit according to the manufacturer’s instructions (QIAGEN). A 5′-upstream region of the ICAD gene was amplified by PCR using primers based on the genomic DNA sequence of the human ICAD gene (GenBank accession No. NM_004401). Primers containing EcoRI sites were used as follows: forward (5′-GAATTCAGGCTGGTCTCAAACTACTG-3′) and reverse (5′-GAATTCGATCTCGCCAGATTCTGGTA-3′). The PCR reaction was carried out at 94°C for 1 min, 55°C for 1.5 min, and 72°C for 2 min, for total of 35 cycles. Each PCR product was purified and subcloned into a pGEM-T Easy vector (Promega), followed by sequencing using an ABI PRISM 310 automated DNA sequencer (Applied Biosystems).
5′ rapid amplification of cDNA ends (5′-RACE) assay
The 5′ ends of the ICAD transcript were cloned by 5′-RACE using a Gene Racer RACE-Ready cDNA kit in accordance with the manufacturer’s instructions (Invitrogen). Double-stranded cDNA molecules prepared from human liver poly(A)+ RNAs ligated by exposure to the adaptor were amplified in a primary PCR reaction using the adaptor primer 1 and the ICAD gene-specific antisense primer ICA1AS (5′-GTGCTGTTCGCGGCTGTAGTT-3′, nt −147 to −173), followed by a secondary PCR reaction using nested oligonucleotides, the adaptor primer 2, and the antisense ICAD specific primer ICA2AS (5′-CACGGTGACTGGTGTCAGGGACTTATC-3′, nt −228 to −254). PCR products were purified and cloned, followed by nucleotide sequencing as described above.
Plasmid constructions
The 1-kb sequence containing the human ICAD promoter was excised by digestion of the above-mentioned pGEM-T vector with SacI and HindIII, and then cloned into the firefly-luciferase-expressing reporter plasmid pGL3-Basic (Promega), resulting in pLUC1005. A series of constructs with 5′ end-deletions of the ICAD promoter were created by PCR amplification using the reverse primer (5′-CAAGCTTGCCTCCACAAGGTGGGACCTG-3′) and the following forward primers: 5′-GGCTAGCCAGTACCCATTTCTGAAGAAG-3′ (nt −936 to +71), 5′-GGCTAGCCCTCATTTGGGTCCATTTTCC-3′ (nt −622 to +71), 5′-GGCTAGCCAGCTTTTTCAGACAGAATGG-3′ (nt −272 to +71), 5′-GGCTAGCCAGCTTTTTCAGACAGAATGG-3′ (nt −205 to +71), 5′-GGCTAGCCAGCTTTTTCAGACAGAATGG-3′ (nt −145 to +71), 5′-GGCTAGCCAGCTTTTTCAGACAGAATGG-3′ (nt −106 to +71), 5′-GGCTAGCCTATTTAGTTTGGTTAGTAAT-3′ (nt −90 to +71), and 5′-GGCTAGCCCAGATGGTAAATATACACAA-3′ (nt −43 to +71). Each SacI/HindIII fragment was inserted into the pGL3-basic vector to yield pLUC(−936/+71), pLUC(−622/+71), pLUC(−272/+71), pLUC(−205/+71), pLUC(−145/+71), pLUC(−106/+71), pLUC(−90/+71) and pLUC(−43/+71).
Transfection and reporter assay
Huh-7 cells were seeded at 5 × 104 cells/well in 24-well plates and maintained at 37°C in a 5% CO2/95% atmosphere. DNA transfection of cells with each ICAD-promoter-luciferase construct (1 μg) with an internal control vector pRL-TK (0.1 μg) (Promega) was performed with Trans IT LT-1 (Mirus) during 6-h of incubation. Cells were then rinsed with phosphate-buffered saline (PBS) 48 h after transfection, and luciferase activity was measured in the cell lysate using dual luciferase assay reagents (Promega) [17]. Firefly luciferase activity was standardized according to Renilla luciferase activity.
