Abstract
The global spread of epidemic lung diseases, including COVID-19, underscores the need for efficient diagnostic methods. Addressing this, we developed and tested a computer-aided, lightweight Convolutional Neural Network (CNN) for rapid and accurate identification of lung diseases from 29,131 aggregated Chest X-ray (CXR) images representing seven disease categories. Employing the five-fold cross-validation method to ensure the robustness of our results, our CNN model, optimized for heterogeneous embedded devices, demonstrated superior diagnostic performance. It achieved a 98.56% accuracy, outperforming established networks like ResNet50, NASNetMobile, Xception, MobileNetV2, DenseNet121, and ViT-B/16 across precision, recall, F1-score, and AUC metrics. Notably, our model requires significantly less computational power and only 55 minutes of average training time per fold, making it highly suitable for resource-constrained environments. This study contributes to developing efficient, lightweight networks in medical image analysis, underscoring their potential to enhance point-of-care diagnostic processes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Epidemic lung diseases, such as lung opacity, viral pneumonia, fibrosis, bacterial pneumonia, COVID-19, and tuberculosis, have emerged as significant global health concerns, leading to substantial morbidity and mortality worldwide. The infected individuals experience various symptoms, including mild to moderate manifestations like fever, coughing, and shortness of breath. However, some individuals develop severe pulmonary conditions that can result in fatality [1, 2]. The outbreak of COVID-19 has been particularly alarming, with a substantial number of cases exhibiting intense chest congestion and a significant decline in oxygen levels, leading to severe cardiovascular complications [3,4,5]. Conversely, pneumonia is a lung disease distinguished by inflammation in the microscopic air sacs within the lungs. Pneumonia can be caused by various factors, including viral infections (such as influenza, COVID-19, or respiratory syncytial virus), common cold viruses, and bacterial infections [6,7,8]. Rapid prognosis and appropriate treatment are critical for addressing these epidemic lung diseases and minimizing their impact on public health [9, 10].
Timely identification and accurate diagnosis of lung diseases are paramount in ensuring proper and effective treatment. Healthcare professionals play a vital role in this process as they are equipped to administer targeted therapies based on the specific underlying cause of the infection [11]. These therapies may include antibiotics for bacterial infections, antiviral medications for viral infections, or other interventions tailored to the particular disease. The primary objective of treatment is to resolve the infection, alleviate symptoms, and prevent complications that may arise from the condition [12]. Complications can range from respiratory failure and pneumonia to long-term lung damage. So, by promptly identifying lung disease and providing appropriate treatment, healthcare professionals can minimize the risk of these complications and significantly reduce the mortality rate associated with these conditions [13, 14].
Chest X-ray (CXR) imaging is a widely employed diagnostic tool that plays a critical characteristic in evaluating the condition of the lungs and detecting abnormalities. It provides valuable information into the presence and extent of various lung diseases and abnormalities, allowing clinicians to make informed decisions regarding patient care [15]. However, accurately distinguishing between different epidemic lung diseases based solely on visual inspection of CXR images can be intricate. It requires specialized expertise and in-depth knowledge of each disease’s radiological features and patterns [16]. Unfortunately, these diseases’ radiological features and patterns often overlap, making it difficult to differentiate them accurately using visual examination alone, and this can lead to diagnostic errors and delays in initiating appropriate treatments [17, 18].
Machine learning (ML) [19, 20], deep learning (DL) [21,22,23], convolutional neural networks (CNNs) [24] and artificial intelligence (AI) [25, 26] have made significant advancements in medical image breakdown, including the analysis of complex medical images such as CXR. These techniques with algorithms have shown great potential in extracting meaningful patterns and features from medical images that may not be readily apparent to human observers, leading healthcare professionals to more accurate and objective diagnoses and reducing diagnostic errors [27,28,29] They can also assist healthcare professionals by highlighting regions of interest, providing quantitative measurements, and suggesting potential diagnoses based on learned patterns. Thus, using those in medical image analysis can improve the efficiency of the diagnostic process, allowing for faster interpretation and decision-making by healthcare professionals. It is vital to mention that those technologies are indicated to augment the expertise of healthcare professionals rather than replace them. Combining human expertise and advanced algorithms can lead to more accurate diagnoses and better patient care in the context of epidemic lung diseases [30].
Today, there is a particular emphasis on point-of-care medicine, which involves providing medical care and diagnostic testing directly or near the location where the patient is being treated [31, 32]. Portable radiographic machines or handheld devices can be used to acquire images, eliminating the need for patient transportation to centralized radiology departments. This saves time and reduces the burden on patients, especially those who may be critically ill or have limited mobility. This approach aims to deliver immediate healthcare services, enabling access to imaging, as it reduces the time between image acquisition and interpretation [33, 34]. This is particularly valuable in critical situations or emergencies where a prompt diagnosis is crucial for initiating appropriate diagnostic decisions. Such a point-of-care can improve healthcare systems’ efficiency and is particularly beneficial in environments with limited resources or remote or underserved areas [35, 36].
This research constitutes precious contributions to the field of medical imaging and disease diagnosis:
-
We merged multiple datasets to create a more diverse and comprehensive collection of CXR images, effectively forming a seven-category classification problem that enhances the robustness and generalizability of our approach. We developed a lightweight Convolutional Neural Network (CNN) model suitable for embedded applications, making it appealing, particularly in point-of-care environments. The model’s lightweight architecture ensures it can be efficiently utilized on resource-constrained systems such as embedded devices or portable diagnostic tools without compromising its diagnostic efficacy.
-
We address the prevalent issue of category imbalance in medical image identification and adopt the enhanced focal loss (FE) as the pivotal loss function during the training phase to mitigate the bias towards a particular category. The enhanced FE emphasizes difficult-to-categorized examples, reducing the bias towards well-represented categories. This approach improves the model’s efficacy in accurately identifying rare or underrepresented diseases.
-
We compared the performance of our model against well-known transfer learning architectures widely used in medical image analysis, such as ResNet50, NASNetMobile, Xception, MobileNetV2, DenseNet121, and ViT-B/16. We employed a five-fold cross-validation (CV) technique to ensure the robustness and reliability of our results. The results of our experiments demonstrated that our model outperforms existing methods in disease categorization from CXR images across critical metrics such as accuracy, precision, and recall, thereby underscoring its potential to significantly aid healthcare professionals in delivering timely and precise diagnoses and treatments.
The remainder of the paper is organized as follows: Section 2 examines related works, while details of the materials are presented in Section 3. Section 4 explains the methodology of the system design implementation, and Section 5 shows the experimental outcomes. Section 6, we discuss our work outcomes. Furthermore, the article concludes with a proposal for future works in Section 7.
2 Related work
In recent years, the prognosis and diagnosis of lung diseases have been the subject of extensive research and development efforts by medical experts, researchers, and scientists worldwide. Over the years, medical imaging technologies, ML, AI, and CNN advancements have improved lung disease categorization accuracy, speed, and efficiency [37]. However, it is essential to note that research in this area continues to evolve, with ongoing efforts to develop even more sophisticated and practical diagnostic solutions.
