Abstract
Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-TOF MS) has recently emerged as an alternative to morphological and molecular tools to identify tick species. In this study, we set out to evaluate and confirm the ability of MALDI-TOF MS to identify different species of ticks collected in the Democratic Republic of the Congo and preserved in 70% ethanol. A total of 575 ticks, of which 530 were collected from domestic pigs and 45 from wild animals, were subjected to MALDI-TOF MS analysis to evaluate the intraspecies reproducibility and interspecies specificity of MS profiles obtained from the different species. Morphologically, the ticks belonged to seven different species, namely Rhipicephalus complanatus, Rhipicephalus congolensis, Haemaphysalis muhsamae, Ixodes cumulatimpunctatus, Amblyomma exornatum, Amblyomma compressum and an unidentified Rhipicephalus sp. A total of 535/575 (93%) of the spectra obtained were of good enough quality to be used for our analyses. Our home-made MALDI-TOF MS arthropod database was upgraded with spectra obtained from between one and five randomly selected specimens per species. For these reference specimens, molecular identification of the ticks was also made using 16S, 12S rDNA genes and the Cox1 mtDNA gene sequencing. The remaining good quality spectra were then queried against the upgraded MALDI-TOF MS database, showing that 100% were in agreement with the morphological identification, with logarithmic score values (LSVs) between 1.813 and 2.51. The consistency between our morphological, molecular and MALDI-TOF MS identification confirms the capability and precision of MALDI-TOF MS for tick identification.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ticks are obligate blood-feeding arthropods that parasitise a large number of vertebrates, including mammals, birds, reptiles and amphibians (Mediannikov and Fenollar 2014; Parola and Raoult 2001). They are distributed into three families: Argasidae (soft ticks), Ixodidae (hard ticks) and Nuttalliellidae, representing a total of at least 898 recognised species (Dantas-Torres et al. 2012). In Africa, seven genera and 16 species of ticks are of major veterinary importance (Walker 2003). As the world’s second most important vectors after mosquitoes (Ghosh and Nagar 2014), ticks are capable of transmitting a wide variety of pathogens to humans and animals (Merino et al. 2013), such as protozoa, bacteria and viruses (Aktas 2014), which cause most emerging zoonotic infectious diseases (Ghosh and Nagar 2014).
Despite the important role that ticks play in animal and human health, tick-borne pathogens infecting humans, domestic and wild animals are poorly known in the Democratic Republic of the Congo (DRC), in Central Africa. However, some agents of tick-borne diseases such as Crimean-Congo haemorrhagic fever virus (CCHFV), Anaplasma, Rickettsia, Theileria and Babesia have been reported in the DRC in humans (Grard et al. 2011; Simpson et al. 1967; Woodall et al. 1967), domestic animals (Dahmana et al. 2019; Kalume et al. 2009, 2013; Sas et al. 2017) and tick vectors (Rhipicephalus sanguineus s.l. and Amblyomma compressum) (Mediannikov et al. 2012; Sanogo et al. 2003).
Controlling ticks and tick-borne diseases through entomological surveys relies on the accurate identification of tick species and determination of their infectious status (Parola and Raoult 2001). Tick identification occurs mainly by observing morphological characteristics or by using molecular methods (Mediannikov and Fenollar 2014; Walker 2003). However, the expertise required for the morphological identification of ticks, the impossibility of distinguishing between species of the same complex, immature or damaged specimens, the outdated of identification key, as well as the cost and length of molecular biological assays (between seven and eight hours for a sample), are limiting factors for these methods (Diarra et al. 2017).
Recently, matrix-assisted laser desorption/ionization mass spectrometry (MALDI-TOF MS) has emerged as an alternative tool for the accurate and rapid identification of many arthropod species (Yssouf et al. 2016), including mosquitoes (Diarra et al. 2019; Lawrence et al. 2019; Tandina et al. 2018; Vega-Rua et al. 2018), ticks (Boucheikhchoukh et al. 2018; Boyer et al. 2019; Diarra et al. 2017; Rothen et al. 2016), fleas (Yssouf et al. 2014; Zurita et al. 2019), sand flies (Arfuso et al. 2019; Dvorak et al. 2014; Lafri et al. 2016), triatomine bugs (Laroche et al. 2018), and lice (Ouarti et al. 2020). The effectiveness and precision of MALDI-TOF MS in identifying ticks using their legs has been reported in several studies (Boucheikhchoukh et al. 2018; Boyer et al. 2019; Diarra et al. 2017; Kumsa et al. 2016; Rothen et al. 2016; Yssouf et al. 2013). Previous studies have demonstrated the ability of MALDI-TOF MS to accurately identify several tick species from western, eastern and northern Africa (Boucheikhchoukh et al. 2018; Diarra et al. 2017; Kumsa et al. 2016). The objective of this work was to confirm the ability of MALDI-TOF MS to identify seven species of ticks collected in the DRC, stored in alcohol, and absent in our home-made MALDI-TOF MS database.
Material and methods
Study areas
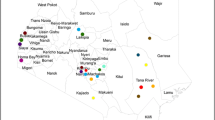
The study was carried out at the slaughterhouse of the International Aeronautic Transit (IAT) market (0° 30′ N 25° 10′ E) in the city of Kisangani (0° 30′ N; 25° 11′ E) in the province of Tshopo, DRC. Tshopo is one of the 26 provinces of the DRC located in the northeast of the country and its capital city is Kisangani. About 75.5% of the population of this province lives below the poverty line. The economy is based on artisanal mining, small business and food crop agriculture, which mainly produces cassava, plantain banana, sweet potato and rice. Perennial agriculture, although in decline, is also practiced, involving coffee, cocoa, rubber and oil palm (PUND 2009). The city of Kisangani is divided into six municipalities, namely Makiso, Tshopo, Mangobo, Kabondo, Kisangani (on the right bank of the Congo River) and Lubunga (on the left bank). The IAT market slaughterhouse is in the municipality of Makiso (Fig. 1). This market is supplied by products from different cities or provinces of the DRC (Kinshasa, Mbandaka, Bumba, Basoko, etc.) via the Congo River, which plays a key role in the city’s economy. However, the domestic animals sold and butchered at the IAT market slaughterhouse mostly originate from the city of Basoko, which is surrounded by lowland tropical rainforest, and located 200 km north of Kisangani on the right bank of the Congo River (Steve et al.; unpublished).