Chromatin immunoprecipitation
Chromatin immunoprecipitation assays were performed using the ChIP assay kit (Upstate). Briefly, cells in 100-mm dishes were grown to 70% confluency over 48 h. The chromatin from formaldehyde-fixed cells was sonicated and immunoprecipitated using mouse monoclonal anti-c-Myc or anti-N-Myc antibodies (Santa Cruz). The chromatin immunoprecipitate was analysed by PCR with the following primer pairs: F1 (5′-CGAGCTCGGTATACATGCGTGTGCATCG-3′) and R1 (5′-CAAGCTTGCCTCCACAAGGTGGGACCTG-3′) for amplifying the region from nt −272 to −71 containing potential Myc-binding sites; and F2 (5′-GAGATCAAAACTGCAGTGAG-3′) and R2 (5′-CACTGTTGGAGATTGTTCAG-3′) for amplifying the region from nt −789 to −451 that does not contain Myc-binding sites.
Western blotting
Cells were washed with PBS and lysed in SDS sample buffer. Cell lysate samples were separated by 10% SDS-polyacrylamide gel electrophoresis and electrotransferred to a polyvinylidene difluoride membrane (Immobilion; Millipore). After blocking in nonfat milk solution (Blocking One; Nakaraitesk), the membranes were probed with monoclonal antibody against the ICAD protein (Santa Cruz), c-Myc (Sigma), or N-Myc (Santa Cruz), as the primary antibody for 1 h. After being washed, the membranes were incubated with horseradish peroxidase-conjugated anti-mouse immunoglobulin as the secondary antibody, followed by visualization with SuperSignal West Pico Chemiluminescent Substrate (Pierce Biotechnology).
siRNA experiments
siRNAs for human c-Myc, N-Myc and the negative control were purchased from B-Bridge International. Each siRNA consisted of three different target sequences as follows: 5′-GAGGAGACAUGGUGAACCA-3′, 5′-GAGAAUGUCAAGAGGCGAA-3′, and 5′-GAGAAUGUCAAGAGGCGAA-3′ for c-Myc siRNA; 5′-CGGAGAUGCUGCUUGAGAA-3′, 5′-CGGAGUUGGUAAAGAAUGA-3′, and 5′-CAGCAGUUGCUAAAGAAAA-3′ for N-Myc siRNA; and 5′-ATCCGCGCGATAGTACGTA-3′, 5′-TTACGCGTAGCGTAATACG-3′, and 5′-TATTCGCGCGTATAGCGGT-3′ for the negative control. Huh-7 cells were transfected with 100 pmol/ml siRNA using FuGENE6 (Roche) in Opti-MEM I (Invitrogen), after which the medium was replaced by standard DMEM after 6 h of transfection.
RNA preparation and real-time quantitative RT-PCR
Total RNA was extracted using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. First-strand cDNAs were synthesized using the SuperScriptIII First-Strand Synthesis kit (Invitrogen) and then used as templates for real-time PCR. Quantitative PCR was performed using the ABI Prism 7700 sequence detection system using Taq-Man Gene Expression assays (Applied Biosystems). The standard curve was created using serially diluted total RNA obtained from Huh-7 cultures and β-actin was chosen as the internal standard to control for variability in amplification. Amplification was performed at 95°C for 10 min, followed by 40 cycles of amplification at 95°C for 15 s and 60°C for 60 s.
Results and discussion
Localization of transcription start sites of the human ICAD gene
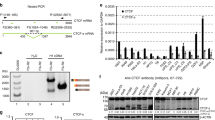
Precise localization of transcription start sites of the ICAD gene was examined in human hepatoma Huh-7 cells by 5′-RACE analysis. A fragment of approximately 100 bp was obtained (Fig. 1a) and cloned, after which sequencing analysis of several cDNA clones identified two of 5′ termini (Fig. 1b). These were located 71 and 68 nucleotides upstream from the translation start codon of the ICAD gene. From a database search of the DBTSS [18], we found that these are two of multiple sites for putative transcription initiation for the human ICAD gene. Although transcription may be initiated at both sites, the upper site was designated position +1 in this study.