The authors in [38] proposed a Conditional Generative Adversarial Network (cGAN) with a ResNet-50 fine-tuned deep transfer learning model designed to identify six distinct categories: COVID-Mild, COVID-Medium, COVID-Severe, normal, pneumonia, and tuberculosis. The dataset utilized in their experiments comprised 40 COVID-Mild, 42 COVID-Medium, 36 COVID-Severe, 348 normal, 500 pneumonia, and 263 tuberculosis images derived from CXR images. The authors trained their model for 30 epochs using the categorical cross-entropy (CE) loss function. As an outcome, the model has reached an accuracy score of 93.67%. Also, in [39], the authors suggested a custom CNN model named DWTMBConvNet. This model aims to detect normal tuberculosis, COVID-19, bacterial, and viral pneumonia. The dataset employed in their experiments comprised 3593 normal, 1837 tuberculosis, 2098 COVID-19, 2786 bacterial pneumonia, and 1505 viral pneumonia images derived from chest radiographs. The model was trained for 500 epochs with a categorical CE loss function. As a result, the model has achieved an accuracy score of 95.50%.
In [40], the authors presented the FC-DenseNet103 model to identify five distinct categories: normal, bacterial pneumonia, viral pneumonia, COVID-19, and tuberculosis. The dataset employed in their investigations contained 191 normal, 54 bacterial pneumonia, 20 viral pneumonia, 180 COVID-19, and 57 tuberculosis images derived from CXR images. The authors trained their model for 100 epochs using the categorical CE loss function. As an effect, the model has attained an accuracy rate of 88.90%. Additionally, the authors proposed a ResNet-18 model to identify four distinct categories: normal, COVID-19, viral pneumonia, and tuberculosis. Specifically, the dataset contained 180 COVID-19 images, 191 normal images, 54 bacterial pneumonia images, and 57 tuberculosis images. The ResNet-18 model was trained for 100 epochs with a categorical CE loss function. The accuracy rate achieved by their trained model was reported to be 91.90%.
Wang et al. [41] introduced a custom CNN model named CoroDet designed explicitly to identify four categories: COVID-19, normal, viral pneumonia, and bacterial pneumonia. The dataset employed in their experiments comprised 500 COVID-19 images, 800 normal images, 400 bacterial pneumonia images, and 400 viral pneumonia images derived from chest radiographs. The authors trained their CoroDet model for 50 epochs, optimizing it to achieve the highest possible accuracy. As a result, the model has achieved an accuracy score of 91.20%. In [42], the authors presented a DCNN model called CoroNet based on the Xception model architecture. CoroNet was designed to identify four different categories: COVID-19, normal, viral pneumonia, and bacterial pneumonia. The model consisted of 33 million parameters. For their experiments, the authors utilized a dataset comprising a total of 284 COVID-19 images, 310 normal images, 330 bacterial pneumonia images, and 327 viral pneumonia. The authors trained the CoroNet model for 80 epochs during the training process. As a consequence, the CoroNet model reached an accuracy rate of 89.60%.
Karthik et al. [43] presented a custom CNN model to identify four distinct categories: COVID-19, normal, viral, and bacterial pneumonia. The dataset operated in their investigations contained a total of 558 COVID-19 images, 1583 normal images, 2780 bacterial pneumonia images, and 1493 viral pneumonia images derived from CXR images. The authors trained their custom CNN model for 70 epochs and reached an accuracy rate of 97.94%. In [44], the authors proposed a modified VGG19 model to identify images into four categories: COVID-19, normal, pneumonia, and lung cancer. Specifically, the dataset included 4320 COVID-19 images, 3500 normal images, 5856 pneumonia images, and 20,000 lung cancer. The model was trained for 500 epochs using the categorical CE loss function. The accuracy rate reached by their trained model was reported to be 98.05%.
Ibrahim et al. [45] proposed a CNN model based on the pre-trained AlexNet model. The model was designed to identify four categories: COVID-19, normal, viral pneumonia and bacterial pneumonia. The authors employed a dataset comprising 371 COVID-19 images, 2882 normal images, 4237 viral pneumonia images and 4078 bacterial pneumonia images. The model was trained for 20 epochs using the categorical CE loss function. The suggested CNN model earned an accuracy rate of 93.42%. At the same time, in [46] suggested a CNN model named CoviXNet. CoviXNet was developed to identify three categories: normal, COVID-19, and pneumonia. For their investigations, the authors employed a dataset comprising a total of 1281 COVID-19 images, 3270 normal images and 1656 pneumonia images. The CoviXNet model earned an accuracy rate of 96.61%. In [47] proposed a Convolutional CapsNet model for multi-category identification (COVID-19, no-findings, and pneumonia). The authors used a dataset comprising 231 COVID-19 images, which were increased to 1050 with the data augmentation process, 1050 no-findings, and 1050 pneumonia images. The model was trained for 50 epochs and achieved an accuracy of 84.22%.
Gupta et al. [48] evaluated four pre-trained CNN models (VGG-19, InceptionV3, MobileNetV2, and DenseNet). In addition to these individual models, the researchers also developed four hybrid models by combining different CNN architectures VID (VGG-19, Inception, and DenseNet), VMI(VGG-19, MobileNet, and Inception), VMD (VGG-19, MobileNet, and DenseNet), and IMD(Inception, MobileNet, and DenseNet). To evaluate the performance of these models, the researchers used a dataset consisting of 1500 images of pneumonia, 1500 images of COVID-19, and 1500 images of normal cases. Each CNN model was trained with the same hyperparameters for 10 epochs using the categorical CE loss function. The results indicated that the VMD hybrid model performed the best among all the models considered. The VMD model achieved an overall testing accuracy of 97.30%. In [49] introduced a custom CNN model named DarkCovidNet model for multi-category identification (COVID-19, no-findings, and pneumonia). The authors used a dataset comprising 127 COVID-19 images, 500 no-findings, and 500 pneumonia images. The model was trained for 100 epochs and achieved an accuracy of 87.02%.