Tick collection and morphological identification
A total of 530 specimens of ticks were collected from 191 randomly selected domestic pigs at the IAT market slaughterhouse in Kisangani between September 2015 and September 2016. Forty-five ticks were collected from wild pangolins (Phataginus spp.) and monitor lizards (Varanus niloticus) in 2010 (Boyekoli Ebale Congo expedition collection). The ticks of wild animals protected by CITES and/or Congolese law were obtained, without financial compensation, from hunters and/or bushmeat sellers during the expedition and were covered by UNIKIS and LEGERA permits regarding the collection of biological samples. No IUCN Red List protected animals were actively trapped. Ectoparasites were removed with tweezers from fresh animal carcasses brought to one of the neighbouring bushmeat markets or encountered in the field. To collect ticks from pigs, the slaughterhouse was visited (48 visits) between 6 and 9 a.m., and visible ticks were carefully removed manually. Ticks collected from the same animal were stored in an Eppendorf vial (1.5 ml) in 70% ethanol with a host unique identifier code. In addition, data about the host such as weight, date, origin, sex, and ectoparasite type were noted in a field book and reported on digital data sheets. Approximately 20 tick specimens were sent to prof. Horak’s team at the University of Pretoria (South Africa) for morphological identification. In the DRC, the identifications made by prof. Horak’s team were extrapolated to the remaining specimens, depending on whether they were collected from the same host, in order to identify them at the species level. The ticks were transferred from IRSNB (Brussels, Belgium) to the University Hospital Institute Méditerranée infection of Marseille (Marseille, France) for analysis. In Marseille, the morphological identification of each specimen entered in the MALDI-TOF MS database was checked using a Leica EZ4 binocular microscope (Zeiss Axio Zoom.V16, Zeiss, Marly le Roi, France) and appropriate identification keys (Apanaskevich et al. 2013; Elbl and Anastos 1966a; Matthysse and Murray 1987; Walker et al. 2000). Ticks which could not be identified using morphology nor MALDI-TOF MS by the entomological team in Marseille, including two Rhipicephalus ticks, were sent to prof. Lorenza Beati, curator of the National Tick Collection, Institute for Coastal Plain Science, Georgia Southern University, USA, for identification.
Sample preparation for MALDI-TOF MS and spectral analysis
After rinsing each tick specimen with distilled water and drying with sterile filter paper, all four legs on the same side of the tick were cut out with a sterile surgical blade and the tick’s body was longitudinally cut in two halves. The four legs were used for MALDI-TOF MS analysis, the half part of tick without legs was selected for molecular analysis, and the second half with the remaining legs was stored at −20 °C. Eppendorf tubes containing tick legs collected from pigs between September 2015 and September 2016 were dried at 37 °C overnight in a dry bath. The legs were then crushed using a TissueLyser device (QIAGEN, Hilden, Germany), as described previously (Nebbak et al. 2017; Diarra et al. 2017). As for ticks collected in 2010 from wild animals, the legs were crushed using the TissueLyser (QIAGEN) over six cycles of 30 ms−1 for 60 s in 20 μl of a mixture of 70% formic acid and 50% acetonitrile (Fluka, Buchs, Switzerland). To control matrix quality, sample loading, and performance of the MALDI-TOF mass spectrometer, the matrix solution alone was used as negative control and a protein extract from laboratory-reared Amblyomma variegatum legs was used as positive control. The MALDI-TOF MS spectra obtained from the tick legs were evaluated by analysing the average spectra obtained from the four spectra of each sample tested using Flex analysis 3.3, MALDI-Biotyper v.3.0. and ClinPro-tools 2.2 software. In order to verify and select MS spectra of excellent quality (i.e., peak intensity > 3000 arbitrary units, without smoothing and right baseline subtraction), the average spectra (main spectrum profile, MSP) obtained from the four spectra of each sample were visualised using flexAnalysis v.3.4 software (Nebbak et al. 2017). Only MS spectra with excellent quality (i.e., peak intensity > 3000 arbitrary units, without smoothing and right baseline subtraction) were selected for further analysis. The assessment of the reproducibility and specificity of the MS spectra and the ability of MALDI-TOF MS to distinguish between male and female tick specimens was performed by principal component analysis (PCA) and dendrogram analysis (cluster analysis). The PCA and dendrograms were performed using ClinPro-tools 2.2 and MALDI-Biotyper v.3.0. software, respectively, as previously described (Boyer et al. 2019).
Database upgrading and blind tests
The spectra obtained from the legs of one to five specimens per tick species identified morphologically and molecularly were added to our laboratory’s reference spectra database, after controlling their quality, reproducibility and specificity. To create a database, MSPs reference spectra were created by combining the results of different spectra from same specimen of each species using the automated function of MALDI-Biotyper v.3.0 software (Bruker Daltonics). MSPs were created based on an unbiased algorithm using peak position, intensity and frequency data. Our entire database is currently not open access, although we are working to make it accessible to all researchers. However, it is available as part of collaborations with other researchers. In this context, the spectra files are available on request and transferable to any Bruker MALDI-TOF MS device. The reference spectra created in this study are provided in Supplementary Data 1. All good quality spectra were then blind tested against the reference spectra database. The identification level was determined using the logarithmic score values (LSVs) assigned by the MALDI-Biotyper v.3.0 software (Bruker Daltonics) (Yssouf et al. 2013). This value, ranging from 0 to 3, assesses the similarity between a tested spectrum and the reference spectra by comparing the position of the peaks and their intensity. The results of tick identification were considered reliable and relevant when the LSV was greater than or equal to 1.8 (Nebbak et al. 2017. A minimum difference of 0.2 between the best species match score and the second species match score was considered as an additional criterion for the validation of our identification (Kumsa et al. 2016).
