Abstract
The purpose of this study was to investigate tick species around Mount Fanjing and analyze bacterial communities in two species – Rhipicephalus microplus and Haemaphysalis longicornis – parasitizing cattle in Tongren, Guizhou province, Southwest China, using high-throughput sequencing methods. In April 2019, ticks were collected from five sites in Jiangkou County, Yinjiang County, and Songtao County. In total, 296 ticks were collected, comprising two genera and three species: H. longicornis, Haemaphysalis flava, and R. microplus. Rhipicephalus microplus was the most representative species (57.4%) within the collected group, being the dominant species in Tongren City, followed by H. longicornis (39.5%) and H. flava (3.0%). Beta-diversity analysis revealed differences in bacterial community composition among the tick species. The bacterial community structure of R. microplus collected in the three counties was highly similar. Chlorella and Bacillus were highly abundant in H. longicornis. Rickettsia was detected at high relative abundance in R. microplus but in low relative abundance in H. longicornis, suggesting that Rickettsia is more associated with R. microplus than with H. longicornis. More in-depth investigations are needed to determine the pathogenic risk of Rickettsia and its relationship with the host. This is the first survey on tick-borne bacterial communities in this area, which is of great significance for the prevention and control of tick-borne diseases locally.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ticks are hematophagous ectoparasites that parasitize almost all kinds of vertebrates, including humans, livestock, and wild animals (Zhang et al. 2021a; Sonenshine 2018). Ticks are responsible for spreading a variety of pathogens that threaten their hosts, such as viruses, rickettisae, bacteria, fungi, and protozoa (Brites-Neto et al. 2015), being the second most important disease vectors to humans after mosquitoes, and the most important disease vectors to livestock and wild animals (de la Fuente et al. 2008). Over 900 tick species have been described worldwide (Estrada-Peña 2015), and 124 tick species in total have been reported in China (Chen and Yang 2021), among which 110 are hard ticks (Ixodidae) and 14 soft ticks (Argasidae). In China, it has been proposed that approximately 130 animal species from 20 orders are potentially parasitized by ticks. Moreover, as ticks have a wide geographical distribution (Zhang et al. 2019a), tick-borne diseases occur in several locations throughout China (Huang et al. 2020; Ni et al. 2020; Fang et al. 2015; Wu et al. 2013).
Guizhou province is located in the hinterland of Southwest China, with a warm and humid climate conducive to ticks. In total 19 tick species have been reported in the province, which belong to two families and five genera (Zhang et al. 2019b). Tongren, located in the northeastern part of Guizhou province, with typical karst landforms, is divided east and west by the Wuyi Mountains with Mount Fanjing as the main peak. Mount Fanjing is located in national conservation and a 5A-level tourist attraction, being also listed as a world natural heritage. It is a suitable habitat for ticks due to its unique geographical environment and diverse animal and plant resources. Thus, bacteria carried by local ticks may threaten livestock, wild animals, and tourists. However, the geographical distribution of tick species in Guizhou province as well as the diversity of tick-borne pathogens have yet to be well known, with only a few studies reporting the prevalence of tick-borne diseases, such as Lyme disease and Q fever (Wu et al. 2013).
Therefore, the purposes of this study were to (i) investigate the distribution of tick species parasitizing cattle in three counties in the vicinity of Mount Fanjing (i.e., Jiangkou County, Yinjiang County, and Songtao County) and (ii) characterize bacterial communities by 16S rDNA high-throughput sequencing of the most prevalent tick species collected in the corresponding geographical area. As R. microplus and H. longicornis were the main tick species collected in the study period and location, the diversity and composition of bacterial communities in these two tick species were the focus of the present work.
Materials and methods
Geographical location and sample collection
Mount Fanjing is located at the junction of the three counties Yinjiang, Jiangkou, and Songtao, between 27°49’50”–28°1’30”N and 108°45’55”–108°48’30”E. In April 2019, five locations were selected in the vicinity of Mount Fanjing for tick collection as there are suitable environments for ticks. Ticks were collected on the body surface of herding cattle with the owner’s consent. They were collected using forceps by first anesthetizing the ticks with 75% ethanol, then holding the capitulum of the ticks with forceps and gently shaking them from side to side until the capitulum came off. Collected ticks were placed in prepared 15-mL cryotubes in liquid nitrogen tanks, sent to the laboratory, and placed at − 80 °C until later use. Distribution of the sample collection sites (Figure S1) was as follows – Jiangkou County: (i) Kaiwen Village, Taiping Town, (ii) Banpo Village, Bapan Town; Yinjiang County: (iii) Heshui Village, Heshui Town; Songtao County: (iv) Dapingcha Village, Shichang Township, (v) Qiandong Caohai, Panshi Town.
Tick identification
Morphological features of ticks were identified based on taxonomic and morphological criteria (Teng and Jiang 1991; Chen and Yang 2021) using an SZX7 stereomicroscope (Olympus, Chaoyang, Beijing, China) and a Smartzoom 5 digital microscope (Zeiss, Pudong, Shanghai, China). Observed structures included the shape of basis capituli, porose area, palp, scutum, and peritreme, as well as the dentition formula of hypostome, the presence or absence of festoon, among other structures.