Table 1 summarises state-of-the-art literature using CNN methods to identify multi-category lung diseases using CXR images. In accordance with state-of-the-art publications, previous researchers have utilized DL techniques to identify lung diseases using CXR images that are not as effectively handled by standard CE loss. However, many existing studies in lung disease identification have been constrained by the limited scope of their datasets, focusing on a narrow range of three to four disease categories. This reliance on smaller datasets raises concerns about these models’ broader applicability and generalizability to more diverse and expansive datasets sourced from varied medical contexts. The accuracy and reliability of these models on larger, more heterogeneous datasets remain uncertain, as there is no assurance that performance levels observed on smaller datasets will be held. Furthermore, a critical challenge with training CNNs on smaller datasets is the increased risk of overfitting. In this case, the model becomes overly tailored to the specific characteristics of the training data, potentially leading to suboptimal performance when exposed to new, unseen data. This issue underscores the need for models that can generalize better across different data sets. Motivated by these limitations, our study aims to overcome these challenges by developing a lightweight CNN architecture for heterogeneous embedded devices, enabling its widespread application, particularly in the point-of-care setting, where quick and accurate disease identification is essential. To augment the diversity and comprehensiveness of our training dataset, we amalgamated multiple CXR image datasets, resulting in an enriched dataset that represents a broader array of lung disorders across seven distinct categories. This approach ensures a more robust and representative model capable of handling diverse clinical systems. Moreover, our study also considers the issue of category imbalance, a common challenge in medical image analysis where specific disease categories are underrepresented. We addressed this by employing and assessing the efficacy of FL as a loss function during model training. This methodology is designed to reduce bias towards predominant categories, enhancing the model’s ability to identify less common lung diseases accurately. Additionally, our study employed the five-fold CV technique throughout the model training and testing phases, strengthening our findings and ensuring the model’s efficacy and reliability across various subsets of the dataset. Finally, our implementation considers the unique computational constraints of the NVIDIA Jetson Xavier platform, ensuring that the model remains efficient and deployable in real-world medical settings.
3 Materials
In Fig. 1, the comprehensive step-by-step process for the categorization of lung diseases and a healthy class using CXR images is illustrated. Initially, a diverse collection of CXR images is gathered from publicly accessible sources, and these images are aggregated to form an extensive dataset encompassing seven distinct categories, each corresponding to specific lung diseases. Then, to ensure the reliability of the designed categorization CNN model, this collection undergoes the five-fold CV process. The entire collection is split into five-folds where four-folds are used for training, and one different fold is used for testing the model’s performance. This process is repeated five times, with each fold used for testing exactly once. Following that were created pre-trained models (ResNet50, NASNetMobile, Xception, MobileNetV2, DenseNet121 and ViT-B/16) and the lightweight proposed CNN. Finally, the performance evaluation phase commences, where the models’ categorization accuracy and efficacy are averaged across all five-folds assessed using diverse performance metrics. Using the five-fold CV in this process ensures that the evaluation of each model accounts for variability within the collection, thereby providing a more reliable assessment of their categorization capabilities.
3.1 Data collection
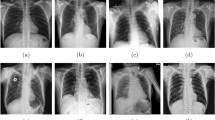
Our work focused on multi-category identification and combined 4 medical datasets with CXR images. These datasets were obtained from combined open databases of images [50]. Overall, we gathered 29,131 images, encompassing both normal cases and six different lung diseases. The first medical dataset we employed was sourced from [51, 52]. This dataset specifically included 3616 cases of COVID-19-positive images, 6012 images displaying lung opacity, 10,192 normal images, and 1345 images of viral pneumonia. For our study, all categories within this dataset were utilized. The second medical dataset we incorporated was obtained from [53]. This dataset contained 2780 images of bacterial pneumonia and 1493 images of viral pneumonia. However, we only utilized the bacterial pneumonia images for our analysis. The third medical dataset we integrated was acquired from [54]. Within this dataset, there were 3500 images of tuberculosis and an equal number of 3500 normal CXR images. For our study, we solely utilized the tuberculosis images. The fourth medical dataset we included in our study was sourced from [55]. From this dataset, 1686 fibrosis images were the only ones employed in our analysis. Figure 2 demonstrates sample CXR images from the dataset, representing distinct categories. Furthermore, Table 2 provides a summary of the dataset’s distribution per category, indicating the number of CXR images for each. Finally, Fig. 3 represents the distribution of CXR images per category from the dataset collection.
3.2 Data pre-processing
Most CNNs for image identification assignments are trained on images with a standardized resolution of \(224 \times 224\) pixels [56]. This resolution has become a common practice in the field of computer vision due to its balance between computational efficiency and preserving sufficient image details for accurate categorization. Therefore, all the images in the collection dataset were resized to \(224 \times 224\) pixels.
Additionally, the images are represented as a matrix of integers, where each element corresponds to a pixel’s intensity value. In this representation, the pixel values range from 0 (representing black) to 255 (representing white) for an 8-bit grayscale image. In the context of reducing computational difficulties during training, pixel values are normalized to a range of 0 to 1. This normalization is achieved by dividing each pixel value by 255. The normalization technique is employed to improve the stability, convergence, and compatibility of CNNs during training [57].
3.3 Collection splitting
The data collection in this work consists of a total of 29,131 CXR images. This collection undergoes the five-fold CV process to ensure the reliability of the designed categorization CNN model. The entire collection is split into five equal parts or folds. In each iteration of the process, four-folds are used to train the model, and one distinct fold is used to test its performance. This process is repeated five times, ensuring that each fold serves as the test set exactly once. The five-fold CV method has several advantages. Firstly, it maximizes both the training and testing data that can be used, and the model is tested across the entire collection, which increases the generalization of the findings. Secondly, it provides a more accurate estimate of the model’s performance on unseen data, reducing the variability that might result from a single random split. This validation process results in a well-tested and robust model against overfitting, ensuring its reliability across different subsets of the data. During the training phase, the neural network optimizes its weights by minimizing the predicted and actual output discrepancies. This process involves iteratively adjusting the network’s parameters to improve its ability to categorize CXR images accurately. The training set plays a crucial role in this optimization process, as it provides a large and diverse set of images for the network to learn from and adjust its internal representations accordingly. On the other hand, the test set serves as a critical benchmark for assessing the trained network’s performance. Table 3 illustrates the distribution of the number of CXR images of each disease category in each fold in the five-fold CV process. It contains a different CXR image collection that is not utilized during the training process. This ensures the network is evaluated on unseen data, simulating real-world scenarios where the model encounters new cases.
4 Methodology
4.1 Handling category imbalance
Generally, in medical image identification tasks, the problem involves multi-categorization, where the goal is to identify an observation sample into one of several possible categories. CNNs are commonly used in these tasks, and the standard loss function employed is the CE loss [58]. The CE loss function for multi-categorization identification can be defined as follows:
where N is the number of categories, \(y_{c}\) is an indicator variable (1 if the sample belongs to category c, 0 otherwise), and \(p_{c}\) is the predicted probability of the observation sample belonging to category c.
However, the standard CE loss is not optimal when dealing with category imbalance in medical image identification. Lin et al. (2017) [59] proposed an FL function as an alternative to the CE loss. The FL aims to address the issue of imbalanced categories by assigning different weights to positive and negative samples. The FL is computed using the following equation:
Here, \(p_{c}\) is the predicted probability, \(a_{c}\) is a weighting factor that is applied when the category is 1 (in binary identification), and \(\gamma \) is a hyperparameter known as the modulating factor. The FL places emphasis on difficult-to-categorize examples, reducing the bias towards well-represented categories. This helps improve the model’s performance in identifying rare or underrepresented diseases.