(a) Mapping of the transcription start sites by 5′ RACE analysis. Following nested PCR reaction, a PCR product of 100-bp was detected by 1.5% agarose gel electrophoresis (lane 2). Lane 1: no template control; lane M: 100-bp DNA ladder marker. (b) The 5′ flanking region of the human ICAD gene. The nucleotide sequence is numbered from the major transcription site, which is indicated by an arrow. Putative binding sites for transcription factors analyzed by the TRANSFAC database and search program were shown with underlines
Characterization of the ICAD gene promoter
To examine potential regulatory sequences involved in ICAD gene expression, a 1 kb 5′-upstream region of the ICAD gene was sequenced and analyzed. The overall GC content of the 1-kb genomic DNA fragment was 46%, while GC content among the 200 bp proximal to the ATG start codon was approximately 62%. A search of transcription consensus motifs using the TRANSFAC database [19] demonstrated that none of the major eukaryotic promoter elements, TATA-, CCAAT-, or GC box, were located within 200 bp upstream of the transcription start site, although a TATA-like element was found more than 400 bp upstream from the start sites, indicating that the human ICAD gene has a TATA-less promoter. No CpG island, which is frequently observed in the TATA-less promoter, was found by screening with the CpG island searcher (http://cpgislands.usc.edu/). Absence of a TATA-like sequence upstream of the transcription start site has also been reported for the mouse ICAD gene, suggesting that TATA-less promoters might be a common feature in transcriptional regulation of the ICAD gene.
As shown in Fig. 1b, the 1-kb sequence contains potential binding sites for several transcription factors. The ability of the 5′-upstream region of the ICAD gene to function as a promoter was assessed by its capacity to drive the expression of a luciferase reporter gene. A series of constructs in which genomic DNA fragments were fused to a promoterless firefly luciferase gene of the pGL3-basic vector were generated with the 3′ end always terminating +71 bp from the transcription start site. The 5′ ends began at bases −936, −662, −272, −205, −145, −106, −90, and −43 (Fig. 2). Promoter activity was assessed by measuring luciferase activity in transiently transfected Huh-7 cells. Luciferase activity was normalized to Renilla luciferase activity. As shown in Fig. 2 on the right, the region extending from −936 to +71 had promoter activity since luciferase activity of the reporter gene was 45-fold that of the empty vector. A 5′ deletion of the promoter sequence to −662(pLuc(−662/+71)) enhanced promoter activity, and successive removal of nucleotides from −272 to −106 also enhanced promoter activity, suggesting the possibility of negative regulatory elements within the −936/−622 and −272/−106 regions. When the sequence from −106 to −90 was removed (pLuc(−90/+71)), promoter activity fell to parallel that of the pGL3 basic vector. This suggests the presence of a critical element in the region extending from −106 to −90, thus, we have identified the region 106 bp upstream of the transcription start site as the basal promoter of the human ICAD gene.