Molecular identification of ticks
The half-ticks were incubated individually at 56 °C overnight in 180 μl of G2 lysis buffer (Qiagen, Hilden, Germany) and 20 μl of proteinase K (Qiagen), followed by extraction using the EZ1 DNA tissue kit (Qiagen) according to the manufacturer’s recommendations. The 15 tick specimens whose spectra were entered in our database as well as 43 ticks with LSVs > 1.8 and five specimens with LSVs < 1.8 randomly selected were subjected to standard PCR, followed by sequencing using the 16s, 12s rDNA genes and the mDNA Cox1 gene (Boyer et al. 2019; Diarra et al. 2017). The forward and reverse reads of the targeted mtDNA sequences were assembled and analysed using ChromasPro v.1.34 software (Technelysium, Tewantin, Australia) and submitted for analysis to the NCBI BLAST website (http://blast.ncbi.nlm.nih.gov).
The different mtDNA gene sequences were aligned with BioEdit, and phylogenetic trees were inferred using TOPALi v.2.5 software (Biomathematics and Statistics Scotland, Edinburgh, UK) (Laroche et al. 2017b). The model Maximum Likelihood (ML) phylogenetic tree proposed default in TOPALi v.2.5 software was used for the construction of phylogenetic trees. Node numbers are percentages of the bootstrap values obtained by repeating 100 iterations of the analysis to generate a majority consensus tree (only those with a value equal to or greater than 80 were retained).
Results
Tick collection and morphological identification
A total of 575 ticks were collected, including 530 (92.2%) which were collected from 191 domestic pigs and 45 (7.8%) which were collected from five wild animals. Of the 530 ticks collected from domestic pigs, 452 (85.3%) were males and 78 (14.7%) were females. Of the ticks collected from wild animals, 28 (62.2%) were males, 10 (22.2%) were nymphs and seven (15.6%) were females. The morphological identification performed by the DRC team initially identified four tick species (with the limits of the method described above); namely Rhipicephalus complanatus (456; 79.3%), Rhipicephalus congolensis (61; 10.6%), Haemaphysalis muhsamae (one; 0.2%), and Ixodes cumulatimpunctatus (12; 2.1%). The two species collected from wild animals included Amblyomma exornatum (35; 6.1%) and A. compressum (10; 1.7%) (Table 1). Five specimens found on pigs were also re-examined following MALDI-TOF MS and molecular identification, and were found to belong to the Rhipicephalus genus, although it was not possible to identify them to the species level.
MALDI-TOF MS identification
All 575 ticks were subjected to MALDI-TOF MS analysis. Of these, 535/575 (93%) had good-quality spectra (Fig. 2A), including 517/530 (97.5%) ticks collected between September 2015 and September 2016, and 18/45 (40%) of those collected in 2010. Good quality spectra obtained from the legs of 15 ticks (5 Rh. complanatus, 4 Rh. congolensis, 3 I. cumulatimpunctatus, 2 A. exornatum and 1 A. compressum) were added to our reference database MALDI-TOF MS (Table 1). A comparative analysis of MS spectra for intra-species reproducibility and inter-species specificity was performed. The visualisation of two MS spectra per tick species showed perfect intra-species reproducibility and inter-species specificity (Fig. 2A). Similarly, the dendrogram made with the MS spectra of between one and five specimens per morphologically-identified tick species showed that specimens of the same species grouped on the same branch, confirming intra-species reproducibility and inter-species specificity (Fig. 2B). A comparison of MS spectra from male and female specimens was performed only for Rh. complanatus and Rh. congolensis species for which both sexes were available. The PCA and cluster analysis shows that there was no difference between the MS spectra of males and females (Fig. 3A, B and Supplementary Fig. 1). The remaining 520 good quality spectra of ticks were queried against our upgraded database. This blind test showed that 434/439 (98.9%) Rh. complanatus, 56/56 Rh. congolensis, 9/9 I. cumulatimpunctatus, 5/5 A. compressum and 12/12 of exornatum were correctly identified by MALDI-TOF MS, with LSVs between 1.813 and 2.51 (Table 1). Thus, 99% (515/520) of the ticks which were blind tested presented a concordance with the morphological identification and were considered correctly identified.
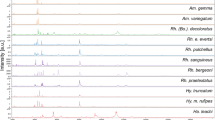
Comparison of MALDI-TOF MS spectra from different species of ticks collected from domestic animals between September 2015 and September 2016 and from wild animals in 2010 in the Democratic Republic of the Congo (DRC) using Flex analysis v.3.3 and MALDI-Biotyper v.3.0 software. A Representative MS spectra of Amblyomma compressum (A, B), A. exornatum (C, D), Haemaphysalis mushamae (E, F), Ixodes cumulatimpunctatus (G, H), Rhipicephalus congolensis (I, J), Rhipicephalus complanatus (K, L) and Rhipicephalus sp. (M, N). Abbreviations: m/z, mass-to-charge ratio; B MALDI-TOF MS MSP dendrogram of the seven tick species. Between one and five specimens per species were used to construct the dendrogram. The dendrogram was created using Biotyper v.3.0 software and the distance units correspond to the relative similarity of MS spectra
Comparison of male and female MALDI-TOF MS spectra from two different tick species collected from domestic animals between September 2015 and September 2016 in the Democratic Republic of the Congo (DRC) using the principal component analysis tool of ClinProTools v.2.2 software. A PCA of males (green) and females (red) of Rhipicephalus complanatus; B PCA of male (green) and females (red) of Rhipicephalus congolensis
The morphology of the five ticks which were initially morphologically identified as Rh. complanatus that were not correctly identified by MALDI-TOF MS (LSV < 1.8) MALDI-TOF MS were rechecked, using the remaining part of the body. This revealed that one tick was in fact Hae. muhsamae and that four identical specimens belonged to another Rhipicephalus sp., which could not be definitively identified to the species level. The spectra of these five specimens did not match any of the ticks in our MALDI-TOF MS database and had LSVs ranging from 1.00 to 1.327. The spectrum of Hae. muhsamae was added to the MALDI-TOF MS database. The spectrum of one Rhipicephalus sp. was also temporarily added to the database as a potential reference spectrum for the remaining Rhipicephalus sp. The blind test showed that the three other unidentified Rhipicephalus sp. indeed matched with the spectrum of the Rhipicehalus sp. introduced in the database, with LSVs ranging from 2.05 to 2.22. The mean differences between the first species match scores and the second species match scores were 0.66 ± 0.11, 0.62 ± 0.10, 0.63 ± 0.18, 0.86 ± 0.15 and 0.85 ± 0.10 for Rh. complanatus, congolensis, I. cumulatimpunctatus, A. exornatum and A. compressum, respectively (Table 1). We had 100% agreement between our morphological identification and MALDI-TOF MS for the specimens tested.