Nevertheless, the classical FL function is primarily designed for binary issues in target identification. In the case of multiple lung disease categories that require identification in a model, enhancements to the FL function are required. The enhanced FL function, used in the lung disease recognition model, can be calculated using the following equations:
where \(a_{c}\) is the weighting factor, \(p_{c}\) represents the predicted probability distribution, \(\gamma \) is the modulating parameter, and N symbolises the whole number of categories (lung disease types). Our enhanced version of the FL for multi-categorization, particularly in lung disease identification from CXR images, incorporates elements tailored to the specific challenges of this application. Our approach optimizes the FL parameters to align with medical imaging datasets’ intricacies and category imbalances. The customization enhances the model’s sensitivity and specificity for each lung disease category, ensuring a balanced performance across all categories. Additionally, our implementation of the FL is integrated with the architecture and optimization constraints of the NVIDIA Jetson Xavier platform. This integration aims to achieve high accuracy in a resource-constrained environment, which differs from most existing works that do not focus on edge computing applications.
4.2 Proposed lightweight model design
The proposed CNN architecture for lung diseases and healthy identification using the CXR images is displayed in Fig. 4. The proposed lightweight model starts with an input layer, specifying the input shape of the images, which is (224, 224, 3). This means the images have a height and width of 224 pixels with three colour channels (RGB). The first convolutional block consists of 64 filters, a kernel size of \(5 \times 5\), and “same” padding. The “same” padding ensures that the output feature map has the same spatial dimensions as the input. BatchNormalization follows this to normalize the activations, ReLu activation function to introduce non-linearity and a MaxPooling2D layer with a pool size of \(2 \times 2\) for downsampling the spatial dimensions. The second convolutional block includes 32 filters, a kernel size of \(3 \times 3\), and “same” padding. Again, “same” padding is used to maintain the spatial dimensions. Batch normalization, activation function, and max pooling are applied in the same manner as in the previous block. The third convolutional layer consists of 16 filters, a kernel size of \(3 \times 3\), and “same” padding. Batch normalization, activation function, and max pooling are performed. The “same” padding ensures that the spatial dimensions remain the same.
After the convolutional layers, a Flatten layer is added to flatten the output from the previous layers into a one-dimensional vector. This prepares the data for the fully connected layers. The model then proceeds to the fully connected layers. The first fully connected layer has 256 units. BatchNormalization is applied to normalize the activations, followed by a ReLu activation function to introduce non-linearity. During training, a Dropout layer with a rate of 0.3 is added to prevent overfitting. Next is another fully connected layer with 128 units. Again, BatchNormalization, ReLu activation function and Dropout are applied to the layer. Finally, the model concludes with an output layer consisting of 7 units corresponding to the identified disease categories. The output layer is activated using the SoftMax function, which generates probability scores for each category, representing the likelihood of the input image belonging to each category.
The convolutional layers with BatchNormalization and ReLu activation function perform the initial feature extraction. These layers apply a set of learnable filters to the input images, extracting different visual features at multiple levels of abstraction. The filters capture patterns such as edges, textures, and shapes. The MaxPooling2D layers downsample the feature maps, reducing their spatial dimensions while retaining the most salient features. MaxPooling2D helps capture the most important features while reducing computational complexity and spatial dimensions. The Flatten layer reshapes the output from the convolutional layers into a one-dimensional vector, preparing it to be fed into the subsequent fully connected layers. The fully connected layers that follow the Flatten layer are responsible for identification. They take the extracted features as input and perform the task of mapping these features to specific categories. The first fully connected layer with 256 units processes the flattened feature vector and applies a linear transformation to generate a set of 256-dimensional feature vectors. It introduces non-linearity with the ReLu activation function to capture complex relationships between the features. The subsequent fully connected layers with 128 units further process the feature vectors, extracting more abstract representations and reducing dimensionality. They again use BatchNormalization, ReLu activation functions, and dropout regularization to enhance the model’s learning capacity, preventing overfitting and improving generalization. The final output layer with seven units performs the categorization task. It produces a set of output activations for each of the seven disease categories. The activation SoftMax function normalizes the outputs into a probability distribution, indicating the likelihood of the input image belonging to each category. Table 4 summarizes the lightweight CNN model that we developed.
Our lightweight CNN’s innovation lies in its structural design, which is tailored for deployment on resource-constrained edge computing devices like the NVIDIA Jetson Xavier. The model is crafted to balance accuracy with computational efficiency, an essential requirement for edge computing applications with high performance without overburdening the platform’s limited computational resources. This optimization is evident in the balance between the number of convolutional layers, batch normalization, and dropout layers, which together contribute to the model’s efficiency. Our work contributes to the growing field of edge computing in medical imaging by demonstrating that a lightweight yet effective CNN can be deployed on platforms like the NVIDIA Jetson Xavier as point-of-care diagnostics and remote healthcare applications.
4.3 Pre-trained CNN models design
This study tested six pre-trained CNN models (ResNet50, NASNetMobile, Xception, MobileNetV2, DenseNet121 and ViT-B/16). Each pre-trained model loads the pre-trained weights trained on the ImageNet dataset, and the top dense layers of the model (used for categorization) are not included. We froze the pre-trained weights and added three fully connected layers. The initial fully connected layer has 256 units, followed by BatchNormalization, a ReLu activation function and a Dropout layer with a rate of 0.3 is added to prevent overfitting. Next is another fully connected layer with 128 units. Similarly, BatchNormalization, ReLu activation function and Dropout are applied to the layer. Finally, the model concludes with the final output layer with seven units performing the categorization task. The activation SoftMax function normalizes the outputs into a probability distribution, indicating the likelihood of the input image belonging to each category.
The six pre-trained model designs are illustrated in Fig. 5. The primary consideration in deciding the hyperparameters was maintaining the high performance of the proposed lightweight and pre-trained CNNs. Table 5 shows each applied model’s total number of parameters, memory (in MB) and floating-point operations (FLOPs).
5 Experimental outcomes
5.1 GPU implementation on NVIDIA Jetson Xavier
Nowadays, many researchers have proposed the usage of parallel processors. An appealing solution that incorporates multi-processors for graphics processing is GPUs, which are employed for high-efficiency computing and can be considered as multiple hardware cores with a software layer that allows parallel computing. A typical example of this architecture is the NVIDIA Jetson AGX Xavier [60], which is a high-performance embedded platform for AI applications. It combines a robust GPU with a custom-built CPU and DL accelerators.