Human ICAD promoter drives reporter gene expression in transiently transfected Huh-7 cells. Deletion constructs of the upstream region of the ICAD gene linked to the firefly luciferase reporter gene (LUC) are illustrated in the left. Huh-7 cells were cotransfected with a firefly luciferase reporter and a Renilla luciferase internal control (pRL-TK). The relative luciferase activity (RLU) was obtained by normalizing the firefly luciferase activity to the Renilla luciferase activity, and is expressed as a percentage of RLU of pLuc(−936/+71). The average values are shown with standard deviation (SD) (n = 4 per construct)
Myc and USF enhance ICAD promoter activity
As indicated in Fig. 1b, a computer-based sequence analysis revealed that the 106 bp region contains potential binding sites for GATA-1, deltaE, Myc, and various upstream stimulatory factors (USF). Within this region, there is a canonical E-box sequence (5′-CACGTG-3′) located between −103 and −98. E box motifs are known to bind to transcription factors of the basic helix-loop-helix/leucine zipper (bHLH LZ) family, including Myc and USF [20–23]. We examined the potential role of E-box-binding proteins, Myc and USF, in potentiating ICAD gene transcription. Myc is bound to a partner protein Max via a bHLH LZ domain and the Myc-Max heterodimer activates transcription by binding to an E-box sequence [24, 25]. Max is present in greater amounts than Myc since the Myc transcript and protein have shorter half-lives compared to Max [26, 27]. Therefore, it is highly likely that Myc is the limiting, regulated component of the heterodimer. To examine the potential role of E box binding proteins, Huh-7 cells were co-transfected with luciferase reporter plasmids and expression vectors for c-Myc, N-Myc, USF1, and USF2 (Fig. 3a). Luciferase activity of the −145/+71 promoter construct increased 2- to 5-fold, and 1.5- to 2-fold, when co-expressed with Myc or USF, respectively. The effect of c-Myc and N-Myc on activity of the −90/+71 construct was less than for the −145/+71 construct. USF expression did not enhance activity of the −90/+71 construct.
(a) ICAD promoter activity after transient expression of c-Myc, N-Myc, USF1 and USF2. Huh-7 cells were cotransfected with either expression vector for c-Myc, N-Myc, USF1 or USF2 driven by the CMV promoter and pLuc(−145/+71), pLuc(−90/+71) or pGL3 basic. Each firefly luciferase reporter plasmid (n = 4) was cotransfected in cells with pRL-TK for normalization of the reporter activity. RLU is expressed as a percentage of that of pLuc(−145/+71) in the absence of expressing plasmids for Myc and USF. (b) Effect of substitution mutation in the E-box element on the ICAD promoter activity. Cells were cotransfected with pLuc(−106/±71) or an E-box-mutant, pLuc(−106/±71)mt and either expression vector for c-Myc or N-Myc. pRL-TK was also used for normalization of the activity. RLU is expressed as a percentage of that of pLuc(−106/±71) without over-expression of Myc
To further investigate involvement of the E box sequence in ICAD promoter activity, a reporter construct with an E box mutation (CCCGCG) was constructed and luciferase activity was examined in transfected cells (Fig. 3b). The E box mutation resulted in marked down-regulation of reporter gene expression and the reporter activity expressed from the E-box mutant was little increased when co-expressed with Myc, suggesting a functionally important role of the −106/−90 region E box motif with regard to basal transcriptional activation of the ICAD promoter. These data indicates that E-box-binding proteins, especially Myc, actively participate in positive regulation of the human ICAD promoter.
Myc binds to the ICAD promoter in vivo
To obtain direct evidence of an interaction between Myc and the ICAD promoter in vivo, we next examined binding of c-Myc and N-Myc to the 5′-upstream region of the ICAD gene within the context of native chromatin in living cells by chromatin immunoprecipitation. An alignment of the sequence around putative Myc binding site of human ICAD promoter and the corresponding part of mouse ICAD sequence demonstrated conservation of Myc binding sequence in human and mouse (Fig. 4a) [16]. Proteins were cross-linked to genomic DNA isolated not only from human (Huh-7) but mouse (IMR-32 and GOTO) cells, followed by immunoprecipitation with normal rabbit IgG or polyclonal antibodies to either c-Myc or N-Myc. IMR-32 and GOTO are neuroblastoma cells with N-Myc gene amplification [28]. The precipitated DNA was then subjected to PCR utilizing primers designed to amplify a 343-bp fragment (−272/+71) or a 338 bp fragment (−789/−451) of the ICAD 5′ flanking region. As shown in Fig. 4b, the 343-bp DNA fragment was observed in the immunoprecipitate from Huh-7 cells following exposure to anti-c-Myc or anti-N-Myc, while amplification of the ICAD promoter fragment was not detected in the negative control immunoprecipitate. Under similar experimental conditions, we did not detect binding of c-Myc and N-Myc to the −789/−451 region, which is not thought to contain a Myc-binding site. Similar results were obtained from IMR-32 and GOTO cells, which were immunoprecipitated with anti-N-Myc antibody. These results demonstrate that Myc forms a complex with the human ICAD promoter in cells, presumably through binding to the E-box sequence.