High-throughput sequencing
Sample grouping
Based on collection site, tick species, and tick size, representants of the same tick species originating from different collection sites within the same geographical area were pooled. All ticks were fully engorged. The total volume of each sample was approximately the size of a soy bean, and samples were divided into four groups (Table 1) with three replicates each. The negative control used ddH2O instead of sample DNA to exclude environmental and reagent contamination, and the rest of the experimental conditions and procedures were consistent with the samples.
Sample preparation
Each sample was washed 3× with 75% ethanol (Lircon, Dezhou, Shandong province, China), and then once with 1× PBS buffer (Life Technologies, Grand Island, NY, USA), and placed in a 2-mL centrifuge tube to prepare for nucleic acid extraction.
DNA extraction and library construction
Samples were ground to powder with stainless steel grinding balls in a Scientz-48 high throughput tissue grinder (Scientz, Ningbo, Zhejiang province, China). Nucleic acid extraction was carried out using E.Z.N.A. Mag-Bind Soil DNA Kit (Omega Bio-Tek, Norcross, GA, USA) following the manufacturer’s instructions, and DNA concentration was quantified in a Qubit v.3.0. fluorometer (Invitrogen, Carlsbad, CA, USA). Primers were universal primers for the V3–V4 hyper-variable region of the bacterial 16S rDNA gene (341 F: CCTACGGGNGGCWGCAG and 785R: GACTACHVGGGTATCTAATCC) (Klindworth et al. 2013). PCR amplifications were performed as follows: initial denaturation at 94 °C for 3 min; followed by five cycles of denaturation at 94 °C for 30 s, annealing at 45 °C for 20 s, extension at 65 °C for 30 s; then 20 cycles of denaturation at 94 °C for 20 s, annealing at 55 °C for 20 s, and extension at 72 °C for 30 s; and a final extension at 72 °C for 5 min. A second bridge PCR was conducted with Illumina-compatible primers to complete library construction, and PCR amplification conditions were as follows: initial denaturation at 95 °C for 5 min; followed by five cycles of denaturation at 94 °C for 20 s, annealing at 55 °C for 20 s, and extension at 72 °C for 30 s; and a final extension step at 72 °C for 5 min.
Library quality control and sequencing
Library quality was determined by electrophoresis on 2% agarose gels. DNA fragments of approximately 400 bp were recovered using magnetic beads. The quality and concentration of prepared libraries were determined using Qubit v.3.0 fluorometer. High-throughput sequencing was performed using the Illumina MiSeq platform by Sangon Biotech (Shanghai). The company also performed a bioinformatic analysis for us.
Bioinformatics analysis
Raw sequence image data obtained by high-throughput sequencing were analyzed by base calling and transformed into raw sequenced reads, and saved as FASTQ files. Adapters were removed using cutadapt v.1.18, and paired-end reads were merged using PEAR v.0.9.8 using the overlapping method. Finally, quality control and filtering were carried out in each sample to obtain clean data. Operational taxonomic unit (OTU) clustering was performed on clean sequences, and chimera sequences were removed to obtain optimal OTU sequences, which were selected as representative sequences with ≥ 97% similarity. Then the generated OTUs were compared with the ribosomal database project (RDP, http://rdp.cme.msu.edu/index.jsp). Sequence classification assignments were performed using the RDP classifier, which was based on Bergey’s taxonomy, and the Naive Bayesian assignment algorithm was used to calculate the probability value of each sequence assigned to various hierarchical levels.
Six metrics (indices) were used for determining alpha-diversity analysis: Shannon, Chao, ACE, Simpson, Shannoneven, and Coverage. The Shannon and Simpson indices indicate the diversity of bacterial communities in the sample, and the Chao and ACE indices indicate the abundance of bacterial communities. The Shannoneven index was used to reflect the evenness of bacterial communities, and the Coverage index reflects the coverage of each sample library.
Data analysis was performed in the statistical language R (v.3.2). Hierarchical clustering was based on the Bray-Curtis distance algorithm, and the package ‘ape’ (v.5.3) of R was used to construct dendrograms.
Results
Tick species identification
In total, 296 ticks were collected from two tick genera and three tick species in the three counties included in the study (Table 2). The ticks were identified morphologically as R. microplus (n = 170), H. longicornis (n = 117), and Haemaphysalis flava (n = 9). The most prevalent tick species was R. microplus (57.4%), followed by H. longicornis (39.5%) and H. flava (3.0%).
16S rDNA sequencing
The effective number of reads (Table 3) in samples was within the range of 30,000–65,000, and the average length was approximately 400 bp. In total, 904 OTUs were identified in all samples. A rarefaction curve was constructed with the number of reads in the horizontal axis and the number of OTUs in the vertical axis (Fig. 1). The observed number of OTUs increased with sequencing depth, and the curve tended to become flat towards its end, indicating that sequencing depth was satisfactory to cover all bacterial species in the sample. Thus, sequencing data could likely reflect the features of the tick-borne bacterial community in the investigated geographical area.