Molecular identification of ticks
Fifty-seven ticks, including 25 Rh. complanatus, 14 Rh. congolensis, 5 I. cumulatimpunctatus, 4 A. exornatum, 3 A. compressum, 2 Hae. muhsamae and 4 Rhipicephalus sp., were subjected to standard PCR and sequenced using the 16s, 12s rDNA gene and the Cox1 mtDNA gene. We obtained sequences for all the specimens using the 16s rDNA gene, while only 15 were obtained with the 12s rDNA gene (3 Rh. complanatus, 3 Rh. congolensis, 2 I. cumulatimpunctatus, 2 A. exornatum, 2 A. compressum, 1 H. muhsamae and 1 Rhipicephalus sp.) (Table 2). We obtained 22 sequences when targeting the Cox1 mtDNA gene, including 9 Rh. complanatus, 2 Rh. congolensis, 4 A. exornatum, 2 A. compressum, 1 Hae. muhsamae and 4 Rhipicephalus sp. Blast results showed that the percentages of sequence identities ranged from 85.3 to 96.6%, 85.6 to 96.8% and 86.7 to 90.6%, respectively, for genes 16s, 12s rDNA and Cox1 mtDNA, with the known species available on GenBank. This DNA-based approach did not allow us to identify the Rhipicephalus sp. ticks that had not been morphologically identified down to the species level. The percentage of sequence identity of these ticks was 92.2, 93.5 and 87.4% with the sequences of Rh. sanguineus (KT382455), Rh. pusillus (AF150022) and Rh. sanguineus (KU364312), respectively, for genes 16s, 12s rDNA and Cox1 mtDNA (Table 2). These BLAST results were confirmed by building phylogenetic trees, and these trees were consistent with the BLAST results (Figs. 4, 5 and 6). The sequences of the three genes of each tick species, excluding I. cumulatimpunctatus, were deposited in GenBank and the accession numbers are available in Supplementary Table 1.
Phylogenetic analysis based on the 410 base pair (bp) fragment of the 16s ribosomal RNA gene of ticks collected from domestic animals between September 2015 and September 2016 and from wild animals in 2010 in the Democratic Republic of the Congo (DRC). The tree was generated using the Maximum Likelihood algorithm (PhyML) with the general time-reversible (GTR) model proposed by TOPALi v.2.5 software. The specimens in our study are shown in bold and are preceded by the GenBank accession numbers of each sequence
Phylogenetic analysis based on the 340 bp fragment of the 12s ribosomal RNA gene of ticks collected from domestic animals between September 2015 and September 2016 and from wild animals in 2010 in the Democratic Republic of the Congo (DRC). The tree was generated using the Maximum Likelihood algorithm (PhyML) with the general time-reversible (GTR) model proposed by TOPALi v.2.5 software. The specimens in our study are shown in bold, and are preceded by the GenBank accession numbers of each sequence
Phylogenetic analysis based on the 720 bp fragment of the Cox1 mtDNA gene of ticks collected from domestic animals between September 2015 and September 2016 and from wild animals in 2010 in the Democratic Republic of the Congo (DRC). The tree was generated using the Maximum Likelihood algorithm (PhyML) with the general time-reversible (GTR) model proposed by TOPALi v.2.5 software. The specimens in our study are shown in bold and are preceded by the GenBank accession numbers of each sequence
Discussion
Recently, MALDI-TOF MS has been proposed as an accurate, rapid and economical tool for the identification of many arthropods of medical interest, including ticks (Yssouf et al. 2016). Although the acquisition of a MALDI-TOF mass spectrometer can be expensive, its use for entomological analyses involves low additional costs. The reagents used for this high-throughput technique are indeed inexpensive, and the sample preparation and the data analyses are simple and fast compared to morphological and molecular methods (Laroche et al. 2017a).
MALDI-TOF MS is a method which can generate a specific protein signature of the analysed organism. It is an approach that allows for the rapid analysis of large sets of samples of a priori unidentified arthropods and provides a specific spectrum for each arthropod species that can be compared against a library of reference spectra of reliably identified specimens (Boucheikhchoukh et al. 2018). MALDI-TOF MS has been used to identify ticks that had been frozen or preserved in alcohol, and collected from various countries (Boucheikhchoukh et al. 2018; Boyer et al. 2019; Diarra et al. 2017; Karger et al. 2012; Kumsa et al. 2016; Yssouf et al. 2013). Currently, the challenges of this technique remain selecting the body part to be used, the methods of arthropod conservation in order to generate sufficiently reproducible and specific spectra, and the need to have a database of arthropod spectra formally identified by morphology and confirmed by molecular biology, if possible (Laroche et al. 2017a).