The Jetson AGX Xavier features a custom-designed 64-bit ARMv8.2 CPU with eight cores, known as Carmel cores and provides high-performance computing capabilities. It incorporates a powerful NVIDIA Volta GPU with 512 CUDA and 64 Tensor cores, which delivers exceptional parallel processing performance and is optimized for DL tasks. Additionally, it includes two NVDLA (NVIDIA Deep Learning Accelerator) engines, which is a modular architecture designed for the efficient execution of neural networks, providing high performance and energy efficiency. Furthermore, it has 16 GB of LPDDR4x RAM, which offers high-bandwidth memory access for running computationally intensive tasks and large-scale models. It has 32 GB of eMMC (embedded MultiMediaCard) storage for storing the operating system, applications, and model data.
Also, it provides various connectivity options, including Gigabit Ethernet, USB 3.1 ports, PCIe Gen3 x16, HDMI, and DisplayPort and allows for flexible integration with peripherals and expansion modules. The Jetson AGX Xavier is compatible with NVIDIA’s CUDA parallel computing platform and popular DL frameworks, such as TensorFlow, PyTorch, and Caffe. Finally, it is designed to be energy-efficient while delivering high-performance computing, and its power consumption ranges from 10 to 30 watts, depending on the workload [61].
In this study, we have undertaken a comprehensive experimental comparative analysis using the NVIDIA Jetson AGX Xavier, leveraging its advanced GPU capabilities for AI medical inference tasks. The Jetson AGX Xavier’s integrated Volta GPU with Tensor Cores provides the computational power necessary for high-throughput and low-latency inference, a critical requirement for real-time applications. Our experiments evaluated the models based on several criteria, including inference speed, FPS and efficiency. These comparative results underscore the suitability of the Jetson AGX Xavier for real-time, on-device AI applications, especially in point-of-care environments.
5.2 Performance metrics
In this study, the performance metrics of the proposed lightweight CNN model and the six pre-trained CNNs were assessed utilizing various metrics, including accuracy, precision, recall, F1-score, and area under the receiver operating characteristic curve (AUC) [62]. Accuracy is a metric that calculates the proportion of correct predictions made by a model out of the total number of predictions. It calculates the total number of outcomes from a CNN model that are correctly predicted. Precision is a metric that calculates the number of valid positive predictions a model makes. It focuses on the accuracy of the positive predictions made and supplies insights into the model’s capability to avoid false positives. Recall, also named sensitivity or true positive rate, is a metric of how satisfactorily a model can identify correct positives. It calculates the proportion of actual positive samples rightly identified by the model. The F1-score is a combined evaluation metric considering precision and recall. It balances precision and recall by considering the harmonic mean of the two measures. Equations (5)–(8) were operated to calculate accuracy, precision, recall, and F1-score, where tp, fn, fp, tn represent the number of true positives, false negatives, false positives and true negatives.
5.3 Training and validation phase
Every network is trained for 30 epochs per fold with a constant learning rate of 0.0001, and the Adam algorithm is used to optimise the network’s parameters [63]. A batch size of 16 was used, striking a balance between computational load and learning efficiency. This size is small enough to ensure that training is manageable on the NVIDIA Jetson Xavier, which is crucial for a lightweight network, while still large enough to facilitate effective model training. The parameters \(\alpha \) and \(\gamma \) are for the FL function. \(\alpha \) balances the importance of positive against negative examples in the loss calculation, with a value of 0.5 indicating equal importance. \(\gamma \), set at 2, adjusts the focus on hard-to-identify examples, ensuring that the model does not become complacent with easily classified examples [59]. Table 6 shows the hyperparameter settings for lightweight networks training.
The average total training time taken for 30 epochs and the testing time per fold is shown in Table 7. Table 7 shows that the proposed lightweight CNN model generally has lower training and testing times than other models, indicating its efficiency.
The average training and validation accuracy curves for the five-fold CV of the CNN models with CE and FL are depicted in Fig. 6. We notice that in all models, FL is the fastest while CE is the slowest. The key reason for this is that FL helps address the issue of category imbalance, which is ordinary in medical assignments. The FL provides more weight to hard-to-category examples, which helps the model focus on learning from the minority categories, leading to faster convergence. Among the models, the lightweight proposed CNN model with FL achieved a remarkable validation accuracy of approximately 98.56%. In comparison, the validation accuracies of the ResNet50, NASNetMobile, Xception, MobileNetV2, DenseNet121, and ViT-B/16 models were 86.25%, 85.43%, 90.11%, 88.19%, 88.69%, and 88.69%, respectively. On the other hand, the lightweight proposed CNN model with CE performed a validation accuracy of 96.57% while ResNet50, NASNetMobile, Xception, MobileNetV2, DenseNet121, and ViT-B/16 models were 85.05%, 84.98%, 88.79%, 87.01%, 88.54%, and 86.65%, respectively. These outcomes demonstrate that the lightweight proposed CNN model surpassed the other models regarding validation accuracy.
Figure 7 presents the average train and validation loss curves for the five-fold CV with CE and FL for the abovementioned models. The lightweight proposed CNN model with FL exhibited a validation loss value of 0.2262, demonstrating its ability to minimize errors during training effectively. In contrast, the ResNet50, NASNetMobile, Xception, MobileNetV2, DenseNet121, and ViT-B/16 models had higher loss values of 0.3026, 0.3207, 0.2868, 0.3264, 0.2908 and 0.3114, respectively. The lightweight proposed CNN model with CE exhibited a validation loss value of 0.2457, while the ResNet50, NASNetMobile, Xception, MobileNetV2, DenseNet121, and ViT-B/16 models had higher loss values of 0.3471, 0.3238, 0.3282, 0.3646, 0.3402 and 0.3785, respectively. Thus, this implies that the lightweight proposed CNN model with FL achieved better optimization and generalization capabilities than the other models. Figures 6 and 7 show that the lightweight proposed CNN model excelled in accuracy and loss minimization. So, high validation accuracy and low loss values suggest it is a promising model for the medical task.
5.4 Testing phase
The evaluation of the lightweight proposed CNN and other lightweight pre-trained CNNs for seven-category identification was assessed using a confusion matrix after the five-fold CV, as depicted in Figs. 8 and 9. In this evaluation, a total of 5825 instances were considered. The proposed CNN with FL miscategorized 84 instances among these samples, indicating its accuracy in identifying the seven categories. Comparatively, the NASNetMobile network exhibited more miscategorizations, with 847 miscategorized instances. Similarly, the ResNet50 network also showed many miscategorized instances, with 797 misclassifications. The DenseNet121 network miscategorized 598 instances. The MobileNetV2 network had a notable number of miscategorizations, with 684 instances being assigned to incorrect categories. Additionally, the ViT-B/16 network miscategorized 655 instances. In contrast, the Xception network exhibited a comparatively lower number of misclassifications, with 573 instances being miscategorized. The proposed CNN with CE misclassified 201 instances, demonstrating its competitive performance. However, the ResNet50, NASNetMobile, Xception, MobileNetV2, DenseNet121, and ViT-B/16 models displayed higher miscategorization rates of 869, 874, 650, 755, 665, and 774, respectively.