Rarefaction curve showing Operational Taxonomic Unit (OTU) abundance in samples included in the study. See Table 1 for details on the sample codes
Analysis of species diversity and structure of bacterial communities in ticks
Alpha diversity
Collectively, the obtained data revealed that the diversity and abundance of microbial communities in each sample differed. Diversity was higher in samples JK-1 (R. microplus) and ST-4 (H. longicornis) and lower in samples JK-2, ST-1, and YJ-1 (all three R. microplus). Abundance was highest in sample JK-1 and lowest in sample JK-3 (R. microplus). Community distribution in samples JK-1 and ST-4 was more uniform than in the other samples. Coverage indices of all tested samples were > 99.7%, which indicated the reliability of our sequencing data pertaining the microbial community and low probability of sequences not being detected (Table 4).
Bacterial community composition
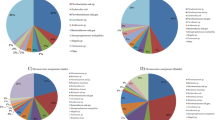
In total, 110 genera, 70 families, 40 orders, 28 classes, and 14 phyla were identified in the 12 samples analyzed. The Venn diagram (with data corresponding to genus level taxa) showed that 33 genera, 42 families, 25 orders, 18 classes, and 12 phyla were shared among the sample groups (Fig. 2). The stacked bar graph and the heat map present the relative abundance of bacterial species in all sample groups, showing only species whose abundance was above 1%. At phylum level, Proteobacteria (65.0%) were the most abundant, followed by Chlorobi (20.5%) and Firmicutes (9.7%); Proteobacteria was the most abundant phylum (> 50%) in nine samples of R. microplus, whereas Chlorobi was the most abundant (> 50%) in two samples of H. longicornis, and Firmicutes (> 80%) in one sample of H. longicornis. At class level, Alphaproteobacteria was detected in high abundance (> 50% in eight samples), with the highest abundance found in sample JK-2 (93%) of R. microplus. At order level, Rickettsiales was detected in all samples, with the highest relative abundance (> 90%) in samples JK-2, YJ-1, and ST-1. At family level, Rickettsiaceae (56.8%), Chlorobiaceae (20.5%), and Bacillaceae (6.1%) were detected at the highest relative abundance on the whole. At genus level (Figs. 3 and 4), Rickettsia was detected in high relative abundance in R. microplus (> 30%), whereas the relative abundance of Chlorobium was highest in samples ST-4 (54.2%) and ST-6 (70.7%), that of Bacillus was highest in sample ST-5 (66.3%). However, the content of Rickettsia in H. longicornis was extremely low (< 2.4%), and in the remaining samples, Bacillus was present at approximately 1%. The top-10 genera, ranked according to their average relative abundance, are Rickettsia (56.8%), Chlorobium (20.5%), Bacillus (6.1%), Acinetobacter (1.6%), Bradyrhizobium (1.1%), Staphylococcus (1.0%), Burkholderia (0.84%), Rhodopseudomonas (0.72%), Corynebacterium (0.43%), and Desulfovibrio (0.38%). As a common endosymbiont of ticks, Coxiella was found in low abundance in seven samples. Only sample JK-1 has a relative abundance of 2.3%, and the rest of samples are < 1% (Table S1).
Venn diagram depicting the frequency of microorganisms identified at genus level in ticks collected in the study. The number depicted in the core indicates the frequency of species commonly present in all samples. See Table 1 for details on the codes in the periphery
Relative abundance of microbial genera in ticks. Genera whose relative abundance was < 1% in all samples were depicted as ‘other’. See Table 1 for details on the sample codes
Heat map of the relative abundance of microorganisms identified at genus level in tick samples evaluated in the study. See Table 1 for details on the sample codes
Beta diversity
Beta-diversity analysis was conducted to determine the dissimilarity of bacterial communities in ticks evaluated in the study. Two main branches (Fig. 5) were identified in dendrograms: R. microplus samples were found in a branch regardless of the site of collection, and H. longicornis samples fell in other branches. Sample ST-5 differed from the other 11 samples in the corresponding branch due to the presence of Bacillus (66.3%), Staphylococcus (11.4%), and very little Chlorobium (0.04%); sample ST-4 was similar to sample ST-6 as the abundance of Chlorobium was 50% and Rickettsia was found in low abundance in both samples, 2.4 and 0.05%, respectively.
Cluster analysis of tick samples evaluated in the study. See Table 1 for details on the sample codes
Discussion
The geographical distribution of the collected ticks was different: in sites i and ii (in Jiangkou County), the tick species were highly similar (mainly R. microplus); in site iii (Yinjiang County), R. microplus was the only species identified, and in sites iv and v (Songtao County) H. longicornis was the dominant species. The environment of the five sampling sites is similar and all locations are near Mount Fanjing, and they all have a subtropical humid monsoon climate. The average annual temperature is 16–17 ℃, the average temperature of the hottest month (July) is about 27 ℃, and that of the coldest month (January) is about 5 ℃. But sampling site v has some uniqueness: it has 230 ha of artificial pasture, stocked with cattle, sheep, rabbits, and other animals; the other sampling sites only stocked cattle. Perhaps this explains the difference in the dominant tick species. Rhipicephalus microplus is the main ectoparasite affecting livestock worldwide (Silva et al. 2016), whereas H. longicornis is a tick species widely distributed in China (Malik et al. 2019).