In this study, we morphologically identified six different species, four of which were collected from domestic pigs (Rh. complanatus, Rh. congolensis, Hae. muhsamae and I. cumulatimpunctatus) and two from wild pangolins and monitor lizards (A. exornatum and A. compressum). Rhipicephalus complanatus is a tick that almost exclusively parasitises domestic and feral pigs. This tick is mainly found in the DRC and neighbouring countries to the west such as Cameroon, Gabon, the Central African Republic and Congo. It has also been reported in Côte d'Ivoire, Liberia, and western Uganda (Walker et al. 2000). Rhipicephalus congolensis is a recently described tick that parasitises red pigs and domestic pigs in the DRC (Apanaskevich et al. 2013). Haemaphysalis muhsamae is a parasite of birds, carnivores, insectivores, rodents and lagomorphs (Elbl and Anastos 1966b; Uilenberg et al. 2013). This tick has been reported in the Guinean and eastern savannah zones in areas with at least 1 000 mm of annual rainfall and at different altitudes (Elbl and Anastos 1966b; Uilenberg et al. 2013). Haemaphysalis muhsamae has not currently been reported on domestic animals, and we are reporting it for the first time on domestic pigs from the DRC. Ixodes cumulatimpunctatus is a non-specific polytropic tick which, in its adult stages live on mammals and in its immature stages, lives on mammals and birds. This tick is widespread throughout the west African lowland rainforest and high altitude wooded savannas from Uganda to Zimbabwe (Cumming 1999; Morel and Mouchet 1965). Amblyomma exornatum ticks are mainly found on reptiles, mostly monitor lizards, lizards and pythons. They can also be found on tortoises, crocodiles, chameleons, and also on mammals (bats, pangolins, rodents) and ungulates (cattle and carnivores) (Nowak-Chmura 2014). Amblyomma exornatum is widespread in Africa and has been reported in Algeria, Senegal, the Côte d’Ivoire, Ghana, Cameroon, Gabon, Congo, Somalia, Kenya, Angola, Tanzania, Botswana, Mozambique and the Republic of South Africa (Nowak-Chmura 2014). Amblyomma compressum is a tick species which has thus far exclusively been reported for pangolin species, including the ground pangolin (Smutsia temminckii), tree pangolin (Manis tricuspis) and giant pangolin (Manis gigantea) (Uilenberg et al. 2013). This tick has been found in Central Africa (DRC, Rwanda, Gabon and Burundi) (Elbl and Anastos 1966a; Uilenberg et al. 2013; Pourrut et al. 2011) and also in West Africa (Ghana, Cote d’Ivoire, Liberia and Nigeria) (Elbl and Anastos 1966a; Mediannikov et al. 2012; Ntiamoa-Baidu et al. 2004; Uilenberg et al. 2013).
In our study, 93% of the ticks submitted to MALDI-TOF MS analysis yielded good quality spectra. The percentage of good quality spectra depends on the storage time of the ticks in alcohol. This percentage higher (97%) for ticks from 2015 and 2016 and lower (40%) for those stored since 2010. The performance of tick identification is not particularly impacted by the alcohol 70% because with an adapted database, we obtained identification percentages very similar to those of fresh ticks, often around 100% (Boucheikhchoukh et al. 2018; Diarra et al. 2017; Kumsa et al. 2016). The impact of alcohol on MS MALDI-TOF profiles, resulting in lower intensity and overall quality than fresh or frozen samples, has been reported in previous studies (Diarra et al. 2017; Kumsa et al. 2016; Nebbak et al. 2017; Rothen et al. 2016). Despite the impact of the preservation method on the quality of the MALDI-TOF MS profiles, MALDI-TOF MS appears to be efficient, fast and less expensive for the identification of ticks if a specific database is created with good quality spectra of specimens preserved under the same conditions, as demonstrated by other studies (Diarra et al. 2017; Kumsa et al. 2016).
In this study, we observed no differences in MS spectra from male and female Rh. complanatus and Rh. congolensis. A previous study reported that it was not possible to definitively distinguish between male and female ticks on the basis of their MS spectra (Yssouf et al. 2013). Nevertheless, differentiation of male and female mosquito MS profiles has been achieved (Fall et al. 2021). More in-depth analysis of fresh tick spectra should be performed in order to assess whether MALDI-TOF MS is actually able to distinguish male and female specimens.
Based on morphological identification, all six of these tick species were not represented in our home-made reference spectra MALDI-TOF MS database. With the exception of Hae. muhsamae, for which we did not have good quality spectra, the reference spectra of five species, namely Rh. complanatus, Rh. congolensis and I. cumulatimpunctatus, collected in 2015 and 2016, and A. exornatum and A. compressum collected in 2015 were added to our MS MALDI-TOF home-made database. In the first step, 515/520 ticks were correctly identified at species level with LSVs > 1.8, which is the threshold for reliable identification of arthropods (Yssouf et al. 2013). The mean differences between the first and second species match scores in our study were > 0.2, which is the minimum difference to distinguish two species (Kumsa et al. 2016). The five unidentified specimens were due to morphological identification errors highlighted by the MALDI-TOF MS tool. MALDI-TOF MS incited us to re-examine the morphology of these ticks which were morphologically identified as Rh. complanatus. We were the able to identify one of the five specimens as Hae. muhsamae. None of the three experts in tick identification involved in this study was able to morphologically identify the remaining four specimens of Rhipicephalus sp. to the species level. However, it should be noted that these ticks were engorged females, whose morphological characteristics were distorted or had disappeared, making identification impossible, highlighting the limits of morphological identification (Walker et al. 2003). After temporarily adding the spectrum of one of these specimens in the database, MALDI-TOF MS confirmed that all four belonged to the same species.