(a) An alignment of the sequence around putative Myc binding site of human ICAD promoter and the corresponding part of mouse ICAD sequence. (b) Binding of Myc proteins in the endogenous ICAD promoter. Crosslinked chromatins isolated from Huh-7, IMR-32 and GOTO cells were immunoprecipitated with indicated antibodies (c-Myc, N-Myc) or an equivalent amount of mouse IgG. Recovered DNAs were purified and PCR-amplified with primers for nt −272/±71 region or for nt −789/−451 region. Input represents 1% of chromatin sample applied for immunoprecipitation
Myc-dependent expression of the ICAD protein
The above data suggests that the ICAD promoter is regulated by endogenous Myc proteins. This assumption is supported by the loss-of-function studies shown in Fig. 5a. We employed siRNA (small interference RNA) transfection to knock-down the expression of c-Myc or N-Myc in cells, after which we analyzed the effect of reduced Myc expression on steady state levels of the ICAD protein by Western blotting. Treatment with siRNA specific for either c-Myc or N-Myc markedly reduced ICAD expression without affecting the expression of β-actin. In contrast, control siRNA did not reduce ICAD levels. We further investigated the effect of over-expression of Myc on ICAD protein levels (Fig. 5b). Transfection of Huh-7 and mouse 3T3 cells with a c-Myc or N-Myc expression vector enhanced ICAD protein levels with an effect that was less pronounced than in the loss-of-function studies, but reproducible nonetheless. These results suggest an important role of Myc proteins in positive regulation of ICAD expression.
Myc-dependent expression of human ICAD. (a) Suppression of Myc expression decreases expression of ICAD protein. Huh-7 cells were transiently transfected with Myc siRNA (c-Myc, N-Myc) or the control siRNA (cont). Three days later, the cells were harvested and subjected to Western blotting. (b) Ectopic expression of Myc increases expression of ICAD protein. Cells (Huh-7 and mouse 3T3) were transfected with the expression vector for c-Myc or N-Myc, and after 3 days the cells were harvested for Western blotting. (c) Comparison of the mRNA expression of c-Myc and ICAD in Huh-7 cells and human oral squamous carcinoma cells (Ca22-9, HSC-2, and HSC-3). Levels of mRNA expression of ICAD and c-Myc were measured by quantitative real-time RT-PCR based on Taq Man chemistry. Results, relative to β-actin mRNA, are depicted as averages with SD (n = 3)
Finally, we examined if there might be a correlation between ICAD and Myc expression levels in cancer cell lines. The mRNA expression of ICAD and c-Myc in human oral squamous cell carcinomas (Ca22-9, HSC-2, and HSC-3), in which ICAD expression has not been investigated to date, as well as in Huh-7 cells during their late exponential phase of growth, was analyzed by quantitative RT-PCR (Fig. 5c). ICAD mRNA was readily detected in all cancer cells but the expression level was not consistent. A relatively high level of expression was observed in Ca22-9 and HSC-3 cells, while HSC-2 cells demonstrated the lowest. Interestingly, expression pattern of c-Myc mRNA in these cell lines showed a tendency similar to that of ICAD, suggesting an important role of Myc in ICAD expression in human cancers. A large scale analysis for investigating correlation between ICAD and Myc expression in a variety of cancer tissues obtained from patients is ongoing.