The ROC curves for the six CNN models after the five-fold CV employed in this study, which involved seven categories, are presented in Figs. 10 and 11. The findings in Figs. 8 and 11 underscore the superiority of the proposed CNN with FL, as it consistently outperformed the other networks by miscategorizing fewer instances, suggesting its potential as an effective choice for accurate categorization tasks.
The AUC curves for the six CNN models with CE and FL after the five-fold CV are presented in Fig. 12. The lightweight proposed CNN model with FL demonstrated a remarkable AUC value of 0.9930, indicating its exceptional performance in distinguishing between the categories. In comparison, the ResNet50, NASNetMobile, Xception, MobileNetV2, DenseNet121, and ViT-B/16 models achieved AUCs of 0.9168, 0.9130, 0.9415, 0.9313, 0.9396, and 0.9332, respectively. On the other hand, the lightweight proposed CNN model with CE achieved an AUC value of 0.9844, showcasing its discriminatory solid power. In contrast, the ResNet50, NASNetMobile, Xception, MobileNetV2, DenseNet121, and ViT-B/16 models achieved AUCs of 0.9123, 0.9129, 0.9345, 0.9257, 0.9353, and 0.9211, respectively. The pronounced ascent in the true positive rate as the false positive rate approaches zero reflects our classifier’s acute sensitivity to positive instances. Such performance underlines a model adept at identifying the features indicative of the positive category and distinguishing them from those of the negative category with minimal error. This suggests that the classifier possesses robust feature extraction abilities and an effective mechanism for category differentiation. Thus, our model is reliable for recognising true positives, making it especially suitable for scenarios where precision is critical, such as medical diagnostics where the cost of false positives is unacceptable.
In Table 8, the results of six distinct CNN models are presented with CE and FL evaluation measures (average accuracy, precision, recall, and F1-score) after the five-fold CV, showcasing their performance in the task of identifying seven different categories in the context of detecting lung diseases. The proposed lightweight CNN with CE achieves an average accuracy of 96.57%, reflecting its ability to make correct predictions across the seven categories. It also attains an average F1-score of 97.23%, indicating a robust balance between precision and recall, which is essential for accurate classification. However, when employing FL, the model’s performance improves even further, achieving an average accuracy of 98.56% and an average F1-score of 99.01%. This enhancement underlines the effectiveness of FL in fine-tuning the model’s predictive capacity, particularly in handling challenging instances within the dataset. Furthermore, the consistent superiority of FL over CE across all models highlights its utility in handling imbalanced datasets by focusing on hard-to-classify examples. This is a critical consideration in medical image analysis, where certain classes may have fewer instances, making it crucial to ensure accurate detection of all categories, including the minority ones.
Tables 9 - 14 compare various performance metrics - precision, recall, and F1-score - for different lightweight models using CE and FL loss after the five-fold CV. In the precision Tables 9 - 10, the proposed lightweight CNN consistently exhibits high precision across all categories, particularly excelling with the FL loss in categories such as bacterial, COVID, and fibrosis. This indicates its effectiveness in accurately identifying positive cases. Interestingly, there is a notable improvement in precision for most models when using FL loss, especially in categories like viral and fibrosis. The recall Tables 11 - 12 reveal a similar trend, with higher recall rates generally observed in the FL loss setting. This is particularly evident in categories such as COVID, and fibrosis, which show improved performance with FL loss, suggesting enhanced sensitivity and reduced false negatives. Finally, the F1-score Tables 13 - 14, which combines precision and recall, demonstrate the overall effectiveness of these models. The proposed lightweight CNN again stands out, especially with FL loss, showing excellent balance in precision and recall. Across most models and categories, the FL loss seems to contribute to a better balance between false positives and false negatives, leading to higher F1-scores.
Given the critical role of inference time in real-time applications, where swift responsiveness is paramount, these measurements provide valuable insights into the model’s efficiency and capacity to operate efficiently in scenarios where rapid analysis and decision-making are necessary. The average inference time and FPS on GPU and CPU are mentioned in Table 15. Of notable significance is the observation that the proposed lightweight model showcases inference times that are notably shorter than pre-trained models. The proposed lightweight model on the CPU achieves an average inference time of 0.0322 ms, highlighting its ability to process input data and produce corresponding predictions rapidly. Similarly, when the model is deployed on a GPU, it demonstrates an even more accelerated performance, with an inference time of merely 0.0165 ms. Our proposed lightweight CNN model exhibits superior FPS rates, reflecting its quick data handling and processing on both platforms. Comparatively, the MobileNetV2, DenseNet121, NASNetMobile, ViT-B/16, ResNet50, and Xception models follow. As shown in Table 15, our proposed lightweight CNN model demonstrates a notable efficiency when run on CPUs, which generally offer lower parallel processing capabilities than GPUs. This efficiency is attributed to the model’s architecture, which is designed with a few layers and parameters. Such a design significantly minimizes computational complexity, ensuring that the model remains computationally manageable while still achieving high levels of accuracy. In contrast, despite their high efficiency on GPU platforms, the comparison networks do not maintain this performance when shifted to CPU environments. These networks are inherently more complex with a larger number of parameters, making them ideally suited for environments that can leverage GPU’s robust parallel processing abilities. However, this complexity hinders CPUs, where the lack of extensive parallelism leads to slower inference times. These results underscore the efficiency of the model’s design, particularly its streamlined architecture and aptitude for swiftly handling data on CPU and GPU hardware platforms.
6 Discussion
In contemporary medical practice, there is a growing focus on point-of-care medicine. The advent of portable radiographic machines and handheld imaging devices has revolutionized this aspect of healthcare. These devices are particularly beneficial in the rapid diagnostic process for patients who are critically ill or those with restricted mobility, for whom transportation to different hospital departments can be both challenging and discomforting. This immediacy not only enhances the efficiency of healthcare delivery but also ensures that patients receive timely medical interventions. Consequently, point-of-care imaging is a key component in the drive towards more immediate, accessible, and patient-centered healthcare services. On the other hand, medical image analysis, particularly in the context of lung disease categorization, is an area of critical significance. The domain of medical images often grapples with category imbalance in datasets, computational efficiency, and the need for accurate, real-time diagnoses. These issues form the backdrop against which our research is framed.
One significant research gap is the need for more efficient and accurate methods for medical image categorization in point-of-care devices. Existing approaches often rely on pre-trained models that cannot be optimized for specific medical tasks in an embedded platform. Additionally, the challenge of category imbalance in medical datasets further complicates the development of robust models. To bridge this gap, we propose a lightweight CNN model with low computational intensity on the NVIDIA Jetson Xavier platform as a point-of-care device. This model is combined with enhanced FL to deal with category imbalance, which is the main contribution of our study.
The problems in the existing literature are multifaceted. While DL has shown great promise in medical image analysis, there is a need for low computationally intensive models that strike a balance between efficiency and accuracy when implemented in an embedded device where computational resources are limited. Furthermore, the importance of addressing category imbalance in medical datasets cannot be understated. Without robust solutions, models struggle to categorize minority classes, leading to misdiagnoses.