In this study, using 16S rDNA sequencing analysis to explore the diversity of bacterial communities in collected ticks, the species Rickettsia, Chlorobium, and Bacillus were detected in all 12 samples. Rickettsia was detected at the highest relative abundance in R. microplus. Lim (2020) applied high-throughput sequencing to characterize the diversity of bacterial communities in ticks and also detected Rickettsia in high abundance in Dermacentor atrosignatus and Dermacentor compactus. Bacteria within the genus Rickettsia, including spotted fever group rickettsiae (SFGR), are mainly transmitted to animals and humans by tick bites; the genus Rickettsia has at least 30 species distributed worldwide, of which 21 are considered pathogens (Satjanadumrong et al. 2019). SFGR are known to cause a variety of natural focal diseases, including the Rocky Mountain spotted fever (RMSF) (Zazueta et al. 2021) in the Americas and the Mediterranean spotted fever in parts of Asia, Europe, and Africa (Satjanadumrong et al. 2019). In Europe, SFGR is chiefly transmitted by Dermacentor (Buczek et al. 2020), whereas in Asia, Dermacentor and Haemophysalis are most frequently associated with rickettsial carriage. SFGR is highly prevalent in northern China. Ten validated SFGR species have been discovered in ticks and vertebrate hosts (Wang et al. 2021). Nine samples (75%) evaluated in this study were shown to contain a high relative abundance of Rickettsia – they may be pathogens or endosymbionts, so we need further investigation to assess the possible risk posed by R. microplus.
In addition, seven samples were shown to contain a low abundance of Coxiella, ticks collected in Jiangkou County (samples JK-1, 2, and 3), Yinjiang County (YJ-1, 2, and 3), and Songtao County (ST-4). These findings differed from those of Guizzo et al. (2017), in which Coxiella accounted for 99% of the bacterial community in R. microplus eggs and 98.3% in R. microplus larvae. These differences may be attributed to the stage of development of ticks and the surveyed geographical location, but of course this requires confirmation. Bacteria within the genus Coxiella establish a symbiotic relationship with ticks and are able to infect ticks at all stages of their lifecycle (Ni et al. 2020). Coxiella burnetii is an obligate intracellular bacterium and the agent of Q fever in humans; Q fever is a zoonotic disease widely prevalent worldwide (except for New Zealand), characterized by high infectivity and long-term environmental persistence (Long et al. 2019; Klemmer et al. 2018). Ticks were not considered vectors of C. burnetii previously (Abdelkadir et al. 2019), but the results of a recent study support the link between ticks and Q fever, with the latter 3× more likely to occur where the former is found (Hussain et al. 2022). Ticks play an important role in the wild and peridomestic cycles of C. burnetii worldwide, having been isolated from at least 40 tick species within the Ixodidae and 14 tick species of the Argasidae (Bolaños-Rivero et al. 2017). Although animals infected with C. burnetii are often asymptomatic, sheep, goats, and cattle may experience abortion, premature birth, stillbirth, and weak offspring (Di Domenico et al. 2014), thus the bacterium can disseminate in the environment through birth products as well as the urine and milk of infected animals (Rodolakis et al. 2007). Moreover, as it is extremely resistant to desiccation and radiation, C. burnetti can persist in soil and other dry surfaces for long periods (Körner et al. 2021). Humans are very sensitive to C. burnetii, especially those who work with livestock and are exposed to birth products, infectious dust particles, contaminated wool, and highly infectious aerosols. Infections by C. burnetii are often occupation-related, being highly prevalent among slaughterhouse workers, livestock handlers, veterinarians, and farmers (Frangoulidis et al. 2021; Klemmer et al. 2018; Esmaeili et al. 2014).
In the present study, Proteobacteria were detected at the highest abundance in R. microplus, whereas Chlorobi and Firmicutes were at the highest abundance in H. longicornis, thus indicating that bacterial communities differ among tick species. Chlorobium was detected in ticks evaluated in the current study, which are bacteria that mainly live in water and are able to perform photosynthesis; the presence of this bacterium in H. longicornis may be a result of tick’s water ingestion habits. For instance, Amblyomma americanum and Ixodes scapularis have been shown to be able to actively ingest liquid water from the environment to compensate for water loss occurring due to excretion processes (Maldonado-Ruiz et al. 2020, 2021; Kim et al. 2017, 2019). Therefore, Chlorobium was likely ingested along with water by H. longicornis. Of course, the possibility that these bacteria originated from the surface of the exoskeleton and the host skin could not be ruled out, as it is quite difficult to completely remove surface contaminants from the tick exoskeleton.