Several studies have shown up- or down-regulation of ICAD in a variety of human cancers [11–14, 29, 30]. Although transcriptional deregulation is presumably involved in this aberrant expression of ICAD, little is known about the transcriptional regulation of ICAD in human cells. This work was done to characterize the human ICAD promoter and to examine transcription factors which may be involved in its regulation. We experimentally determined putative transcription start sites by employing the 5′-RACE method, and identified a promoter region required for basal ICAD gene expression using promoter-reporter constructs with progressive deletions. A combined study of site-directed mutagenesis of a reporter construct and chromatin immunoprecipitation revealed the importance of an E box Myc-binding motif located at position −103 to −98 from the transcription start sites, as well as an in vivo interaction between c-Myc and N-Myc and the proximal ICAD promoter. Furthermore, we demonstrated the functional importance of Myc proteins with regard to transcriptional regulation of the ICAD gene from studies examining ectopic expression of c-Myc and N-Myc, as well as with RNAi technology. We showed that expression levels of ICAD and Myc correlate in some tumors.
c-Myc and N-Myc are transcription factors of the bHLH LZ family that bind to the E box sequence within promoters to control proliferation, cell differentiation, and apoptosis [25, 31, 32]. c-myc and N-myc genes are deregulated in a number of human cancers and influence proliferation and growth. A link between Myc and cancer is well established both in vivo and in vitro, and oncogenic activation of Myc has been observed to promote the development of a number of clinically significant cancers [25, 33]. However, the molecular and cellular mechanisms of Myc-mediated transformation are not fully understood. Although a variety of Myc target genes were identified and recruitment of Myc to the target promoters such as prothymosin and telomerase were shown [34], there has been no direct evidence on involvement of Myc in regulation of ICAD expression. It is known that c-Myc activation usually occurs during the later stages of carcinoma in humans. Conversely, in premalignant cells, c-Myc is a robust stimulator of apoptosis and programmed cell death [32]. One area of investigation into cancer cell death mechanisms centers on the mechanism by which c-Myc stimulates or suppresses apoptosis. For instance, Myc has been reported to potentiate apoptosis through both p53-dependent and -independent mechanisms [35, 36]. Myc controls the balance between pro- and anti-apoptotic factors at the level of the mitochondria, thereby regulating cytochrome C release and activation of downstream caspases [37, 38]. Involvement of Myc in up-regulation of ICAD expression as demonstrated in this study might present a novel mechanism of Myc-dependent inhibition of apoptosis. It is possible that elevated levels of ICAD in cells inhibit activation of endonuclear activity, thereby increasing the threshold for apoptosis signaling.
Kawane et al. have reported on the structure and analyzed the promoter of a murine ICAD gene, by which they demonstrated that a 118-bp flanking region of the ICAD gene is required for its transcription [16]. The mouse sequence shares approximately 82% homology with a corresponding upstream region of the human ICAD gene. The mouse ICAD promoter has a number of potential binding sites for transcription factors, such as Ikaros, c-Rel, Myc, and Gfi-1 [16]. Conservation of the Myc-binding motif among human and murine promoters suggests a functional significance of Myc in transcriptional regulation of ICAD expression.
In conclusion, this is the first report to identify a functional promoter of the human ICAD gene, and to demonstrate that Myc proteins are able to positively regulate ICAD gene expression. Extensive apoptosis research to date has shown that tumor aggression depends on various defects in apoptosis signaling [39]. Further investigation into the molecular events linking Myc expression with ICAD gene regulation may provide insight into Myc’s role in cell proliferation, transformation and apoptosis.