Knowing that the species which were morphologically identified in our study were not yet represented in the GenBank database, we sequenced the specimens chosen for the creation of the MALDI-TOF MS database, as well as others with confirmed MALDI-TOF MS identification, by amplifying part of the 16s and 12s rDNA genes and the Cox1 mtDNA gene, as previously used to discriminate ticks (Boyer et al. 2019; Diarra et al. 2017). This allowed us, on the one hand, to evaluate the percentage of similarity of sequences from the same tick species for the three genes, and, on the other, to compare the identity of all these sequences with those available in the GenBank database to reinforce the results. The high percentage of similarity between the sequences of different specimens of the same species (97% to 100%) using the three genes supports the robustness of our morphological and MALDI-TOF MS identification. However, the low identity of our sequences (between 85 and 95%) with the reference sequences of the 16s, 12s rDNA and Cox1 mtDNA genes of the tick species available in the GenBank database could be due to the absence of sequences of the tick species from our study in this database. The phylogenetic trees constructed with the sequences of the three genes clearly show that Rhipicephalus sp. form a separate group from Rh. complanatus (Figs. 4, 5 and 6), and thus have different genomic sequences. Similarly, the three genes used did not allow us to identify the four specimens of Rhipicephalus sp., which we could not identify down to the species level by either morphology or MALDI-TOF MS, thus showing the limitations of this technique, namely the insufficient number of reference sequences and the reliability of the morphological identification of tick species whose sequences already available on GenBank (Yssouf et al. 2016).
MALDI-TOF MS has been used in clinical laboratories in developed countries and capacity building was performed in Senegal, where staff and students was trained to use this technique for bacterial and entomological identifications. It can indeed be used in medical entomology, with almost zero additional costs, for the rapid and accurate identification of arthropods and determination of their trophic preferences and infection status (Yssouf et al. 2016). It is a tool that also makes it possible to correct morphological identification errors, as was the case in this study, where the four specimens of Rhipicephalus sp. were not recognised after the blind test, while a fine analysis of the spectral profiles of these ticks showed that they were reproducible and grouped together.
Conclusion
The results of this study show consistency between our MALDI-TOF MS identification and the morphological and molecular identifications, confirming once again that MALDI-TOF MS is an reliable tick identification tool. Four specimens of Rhipicephalus sp. could not be identified by MALDI-TOF MS but neither could they be identified by morphology or molecular biology, thus demonstrating the importance and interdependance of all these approaches. However, we showed that, for ticks that have been kept in alcohol for a very long time, the MALDI-TOF MS sample preparation had to be adjusted, i.e., a reduction in the quantity of the mix and increased crushing time resulted in an improvement of the quality of the spectra.
References
Aktas M (2014) A survey of ixodid tick species and molecular identification of tick-borne pathogens. Vet Parasitol 200:276–283
Apanaskevich DA, Horak IG, Mulumba-Mfumu LK (2013) A new species of Rhipicephalus (Acari: Ixodidae), a parasite of red river hogs and domestic pigs in the Democratic Republic of Congo. J Med Entomol 50:479–484
Arfuso F, Gaglio G, Abbate JM, Caracappa G, Lupia A, Napoli E, Giarratana F, Latrofa MS, Giannetto S, Otranto D, Brianti E (2019) Identification of phlebotomine sand flies through MALDI-TOF mass spectrometry and in-house reference database. Acta Trop 194:47–52
Boucheikhchoukh M, Laroche M, Aouadi A, Dib L, Benakhla A, Raoult D, Parola P (2018) MALDI-TOF MS identification of ticks of domestic and wild animals in Algeria and molecular detection of associated microorganisms. Comp Immunol Microbiol Infect Dis 57:39–49
Boyer PH, Almeras L, Plantard O, Grillon A, Talagrand-Reboul E, McCoy K, Jaulhac B, Boulanger N (2019) Identification of closely related Ixodes species by protein profiling with MALDI-TOF mass spectrometry. PLoS ONE 14:e0223735
Cumming GS (1999) Host distributions do not limit the species ranges of most African ticks (Acari: Ixodida). Bull Entomol Res 89:303–327
Dahmana H, Amanzougaghene N, Davoust B, Normand T, Carette O, Demoncheaux JP, Mulot B, Fabrizy B, Scandola P, Chik M, Fenollar F, Mediannikov O (2019) Great diversity of Piroplasmida in Equidae in Africa and Europe, including potential new species. Vet Parasitol Reg Stud Rep 18:100332
Dantas-Torres F, Chomel BB, Otranto D (2012) Ticks and tick-borne diseases: a One Health perspective. Trends Parasitol 28:437–446
Diarra AZ, Almeras L, Laroche M, Berenger JM, Kone AK, Bocoum Z, Dabo A, Doumbo O, Raoult D, Parola P (2017) Molecular and MALDI-TOF identification of ticks and tick-associated bacteria in Mali. PLoS Negl Trop Dis 11:e0005762
Diarra AZ, Laroche M, Berger F, Parola P (2019) Use of MALDI-TOF MS for the Identification of Chad Mosquitoes and the Origin of Their Blood Meal. Am J Trop Med Hyg 100:47–53
Dvorak V, Halada P, Hlavackova K, Dokianakis E, Antoniou M, Volf P (2014) Identification of phlebotomine sand flies (Diptera: Psychodidae) by matrix-assisted laser desorption/ionization time of flight mass spectrometry. Parasit Vectors 7:21
ElbL A, Anastos G (1966a) Ixodid ticks (acarina, ixodidae) of central Africa. 1:145