Interestingly, the results of the present study differed from previous studies conducted in other provinces which employed high-throughput sequencing technology. Zhang et al. (2019c) evaluated the bacterial communities of H. longicornis, in which Coxiella was shown to be the dominant genus, and Rickettsia was not detected. Xiang et al. (2017) studied the bacterial communities of saliva obtained from engorged adult R. microplus females in Hunan province and found that Proteobacteria was the dominant phylum, and Acinetobacter, Rickettsia, Escherichia, and Coxiella were the major genera. Zhang et al. (2021b) also detected a high abundance of Rickettsia in H. longicornis in Shandong province. At present, it is not possible to provide a comprehensive explanation of the changes in bacterial richness observed in the current study with those described in previous studies. Multiple factors can determine changes in tick microbiome, including tick species, sample collection site, presence of host blood, and degree of engorgement (Clow et al. 2018; Estrada-Peña et al. 2018; Swei and Kwan 2017; Van Treuren et al. 2015; Moreno et al. 2006). In addition, several bacterial species detected in the study, including Acinetobacter and Staphylococcus, are commonly detected in ticks, indicating that these bacteria may play a biological role in these hosts. Moreover, it is worth noting that species from the genera Rickettsia and Coxiella are common tick endosymbionts (Dall’Agnol et al. 2021; Lim et al. 2020; Maldonado-Ruiz et al. 2020). Most importantly, the high carrying rate of Rickettsia in ticks evaluated in the study sites towards the need to strengthen future investigations to identify vector competence and potential epidemiological risk of tick-borne disease in the surveyed area.
Finally, high-throughput 16S rDNA sequencing has become the standard method for microbial classification and identification, which enables the identification of bacteria found in very low abundance and/or non-cultivable states. However, this method has certain limitations, such as accuracy and comprehensiveness being associated with the reference database (Couper and Swei 2018). Considering only the V3–V4 hypervariable regions of the 16S rDNA gene, sequence length was likely insufficient to enable the identification of bacteria at the species level. 16S rDNA full-length sequencing technology yields sequences of approximately 1542 bp in length, including nine hypervariable regions and 10 conserved regions, which would allow for further classification and identification of bacterial communities at the species level (Dong et al. 2021). Unfortunately, the limited number of samples did not allow further investigation of the identity of these bacteria to establish whether these bacteria are pathogens or not. However, the high-throughput sequencing method employed in the present study enabled the successful exploration of the bacterial communities carried by ticks parasitizing cattle in Tongren, Guizhou province, and highlighted variation in bacterial communities found in R. microplus vs. H. longicornis, the most prevalent tick species in the surveyed areas. Collectively, the results discussed herein provide a scientific basis for strengthening prevention and control measures of ticks and tick-borne diseases in Guizhou province.
Conclusions
The present study constitutes a survey of tick distribution affecting cattle in Tongren, Guizhou province, China, as well as of the diversity and composition of microbial communities found in the two tick species most prevalent locally. Regional differences were found in the distribution of tick species as well as in bacterial species carried by these parasites. Future studies should comprise an in-depth analysis of tick microbiota composition at the species level and explore the role of the identified microorganisms in ticks.
Data availability
The datasets generated during and analysed during the current study are available from the corresponding author on reasonable request.
References
Abdelkadir K, Palomar AM, Portillo A, Oteo JA, Ait-Oudhia K, Khelef D (2019) Presence of Rickettsia aeschlimannii, ‘Candidatus Rickettsia barbariae’ and Coxiella burnetii in ticks from livestock in Northwestern Algeria. Ticks Tick-borne Dis 10(4):924–928. https://doi.org/10.1016/j.ttbdis.2019.04.018