References
Raff MC (1992) Social controls on cell survival and cell death. Nature 356:397–400. doi:10.1038/356397a0
Liu X, Zou H, Slaughter C, Wang X (1997) DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell 89:175–184. doi:10.1016/S0092-8674(00)80197-X
Enari M, Sakahira H, Yokoyama H et al (1998) A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature 391:43–50. doi:10.1038/34112
Sabol SL, Li R, Lee TY, Abdul-Khalek R (1998) Inhibition of apoptosis-associated DNA fragmentation activity in nonapoptotic cells: the role of DNA fragmentation factor-45 (DFF45/ICAD). Biochem Biophys Res Commun 253:151–158. doi:10.1006/bbrc.1998.9770
Sakahira H, Enari M, Nagata S (1998) Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature 391:96–99. doi:10.1038/34214
Samejima K, Earnshaw WC (1998) ICAD/DFF regulator of apoptotic nuclease is nuclear. Exp Cell Res 243:453–459. doi:10.1006/excr.1998.4212
Nagata S (2000) Apoptotic DNA fragmentation. Exp Cell Res 256:12–18. doi:10.1006/excr.2000.4834
Nagata S, Nagase H, Kawane K et al (2003) Degradation of chromosomal DNA during apoptosis. Cell Death Differ 10:108–116. doi:10.1038/sj.cdd.4401161
Mukae N, Enari M, Sakahira H et al (1998) Molecular cloning and characterization of human caspase-activated DNase. Proc Natl Acad Sci USA 95:9123–9128. doi:10.1073/pnas.95.16.9123
Leek JP, Carr IM, Bell SM, Markham AF, Lench NJ (1997) Assignment of the DNA fragmentation factor gene (DFFA) to human chromosome bands 1p36.3–>p36.2 by in situ hybridization. Cytogenet Cell Genet 79:212–213
Charrier L, Jarry A, Toquet C et al (2002) Growth phase-dependent expression of ICAD-L/DFF45 modulates the pattern of apoptosis in human colonic cancer cells. Cancer Res 62:2169–2174
Abel F, Sjoberg RM, Ejeskar K, Krona C, Martinsson T (2002) Analyses of apoptotic regulators CASP9 and DFFA at 1P36.2, reveal rare allele variants in human neuroblastoma tumours. Br J Cancer 86:596–604. doi:10.1038/sj.bjc.6600111
Konishi S, Ishiguro H, Shibata Y et al (2002) Decreased expression of DFF45/ICAD is correlated with a poor prognosis in patients with esophageal carcinoma. Cancer 95:2473–2478. doi:10.1002/cncr.10987
Brustmann H (2006) DNA fragmentation factor (DFF45): expression and prognostic value in serous ovarian cancer. Pathol Res Pract 202:713–720. doi:10.1016/j.prp.2006.06.003
Sacco R, Tsutsumi T, Suzuki R et al (2003) Antiapoptotic regulation by hepatitis C virus core protein through up-regulation of inhibitor of caspase-activated DNase. Virology 317:24–35. doi:10.1016/j.virol.2003.08.028
Kawane K, Fukuyama H, Adachi M et al (1999) Structure and promoter analysis of murine CAD and ICAD genes. Cell Death Differ 6:745–752. doi:10.1038/sj.cdd.4400547
Masaki T, Matsuura T, Ohkawa K et al (2006) All-trans retinoic acid down-regulates human albumin gene expression through the induction of C/EBPbeta-LIP. Biochem J 397:345–353. doi:10.1042/BJ20051863
Suzuki Y, Yamashita R, Sugano S, Nakai K (2004) DBTSS, DataBase of Transcriptional Start Sites: progress report 2004. Nucleic Acids Res 32:D78–D81. doi:10.1093/nar/gkh076
Wingender E, Dietze P, Karas H, Knuppel R (1996) TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res 24:238–241. doi:10.1093/nar/24.1.238
Bello-Fernandez C, Packham G, Cleveland JL (1993) The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci USA 90:7804–7808. doi:10.1073/pnas.90.16.7804
Meier JL, Luo X, Sawadogo M, Straus SE (1994) The cellular transcription factor USF cooperates with varicella-zoster virus immediate-early protein 62 to symmetrically activate a bidirectional viral promoter. Mol Cell Biol 14:6896–6906