ElbL A, Anastos G (1966b) IXODID ticks (acarina, ixodidae) of central Africa. IV:148
Fall FK, Laroche M, Bossin H, Musso D, Parola P (2021) Performance of MALDI-TOF mass spectrometry to determine the sex of mosquitoes and identify specific colonies from French polynesia. Am J Trop Med Hyg 22:tpmd20-0031
Ghosh S, Nagar G (2014) Problem of ticks and tick-borne diseases in India with special emphasis on progress in tick control research: a review. J Vector Borne Dis 51:259–270
Grard G, Drexler JF, Fair J, Muyembe JJ, Wolfe ND, Drosten C, Leroy EM (2011) Re-emergence of Crimean-Congo hemorrhagic fever virus in Central Africa. PLoS Negl Trop Dis 5:e1350
Kalume MK, Losson B, Vyambwera CG, Mbegumbaya L, Makumyaviri AM, Saegerman C (2009) Enquête épidémiologique auprès des vétérinaires concernant trois maladies vectorielles des bovins élevés.dans la Province du Nord-Kivu, République Démocratique du Congo. Epidémiol Santé Anim 8:197–216
Kalume MK, Saegerman C, Mbahikyavolo DK, Makumyaviri AM, Marcotty T, Madder M, Caron Y, Lempereur L, Losson B (2013) Identification of hard ticks (Acari: Ixodidae) and seroprevalence to Theileria parva in cattle raised in North Kivu Province, Democratic Republic of Congo. Parasitol Res 112:789–797
Karger A, Kampen H, Bettin B, Dautel H, Ziller M, Hoffmann B, Suss J, Klaus C (2012s) pecies determination and characterization of developmental stages of ticks by whole-animal matrix-assisted laser desorption/ionization mass spectrometry. Ticks Tick Borne Dis 3:78–89
Kumsa B, Laroche M, Almeras L, Mediannikov O, Raoult D, Parola P (2016) Morphological, molecular and MALDI-TOF mass spectrometry identification of ixodid tick species collected in Oromia. Ethiopia Parasitol Res 115:4199–4210
Lafri I, Almeras L, Bitam I, Caputo A, Yssouf A, Forestier CL, Izri A, Raoult D, Parola P (2016) Identification of Algerian field-caught phlebotomine sand fly vectors by MALDI-TOF MS. PLoS Negl Trop Dis 10:e0004351
Laroche M, Berenger JM, Delaunay P, Charrel R, Pradines B, Berger F, Ranque S, Bitam I, Davoust B, Raoult D, Parola P (2017) Medical entomology: a reemerging field of research to better understand vector-borne infectious diseases. Clin Infect Dis 65:S30–S38
Laroche M, Berenger JM, Mediannikov O, Raoult D, Parola P (2017) Detection of a Potential New Bartonella Species “Candidatus Bartonella rondoniensis” in Human Biting Kissing Bugs (Reduviidae; Triatominae). PLoS Negl Trop Dis 11:e0005297
Laroche M, Berenger JM, Gazelle G, Blanchet D, Raoult D, Parola P (2018) MALDI-TOF MS protein profiling for the rapid identification of Chagas disease triatomine vectors and application to the triatomine fauna of French Guiana. Parasitology 145:665–675
Lawrence AL, Batovska J, Webb CE, Lynch SE, Blacket MJ, Slapeta J, Parola P, Laroche M (2019) Accurate identification of Australian mosquitoes using protein profiling. Parasitology 146:462–471
Matthysse JG, Murray HC (1987) The ixodid ticks of uganda. Entomological Society of America
Mediannikov O, Fenollar F (2014) Looking in ticks for human bacterial pathogens. Microb Pathog 77:142–148
Mediannikov O, Davoust B, Socolovschi C, Tshilolo L, Raoult D, Parola P (2012s) potted fever group rickettsiae in ticks and fleas from the Democratic Republic of the Congo. Ticks Tick Borne Dis 3:371–373
Merino O, Alberdi P, Perez de la Lastra JM, de la Fuente J (2013) Tick vaccines and the control of tick-borne pathogens. Front Cell Infect Microbiol 3:30
Morel PC, Mouchet J (1965) Les Tiques du Cameroun (Ixodidae et Argasidae). Annales De Parasitologie 15:477–496
Nebbak A, El HB, Berenger JM, Bitam I, Raoult D, Almeras L, Parola P (2017) Comparative analysis of storage conditions and homogenization methods for tick and flea species for identification by MALDI-TOF MS. Med Vet Entomol 31:438–448
Nebbak A, Koumare S, Willcox AC, Berenger JM, Raoult D, Almeras L, Parola P (2018) Field application of MALDI-TOF MS on mosquito larvae identification. Parasitology 145:677–687
Nowak-Chmura M (2014) A biological/medical review of alien tick species (Acari: Ixodida) accidentally transferred to Poland. Ann Parasitol 60:49–59
Ntiamoa-Baidu Y, Carr-Saunders C, Matthews BE, Preston PM, Walker AR (2004) An updated list of the ticks of Ghana and an assessment of the distribution of the ticks of Ghanaian wild mammals in different vegetation zones. Bull Entomol Res 94:245–260
Ouarti B, Laroche M, Righi S, Meguini MN, Benakhla A, Raoult D, Parola P (2020) Development of MALDI-TOF mass spectrometry for the identification of lice isolated from farm animals. Parasite 27:28
Parola P, Raoult D (2001) Ticks and tickborne bacterial diseases in humans: an emerging infectious threat. Clin Infect Dis 32:897–928
Pourrut X, Emane KA, Camicas JL, Leroy E, Gonzalez JP (2011) Contribution to the knowledge of ticks (Acarina:Ixodidae) in Gabon. Acarologia 51:465–471
Programme-des-nations-unies-pour-le-developpement (PNUD) (2009) Province orientale profil résumé pauvreté et conditions de vie des ménages. http://www.undp.org/content/dam/dem_rep_congo/docs/povred/UNDP-CD-Profil-PROVINCE-Orientale.pdf
Rothen J, Githaka N, Kanduma EG, Olds C, Pfluger V, Mwaura S, Bishop RP, Daubenberger C (2016) Matrix-assisted laser desorption/ionization time of flight mass spectrometry for comprehensive indexing of East African ixodid tick species. Parasit Vectors 9:151
Sanogo YO, Davoust B, Inokuma H, Camicas JL, Parola P, Brouqui P (2003) First evidence of Anaplasma platys in Rhipicephalus sanguineus (Acari: Ixodida) collected from dogs in Africa. Onderstepoort J Vet Res 70:205–212
Sas MA, Mertens M, Kadiat JG, Schuster I, Pongombo CPS, Maloba AGK, Groschup MH (2017) Serosurvey for Crimean-Congo hemorrhagic fever virus infections in ruminants in Katanga province, Democratic Republic of the Congo. Ticks Tick Borne Dis 8:858–861