Bolaños-Rivero M, Carranza-Rodríguez C, Rodríguez NF, Gutiérrez C, Pérez-Arellano JL (2017) Detection of Coxiella burnetii DNA in Peridomestic and Wild Animals and Ticks in an endemic region (Canary Islands, Spain). Vector Borne Zoonotic Dis 17(9):630–634. https://doi.org/10.1089/vbz.2017.2120
Brites-Neto J, Duarte KMR, Martins TF (2015) Tick-borne infections in human and animal population worldwide. Vet World 8(3):301–315. https://doi.org/10.14202/vetworld.2015.301-315
Buczek W, Koman-Iżko A, Buczek A, Buczek A, Bartosik K, Kulina D, Ciura D (2020) Spotted fever group rickettsiae transmitted by Dermacentor ticks and determinants of their spread in Europe. Ann Agric Environ Med 27(4):505–511. https://doi.org/10.26444/aaem/120602
Chen Z, Yang XJ (2021) Systematics and taxonomy of Ixodida. Science Press, Beijing, pp 126–629. (In Chinese)
Clow KM, Weese JS, Rousseau J, Jardine CM (2018) Microbiota of field-collected Ixodes scapularis and Dermacentor variabilis from eastern and southern Ontario, Canada. Ticks Tick-borne Dis 9(2):235–244. https://doi.org/10.1016/j.ttbdis.2017.09.009
Couper L, Swei A (2018) Tick Microbiome characterization by Next-Generation 16S rRNA amplicon sequencing. J Vis Exp 138:58239. https://doi.org/10.3791/58239
Dall’Agnol B, McCulloch JA, Mayer FQ, Souza U, Webster A, Antunes P, Doyle RL, Reck J, Ferreira CAS (2021) Molecular characterization of bacterial communities of two neotropical tick species (Amblyomma aureolatum and Ornithodoros brasiliensis) using rDNA 16S sequencing. Ticks Tick-borne Dis 12(5):101746. https://doi.org/10.1016/j.ttbdis.2021.101746
de la Fuente J, Estrada-Pena A, Venzal JM, Kocan KM, Sonenshine DE (2008) Overview: ticks as vectors of pathogens that cause disease in humans and animals. Front Biosci 13:6938–6946. https://doi.org/10.2741/3200
Di Domenico M, Curini V, De Massis F, Di Provvido A, Scacchia M, Cammà C (2014) Coxiella burnetii in central Italy: novel genotypes are circulating in cattle and goats. Vector Borne Zoonotic Dis 14(10):710–715. https://doi.org/10.1089/vbz.2014.1587
Dong S, Jiao J, Jia S, Li G, Zhang W, Yang K, Wang Z, Liu C, Li D, Wang X (2021) 16S rDNA full-length assembly sequencing technology analysis of intestinal Microbiome in Polycystic Ovary Syndrome. Front Cell Infect Microbiol 11:634981. https://doi.org/10.3389/fcimb.2021.634981
Esmaeili S, Pourhossein B, Gouya MM, Amiri FB, Mostafavi E (2014) Seroepidemiological survey of Q fever and brucellosis in Kurdistan Province, western Iran. Vector Borne Zoonotic Dis 14(1):41–45. https://doi.org/10.1089/vbz.2013.1379
Estrada-Peña A (2015) Ticks as vectors: taxonomy, biology and ecology. Rev Sci Tech 34(1):53–65. https://doi.org/10.20506/rst.34.1.2345
Estrada-Peña A, Cabezas-Cruz A, Pollet T, Vayssier-Taussat M, Cosson JF (2018) High throughput sequencing and network analysis disentangle the Microbial Communities of Ticks and Hosts within and between Ecosystems. Front Cell Infect Microbiol 8:236. https://doi.org/10.3389/fcimb.2018.00236
Fang LQ, Liu K, Li XL, Liang S, Yang Y, Yao HW, Sun RX, Sun Y, Chen WJ, Zuo SQ, Ma MJ, Li H, Jiang JF, Liu W, Yang XF, Gray GC, Krause PJ, Cao WC (2015) Emerging tick-borne infections in mainland China: an increasing public health threat. Lancet Infect Dis 15(12):1467–1479. https://doi.org/10.1016/S1473-3099(15)00177-2
Frangoulidis D, Kahlhofer C, Said AS, Osman AY, ChitimiaDobler L, Shuaib YA (2021) High prevalence and new genotype of Coxiella burnetii in Ticks Infesting Camels in Somalia. Pathogens 10(6):741. https://doi.org/10.3390/pathogens10060741
Guizzo MG, Parizi LF, Nunes RD, Schama R, Albano RM, Tirloni L, Oldiges DP, Vieira RP, Oliveira WHC, de Souza Leite M, Gonzales SA, Farber M, Martins O, da Silva Vaz I, Oliveira PL (2017) A Coxiella mutualist symbiont is essential to the development of Rhipicephalus microplus. Sci Rep 7(1):17554. https://doi.org/10.1038/s41598-017-17309-x
Huang TP, Zhang JB, Sun CY, Liu ZC, He HY, Wu J, Geriletu (2020) A novel arthropod host of brucellosis in the Arid Steppe Ecosystem. Front Vet Sci 7:566253. https://doi.org/10.3389/fvets.2020.566253
Hussain S, Hussain A, Aziz MU, Song B, Zeb J, Hasib FMY, Li J, Rehman A, George D, Cabezas-Cruz A, Sparagano O (2022) First serological evidence of Q fever in large ruminants and its associated risk factors in Punjab. Pakistan Sci Rep 12(1):17278. https://doi.org/10.1038/s41598-022-21405-y