Kiermaier A, Gawn JM, Desbarats L et al (1999) DNA binding of USF is required for specific E-box dependent gene activation in vivo. Oncogene 18:7200–7211. doi:10.1038/sj.onc.1203166
Luscher B, Larsson LG (1999) The basic region/helix-loop-helix/leucine zipper domain of Myc proto-oncoproteins: function and regulation. Oncogene 18:2955–2966. doi:10.1038/sj.onc.1202750
Grandori C, Cowley SM, James LP, Eisenman RN (2000) The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol 16:653–699. doi:10.1146/annurev.cellbio.16.1.653
Adhikary S, Eilers M (2005) Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol 6:635–645. doi:10.1038/nrm1703
Wagner AJ, Le Beau MM, Diaz M, OHay N (1992) Expression, regulation, and chromosomal localization of the Max gene. Proc Natl Acad Sci USA 89:3111–3115. doi:10.1073/pnas.89.7.3111
Skouteris GG, Schroder CH (1996) c-Myc and Max interactions in quiescent and mitogen-stimulated primary hepatocytes. Exp Cell Res 225:237–244. doi:10.1006/excr.1996.0173
Kato H, Okamura K, Kurosawa Y et al (1989) Characterization of DNA rearrangements of N-Myc gene amplification in three neuroblastoma cell lines by pulsed-field gel electrophoresis. FEBS Lett 250:529–535. doi:10.1016/0014-5793(89)80790-2
Masuoka J, Shiraishi T, Ichinose M, Mineta T, Tabuchi K (2001) Expression of ICAD-l and ICAD-S in human brain tumor and its cleavage upon activation of apoptosis by anti-Fas antibody. Jpn J Cancer Res 92:806–812
Yang HW, Chen YZ, Piao HY et al (2001) DNA fragmentation factor 45 (DFF45) gene at 1p36.2 is homozygously deleted and encodes variant transcripts in neuroblastoma cell line. Neoplasia 3:165–169. doi:10.1038/sj.neo.7900141
Secombe J, Pierce SB, Eisenman RN (2004) Myc: a weapon of mass destruction. Cell 117:153–156. doi:10.1016/S0092-8674(04)00336-8
Meyer N, Kim SS, Penn LZ (2006) The Oscar-worthy role of Myc in apoptosis. Semin Cancer Biol 16:275–287. doi:10.1016/j.semcancer.2006.07.011
Donaldson TD, Duronio RJ (2004) Cancer cell biology: Myc wins the competition. Curr Biol 14:R425–R427
Mac SM, D’Cunha CA, Farnham PJ (2000) Direct recruitment of N-Myc to target gene promoters. Mol Carcinog 29:76–86
Sakamuro D, Eviner V, Elliott KJ et al (1995) c-Myc induces apoptosis in epithelial cells by both p53-dependent and p53-independent mechanisms. Oncogene 11:2411–2418
Nilsson JA, Cleveland JL (2003) Myc pathways provoking cell suicide and cancer. Oncogene 22:9007–9021
Juin P, Hueber AO, Littlewood T, Evan G (1999) c-Myc-induced sensitization to apoptosis is mediated through cytochrome c release. Genes Dev 13:1367–1381
Hotti A, Jarvinen K, Siivola P, Holtta E (2000) Caspases and mitochondria in c-Myc-induced apoptosis: identification of ATM as a new target of caspases. Oncogene 19:2354–2362
Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100:57–70
Acknowledgements
The authors gratefully acknowledge Dr. H. Kondo of Osaka University for providing the Myc-expressing plasmids. They also thank M. Matsuda, M. Ikeda, S. Yoshizaki, and T. Shimoji for their technical assistance. This work was partially supported by a grant-in-aid for Scientific Research from the Japan Society for the Promotion of Science, from the Ministry of Health, Labour and Welfare of Japan and from the Ministry of Education, Culture, Sports, Science and Technology, and by the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation of Japan.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Omata, K., Suzuki, R., Masaki, T. et al. Identification and characterization of the human inhibitor of caspase-activated DNase gene promoter. Apoptosis 13, 929–937 (2008). https://doi.org/10.1007/s10495-008-0223-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-008-0223-8