Simpson DI, Knight EM, Courtois G, Williams MC, Weinbren MP, Kibukamusoke JW (1967) Congo virus: a hitherto undescribed virus occurring in Africa. I. Human isolations–clinical notes. East Afr Med J 44:86–92
Tandina F, Niare S, Laroche M, Kone AK, Diarra AZ, Ongoiba A, Berenger JM, Doumbo OK, Raoult D, Parola P (2018) Using MALDI-TOF MS to identify mosquitoes collected in Mali and their blood meals. Parasitology 145:1170–1182
Uilenberg G, Estrada-Pena A, Thal J (2013) Ticks of the Central African Republic. Exp Appl Acarol 60:1–40
Vega-Rua A, Pages N, Fontaine A, Nuccio C, Hery L, Goindin D, Gustave J, Almeras L (2018) Improvement of mosquito identification by MALDI-TOF MS biotyping using protein signatures from two body parts. Parasit Vectors 11:574
Walker JB, Keirans JE, Horak IG (2000) The Genus Rhipicephalus (Acari, Ixodidae): A Guide to the Brown ticks of the World. Press syndicate of university of cambridge. Cambridge University Press, Cambridge
Walker AR, Bouattour A, Camicas J, Estrada-Pena A, Horak IG, Latif AA, Pegram RG, Preston PM (2003) Ticks of domestic animals in Africa: a guide to identification of species. Springer, New York, pp 1–228
Woodall JP, Williams MC, Simpson DI (1967) Congo virus: a hitherto undescribed virus occurring in Africa. II Identification Studies. East Afr Med J 44:93–98
Yssouf A, Flaudrops C, Drali R, Kernif T, Socolovschi C, Berenger JM, Raoult D, Parola P (2013) Matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid identification of tick vectors. J Clin Microbiol 51:522–528
Yssouf A, Socolovschi C, Leulmi H, Kernif T, Bitam I, Audoly G, Almeras L, Raoult D, Parola P (2014) Identification of flea species using MALDI-TOF/MS. Comput Immunol Microbiol Infect Dis 37:153–157
Yssouf A, Almeras L, Raoult D, Parola P (2016) Emerging tools for identification of arthropod vectors. Future Microbiol 11:549–566
Zurita A, Djeghar R, Callejon R, Cutillas C, Parola P, Laroche M (2019) Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry as a useful tool for the rapid identification of wild flea vectors preserved in alcohol. Med Vet Entomol 33:185–194
Acknowledgements
The authors are grateful to the manager of the Kalongo Petrus IAT market slaughterhouse, who kindly helped us to collect our data at the slaughterhouse. We are also grateful to the Centre de Surveillance de la Biodiversité of the University of Kisangani, Royal Institute of Natural Sciences of Belgium (Brussels, Belgium), the Boyekoli Ebale Congo 2010 expedition consortium (Belgian Development Aid, the Belgian Science Policy and the National Lottery of Belgium), and the teams working with profs. Horak (South Africa) and Beati (USA) for their help with the morphological identification of ticks.
Funding
This study was supported by the Institut Hospitalo-Universitaire (IHU) Méditerranée Infection, the National Research Agency under the “Investissements d’avenir” programme, reference ANR-10-IAHU-03, the Région Provence Alpes Côte d’Azur and European ERDF PRIMI funding.
Author information
Authors and Affiliations
Contributions
SN: Designed the study, Collected data (slaughterhouse), Methodology in the field, Wrote the original draft; AZD: Methodology, Data analysis, Wrote the original draft; AL: Designed the study, Collected data (wild animals), Reviewed the manuscript; GG: Reviewed the manuscript, Analysed the data; EV: Reviewed the manuscript, Analysed the data; OM: Reviewed the manuscript; SG: Reviewed the manuscript, Analysed the data; PB: Reviewed the manuscript, Analysed the data; ML: Methodology, Analysed the data, Wrote/reviewed the manuscript; PP: Methodology, Visualised, Supervised, Wrote/reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
There is no IACUC committee in the DRC. However, the Boyekoli Ebale Congo 2010 expedition was approved by the competent authorities (see below). The Boyekoli Ebale Congo 2010 expedition was carried out in the DRC between April and May 2010. Despite the absence of an animal ethics committee in the DRC (or equivalent IACUC), all wildlife sampling was aligned with international research ethics protocols and approved by the competent authorities in the DRC. By “competent authorities”, we refer to the Congolese institutions included in the consortium that validated the research protocols, namely the University of Kisangani (UNIKIN) and the Laboratory of Research in Animal Ecology (LEGERA), details of which are provided in the appendix. By “wildlife samples” we refer to: a) Samples from species not protected by CITES and/or by Congolese law, and ticks obtained from them protected by UNIKIS and LEGERA permits. b) Samples of species protected by CITES and/or by Congolese law, and ticks obtained from them—without financial compensation—from hunters and/or bushmeat sellers during the expedition and protected by UNIKIS and LEGERA permits. No IUCN Red List protected animals were actively trapped. Ticks were removed from the hunted animals with tweezers. Wildlife sampling was carried out under the supervision of the Dean of the Faculty of Biological Sciences of the University of Kisangani, who was a member of the mammalogy research team during the Boyekoli Ebale Congo 2010 expedition. The research was carried out outside the legally recognised parks and reserves in the DRC, outside the protected areas.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ngoy, S., Diarra, A.Z., Laudisoit, A. et al. Using MALDI-TOF mass spectrometry to identify ticks collected on domestic and wild animals from the Democratic Republic of the Congo. Exp Appl Acarol 84, 637–657 (2021). https://doi.org/10.1007/s10493-021-00629-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10493-021-00629-z