Kim D, Maldonado-Ruiz P, Zurek L, Park Y (2017) Water absorption through salivary gland type I acini in the blacklegged tick. Ixodes scapularis PeerJ 5:e3984. https://doi.org/10.7717/peerj.3984
Kim D, Šimo L, Vancová M, Urban J, Park Y (2019) Neural and endocrine regulation of osmoregulatory organs in tick: recent discoveries and implications. Gen Comp Endocrinol 278:42–49. https://doi.org/10.1016/j.ygcen.2018.08.004
Klemmer J, Njeru J, Emam A, El-Sayed A, Moawad AA, Henning K, Elbeskawy MA, Sauter-Louis C, Straubinger RK, Neubauer H, El-Diasty MM (2018) Q fever in Egypt: epidemiological survey of Coxiella burnetii specific antibodies in cattle, buffaloes, sheep, goats and camels. PLoS ONE 13(2):e0192188. https://doi.org/10.1371/journal.pone.0192188
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic acids res 41(1):e1. https://doi.org/10.1093/nar/gks808
Körner S, Makert GR, Ulbert S, Pfeffer M, Mertens-Scholz K (2021) The prevalence of Coxiella burnetii in Hard ticks in Europe and their role in Q fever transmission Revisited-A systematic review. Front Vet Sci 8:655715. https://doi.org/10.3389/fvets.2021.655715
Lim FS, Khoo JJ, Tan KK, Zainal N, Loong SK, Khor CS, AbuBakar S (2020) Bacterial communities in Haemaphysalis, Dermacentor and Amblyomma ticks collected from wild boar of an Orang Asli Community in Malaysia. Ticks Tick-borne Dis 11(2):101352. https://doi.org/10.1016/j.ttbdis.2019.101352
Long CM, Beare PA, Cockrell DC, Larson CL, Heinzen RA (2019) Comparative virulence of diverse Coxiella burnetii strains. Virulence 10(1):133–150. https://doi.org/10.1080/21505594.2019.1575715
Maldonado-Ruiz LP, Park Y, Ludek Z (2020) Liquid water intake of the lone star tick, Amblyomma americanum: implications for tick survival and management. Sci Rep 10(1):6000. https://doi.org/10.1038/s41598-020-63004-9
Maldonado-Ruiz LP, Neupane S, Park Y, Zurek L (2021) The bacterial community of the lone star tick (Amblyomma americanum). Parasit Vectors 14(1):49. https://doi.org/10.1186/s13071-020-04550-z
Malik MI, Nawaz M, Wang YA, Zhang HS, Cao J, Zhou YZ, Hassan IA, Islam MN, Anwar MN, Zhou JL (2019) Localized expression and inhibition effect of miR-184 on blood digestion and oviposition in Haemaphysalis longicornis (Acari: Ixodidae). Parasit Vectors 12(1):500. https://doi.org/10.1186/s13071-019-3754-7
Moreno CX, Moy F, Daniels TJ, Godfrey HP, Cabello FC (2006) Molecular analysis of microbial communities identified in different developmental stages of Ixodes scapularis ticks from Westchester and Dutchess Counties, New York. Environ Microbiol 8(5):761–772. https://doi.org/10.1111/j.1462-2920.2005.00955.x
Ni J, Lin HL, Xu XF, Ren QY, Aizezi M, Luo J, Luo Y, Ma Z, Chen Z, Tan YC, Guo JH, Liu WG, Qu ZQ, Wu ZG, Wang JM, Li YQ, Guan GQ, Luo JX, Yin H, Liu GY (2020) Coxiella burnetii is widespread in ticks (Ixodidae) in the Xinjiang areas of China. BMC Vet Res 16(1):317. https://doi.org/10.1186/s12917-020-02538-6
Rodolakis A, Berri M, Héchard C, Caudron C, Souriau A, Bodier CC, Blanchard B, Camuset P, Devillechaise P, Natorp JC, Vadet JP, Arricau-Bouvery N (2007) Comparison of Coxiella burnetii shedding in milk of dairy bovine, Caprine, and Ovine Herds. J Dairy Sci 90(12):5352–5360. https://doi.org/10.3168/jds.2006-815
Satjanadumrong J, Robinson MT, Hughes T, Blacksell SD (2019) Distribution and ecological drivers of spotted Fever Group Rickettsia in Asia. EcoHealth 16(4):611–626. https://doi.org/10.1007/s10393-019-01409-3
Silva NCS, Vale VF, Franco PF, Gontijo NF, Valenzuela JG, Pereira MH, Sant’Anna MRV, Rodrigues DS, Lima WS, Fux B, Araujo RN (2016) Saliva of Rhipicephalus (Boophilus) microplus (Acari: Ixodidae) inhibits classical and alternative complement pathways. Parasit Vectors 9(1):445. https://doi.org/10.1186/s13071-016-1726-8
Sonenshine DE (2018) Range Expansion of Tick Disease Vectors in North America: implications for spread of Tick-Borne Disease. Int J Environ Res Public Health 15(3):478. https://doi.org/10.3390/ijerph15030478
Swei A, Kwan JY (2017) Tick microbiome and pathogen acquisition altered by host blood meal. ISME J 11(3):813–816. https://doi.org/10.1038/ismej.2016.152
Teng GF, Jiang ZJ (1991) Economic Insect Fauna of China. Fasc 39, Acari: Ixodidae. Beijing, Science Press, pp 43–349. (In Chinese)
Van Treuren W, Ponnusamy L, Brinkerhoff RJ, Gonzalez A, Parobek CM, Juliano JJ, Andreadis TG, Falco RC, Ziegler LB, Hathaway N, Keeler C, Emch M, Bailey JA, Roe RM, Apperson CS, Knight R, Meshnick SR (2015) Variation in the microbiota of Ixodes ticks with regard to Geography, Species, and sex. Appl Environ Microbiol 81(18):6200–6209. https://doi.org/10.1128/AEM.01562-15