Our research is motivated by the urgent need for improved medical image categorization techniques. Accurate and efficient models can aid healthcare professionals in making timely and precise diagnoses, saving lives. Additionally, our motivation extends to the broader field of ML, where optimizing DL models for real-time applications is a pressing concern. So, addressing these issues, our study aims to contribute not only to the field of medical image analysis but also to the broader research on efficient DL methodologies.
In this study, we have presented and discussed the experimental outcomes of our work, with a primary focus on implementing GPU acceleration using the NVIDIA Jetson Xavier platform. Our results demonstrated that the NVIDIA Jetson AGX Xavier, equipped with a custom-designed CPU and a powerful Volta GPU, is suitable for AI applications in medical tasks. The performance evaluation metrics used in this study provided a comprehensive assessment of the models under consideration. We assessed the accuracy, F1-score, precision, recall, and AUC to gauge the models’ capabilities. These metrics are essential, especially in medical tasks where accurate predictions are crucial. The discussion of these metrics revealed that our proposed lightweight CNN model, particularly when trained with FL, outperformed the pre-trained models. The ability to address category imbalance and achieve high validation accuracy demonstrated the potential for accurate medical image categorization.
We observed that the proposed lightweight CNN model with CE or FL exhibited shorter training and testing times during the training and validation phase than pre-trained models - the best efficiency is attributed to using FL, which aids faster convergence by focusing on challenging examples. The superior testing accuracy of 98.56% achieved by the lightweight proposed CNN model with FL showcased its effectiveness. This performance surpassed other pre-trained models, highlighting its suitability for medical image analysis tasks where accuracy is paramount. In the testing phase, the lightweight proposed CNN model consistently outperformed the pre-trained models, as evidenced by the confusion matrix and ROC curve analysis. The proposed model exhibited lower misclassification rates and achieved higher AUC values, indicating their exceptional discriminatory power.
Additionally, we evaluated our models’ inference time and FPS on both GPU and CPU. Our proposed lightweight model demonstrated significantly shorter inference times than pre-trained models in real-time applications, where rapid responsiveness is essential. With its accuracy and energy-efficient design, this efficiency positions our model as a valuable tool for real-world applications in healthcare. In conclusion, our experimental outcomes underscore the potential of our lightweight CNN model, particularly when utilizing FL, as a reliable and efficient solution for medical image analysis tasks, improving healthcare diagnostics systems and decision-making.
A critical distinction between our work and the referenced studies in Table 1 lies in our proposed model’s architectural efficiency and computational performance. Unlike the more complex and parameter-heavy models such as CoroNet [42], based on the Xception architecture, or the DWTMBConvNet [39] custom CNN model, our lightweight CNN framework is designed to achieve high accuracy while maintaining minimal computational overhead. In terms of accuracy, our model achieves competitive results, closely rivalling those of the highest-performing models in the literature, such as the custom CNN model by Karthik et al. [43] and the modified VGG19 approach by Ibrahim et al. [44]. Notably, our model attains this level of accuracy with a significantly reduced computational footprint, emphasizing the effectiveness of our architecture optimization efforts.
Finally, the accuracy and reliability of predictive models, especially in the domain of healthcare and disease diagnosis, can be significantly influenced by a multitude of external factors. Research in various fields has shown that including natural environmental and socioeconomic factors can substantially enhance the performance of predictive models [64]. Specifically, in lung disease diagnosis, integrating factors such as patient demographics, environmental conditions, and socioeconomic status could offer a more holistic approach to diagnosis and treatment planning. These factors could provide insights into the prevalence and severity of lung diseases influenced by geographic, economic, and social determinants of health. So, by incorporating such multidimensional data into the model, it is possible to improve the accuracy of disease detection and tailor the diagnosis to individual patient profiles, taking into account their unique environmental and social contexts. This approach could lead to more personalized healthcare, with treatments and interventions more closely aligned with the patient’s specific circumstances and needs.
7 Conclusions and future work
This work effectively identifies lung diseases and healthy instances utilizing the proposed lightweight CNN and six pre-trained CNNs ResNet50, NASNetMobile, Xception, MobileNetV2, DenseNet121 and ViT-B/16. This study constructed a large dataset with 29,131 CXR images by merging four public databases labelled with seven categories. Our lightweight CNN model is designed to balance effective image categorization with computational efficiency, a crucial aspect in healthcare settings where resources are limited. Considering its proficiency in handling AI tasks, we have chosen NVIDIA Jetson Xavier for implementation. This choice aligns with our aim to make the model suitable for real-world applications, particularly in medical facilities with constrained computational resources. So, to ensure efficient performance on the Jetson Xavier platform, we focus on reducing the model’s complexity and optimizing its use of Xavier’s GPU acceleration capabilities. This approach permits maintaining high performance while managing resource constraints. The proposed lightweight CNN model surpassed the six pre-trained CNNs, achieving an average of 98.56% accuracy, recall, precision, F1-score, and AUC were found to be 0.9891, 0.9910, 0.9901, and 0.9930, respectively, for the seven-categories system. Nevertheless, the pre-trained models ResNet50, NASNetMobile, Xception, MobileNetV2, DenseNet121 and ViT-B/16 also showed poorer categorization accuracies of 86.25%, 85.43%, 90.11%, 88.19%, 89.69%, and 88.69%, respectively. The presented lightweight CNN can be trained in less than sixty minutes to provide an effective detection model. The other advantage is that the total number of parameters is 5.35 times lower, making it highly versatile to deploy in embedded devices. In addition, the proposed lightweight CNN predicted the results within 17.14 s, while the ResNet50, NASNetMobile, Xception, MobileNetV2, DenseNet121 and ViT-B/16 models required 29.36 s, 22.12 s, 20.59 s, 21.48 s, 26.14 s, and 22.52 s, respectively. The proposed CNN produced an AUC of 0.9930, while ResNet50, NASNetMobile, Xception, MobileNetV2, DenseNet121 and ViT-B/16 showed AUCs of 0.9123, 0.9129, 0.9345, 0.9257, 0.9353, and 0.9211, respectively. We aim to broaden the lung disease categorization framework in our future work. Thus, we intend to enhance the model’s ability to discern and differentiate a more comprehensive array of pathological conditions by including a wider spectrum of disease classes and expanding our dataset with patient demographics, environmental data, and socioeconomic indicators. Incorporating a wide range of disease classes and image types from different sources will make the model more adaptable and accurate in various clinical settings. This expansion directly addresses the need for model adaptation to specific scenes, as a broader range of diseases will introduce variability and complexity to the model.
Data availability and access
All the experiments are conducted utilizing publicly accessible datasets.