Wang Q, Guo WB, Pan YS, Jiang BG, Du CH, Que TC, Zhan L, Wu JH, Yu MH, Cui XM, Zhao L, Xu DL, Xia LY, Ye RZ, Li J, Li LF, Wei W, Zhou YH, Jiang JF, Jia N, Cao WC (2021) Detection of Novel spotted Fever Group Rickettsiae (Rickettsiales: Rickettsiaceae) in Ticks (Acari: Ixodidae) in Southwestern China. J Med Entomol 58(3):1363–1369. https://doi.org/10.1093/JME/TJAA294
Wu XB, Na RH, Wei SS, Zhu JS, Peng HJ (2013) Distribution of tick-borne diseases in China. Parasit Vectors 6:119. https://doi.org/10.1186/1756-3305-6-119
Xiang LL, Poźniak B, Cheng TY (2017) Bacteriological analysis of saliva from partially or fully engorged female adult Rhipicephalus microplus by next-generation sequencing. Antonie Van Leeuwenhoek 110(1):105–113. https://doi.org/10.1007/s10482-016-0780-8
Zazueta OE, Armstrong PA, Márquez-Elguea A, Milán NSH, Peterson AE, Ovalle-Marroquín DF, Fierro M, Arroyo-Machado R, Rodriguez-Lomeli M, Trejo-Dozal G, Paddock CD (2021) Rocky Mountain spotted fever in a large Metropolitan Center, Mexico-United States Border, 2009–2019. Emerg Infect Dis 27(6):1567–1576. https://doi.org/10.3201/eid2706.191662
Zhang YK, Zhang XY, Liu JZ (2019a) Ticks (Acari: Ixodoidea) in China: geographical distribution, host diversity, and specificity. Arch Insect Biochem Physiol 102(3):e21544. https://doi.org/10.1002/arch.21544
Zhang GS, Zheng D, Tian YQ, Li S (2019b) A dataset of distribution and diversity of ticks in China. Sci Data 6(1):1–7. https://doi.org/10.1038/s41597-019-0115-5
Zhang RL, Huang ZD, Yu GF, Zhang Z (2019c) Characterization of the bacterial community in Haemaphysalis longicornis (Acari: Ixodidae) throughout developmental stages. Exp Appl Acarol 77(2):173–186. https://doi.org/10.1007/s10493-019-00339-7
Zhang K, Li AJ, Wang Y, Zhang JJ, Chen YJ, Wang HJ, Shi RX, Qiu YH (2021a) Investigation of the presence of Ochrobactrum spp. and Brucella spp. in Haemaphysalis longicornis. Ticks Tick-borne Dis 12(1):101588. https://doi.org/10.1016/j.ttbdis.2020.101588
Zhang RL, Zhang Q, Yu GF, Zhang Z (2021b) Metagenomic deep sequencing obtains taxonomic and functional profiles of Haemaphysalis longicornis that vary in response to different developmental stages and sexes. Exp Appl Acarol 83(2):285–300. https://doi.org/10.1007/S10493-020-00582-3
Acknowledgements
The authors gratefully acknowledge Sangon Biotech (Shanghai) Co., Ltd for the technical support, Tongren Center for Disease Control and Prevention for its help in the sampling process, and farmers for their understanding and cooperation. The authors would like to thank TopEdit (www.topeditsci.com) for its linguistic assistance during the preparation of this manuscript.
Funding
This work was supported by Plan Project of the Science and Technology in Guizhou Province, Construction of Monitoring System and Early Warning Model of Important Vectors and Related Infectious Diseases in Guizhou Province (Qian Ke He (2022) General 178); Independent Research Project of the State Key Laboratory of Infectious Disease Prevention and Control, Investigation on Ticks and Tick-borne Pathogens in Tongren, Guizhou Province (2018SKLID305); Scientific Research Team of Guizhou Provincial Infectious Disease Prevention and Control Talent Base, Center for Surveillance and early warning of vectorial organisms and related infectious diseases (RCJD2107). The Project of National Natural Science Foundation in China, the investigation of tick-transmitted viruses in Guizhou ethnic group autonomous area (81760605).
Author information
Authors and Affiliations
Contributions
XYL, ZJZ and LWQ conceived and designed the experiment. XYL, and ZY performed laboratory work. YFX, LSJ and HY interpreted results. XYL and ZJZ compiled tables and figures. XYL and ZJZ wrote the first draft of the manuscript, and LWQ and LQY contributed to finalizing the paper. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing personal or financial interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xiang, Y., Zhou, J., Yu, F. et al. Characterization of bacterial communities in ticks parasitizing cattle in a touristic location in southwestern China. Exp Appl Acarol 90, 119–135 (2023). https://doi.org/10.1007/s10493-023-00799-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10493-023-00799-y