References
George PM, Spagnolo P, Kreuter M, Altinisik G, Bonifazi M, Martinez FJ, Molyneaux PL, Renzoni EA, Richeldi L, Tomassetti S et al (2020) Progressive fibrosing interstitial lung disease: clinical uncertainties, consensus recommendations, and research priorities. Lancet Respir Med 8(9):925–934
Wijsenbeek M, Suzuki A, Maher TM (2022) Interstitial lung diseases. Lancet 400(10354):769–786
Hamad QS, Samma H, Suandi SA (2023) Feature selection of pre-trained shallow cnn using the qlesca optimizer: covid-19 detection as a case study. Appl Intell 1–23
Chakraborty M, Dhavale SV, Ingole J (2021) Corona-nidaan: lightweight deep convolutional neural network for chest x-ray based covid-19 infection detection. Appl Intell 51(5):3026–3043
Sharma R, Agarwal M, Gupta M, Somendra S, Saxena SK (2020) Clinical characteristics and differential clinical diagnosis of novel coronavirus disease 2019 (covid-19). Coronavirus Disease 2019 (COVID-19) Epidemiology, Pathogenesis, Diagnosis, and Therapeutics, 55–70
Jangam E, Barreto AAD, Annavarapu CSR (2022) Automatic detection of covid-19 from chest ct scan and chest x-rays images using deep learning, transfer learning and stacking. Appl Intell 1–17
Guler SA, Corte TJ (2021) Interstitial lung disease in 2020: a history of progress. Clin Chest Med 42(2):229–239
Jadhav SP, Singh H, Hussain S, Gilhotra R, Mishra A, Prasher P, Krishnan A, Gupta G (2021) Introduction to lung diseases. Targeting cellular Signalling pathways in lung diseases, pp 1–25
Sarvamangala D, Kulkarni RV (2022) Convolutional neural networks in medical image understanding: a survey. Evol Intel 15(1):1–22
Zhao X, Liu B, Yu Y, Wang X, Du Y, Gu J, Wu X (2020) The characteristics and clinical value of chest ct images of novel coronavirus pneumonia. Clin Radiol 75(5):335–340
Troy LK, Grainge C, Corte TJ, Williamson JP, Vallely MP, Cooper WA, Mahar A, Myers JL, Lai S, Mulyadi E et al (2020) Diagnostic accuracy of transbronchial lung cryobiopsy for interstitial lung disease diagnosis (coldice): a prospective, comparative study. Lancet Respir Med 8(2):171–181
Zhang N, Wang L, Deng X, Liang R, Su M, He C, Hu L, Su Y, Ren J, Yu F et al (2020) Recent advances in the detection of respiratory virus infection in humans. J Med Virol 92(4):408–417
Nuutinen M, Leskelä R-L (2023) Systematic review of the performance evaluation of clinicians with or without the aid of machine learning clinical decision support system. Health Technol 1–14
Spagnolo P, Ryerson CJ, Putman R, Oldham J, Salisbury M, Sverzellati N, Valenzuela C, Guler S, Jones S, Wijsenbeek M et al (2021) Early diagnosis of fibrotic interstitial lung disease: challenges and opportunities. Lancet Respir Med 9(9):1065–1076
Das S, Ayus I, Gupta D (2023) A comprehensive review of covid-19 detection with machine learning and deep learning techniques. Health Technol 1–14
Rajagopal R, Karthick R, Meenalochini P, Kalaichelvi T (2023) Deep convolutional spiking neural network optimized with arithmetic optimization algorithm for lung disease detection using chest x-ray images. Biomed Signal Process Control 79:104197
Echtioui A, Ayed YB (2023) Automated detection of covid-19 based on transfer learning. Multimed Tools Appl 1–21
Ravi V, Acharya V, Alazab M (2023) A multichannel efficientnet deep learning-based stacking ensemble approach for lung disease detection using chest x-ray images. Cluster Comput 26(2):1181–1203
Jonathan J, Barakabitze AA (2023) Ml technologies for diagnosing and treatment of tuberculosis: a survey. Health Technol 13(1):17–33
Alyasseri ZAA, Al-Betar MA, Doush IA, Awadallah MA, Abasi AK, Makhadmeh SN, Alomari OA, Abdulkareem KH, Adam A, Damasevicius R et al (2022) Review on covid-19 diagnosis models based on machine learning and deep learning approaches. Expert Syst 39(3):12759
Choudhary T, Gujar S, Goswami A, Mishra V, Badal T (2023) Deep learning-based important weights-only transfer learning approach for covid-19 ct-scan classification. Appl Intell 53(6):7201–7215
Farhan AMQ, Yang S (2023) Automatic lung disease classification from the chest x-ray images using hybrid deep learning algorithm. Multimed Tools Appl 1–27
Zhou D-X (2020) Theory of deep convolutional neural networks: Downsampling. Neural Netw 124:319–327
George GS, Mishra PR, Sinha P, Prusty MR (2023) Covid-19 detection on chest x-ray images using homomorphic transformation and vgg inspired deep convolutional neural network. Biocybern Biomed Eng 43(1):1–16
Askin S, Burkhalter D, Calado G, El Dakrouni S (2023) Artificial intelligence applied to clinical trials: opportunities and challenges. Health Technol 13(2):203–213
Sharma A, Rani S, Gupta D (2020) Artificial intelligence-based classification of chest x-ray images into covid-19 and other infectious diseases. Int J Biomed Imaging 2020:1–10
Rasheed J, Hameed AA, Djeddi C, Jamil A, Al-Turjman F (2021) A machine learning-based framework for diagnosis of covid-19 from chest x-ray images. Interdiscip Sci Comput Life Sci 13:103–117
Demir F, Sengur A, Bajaj V (2020) Convolutional neural networks based efficient approach for classification of lung diseases. Health Inf Sci Syst 8:1–8
Ullah F, Moon J, Naeem H, Jabbar S (2022) Explainable artificial intelligence approach in combating real-time surveillance of covid19 pandemic from ct scan and x-ray images using ensemble model. J Supercomput 78(17):19246–19271
Tahamtan A, Ardebili A (2020) Real-time rt-pcr in covid-19 detection: issues affecting the results. Expert Rev Mol Diagn 20(5):453–454
Suleman S, Shukla SK, Malhotra N, Bukkitgar SD, Shetti NP, Pilloton R, Narang J, Tan YN, Aminabhavi TM (2021) Point of care detection of covid-19: advancement in biosensing and diagnostic methods. Chem Eng J 414:128759
Rahman MA, Hossain MS, Alrajeh NA, Gupta B (2021) A multimodal, multimedia point-of-care deep learning framework for covid-19 diagnosis. ACM Trans Multimed Comput Commun Appl 17(1s):1–24
Boonkaew S, Jang I, Noviana E, Siangproh W, Chailapakul O, Henry CS (2021) Electrochemical paper-based analytical device for multiplexed, point-of-care detection of cardiovascular disease biomarkers. Sens Actuators B Chem 330:129336
Kameda T, Mizuma Y, Taniguchi H, Fujita M, Taniguchi N (2021) Point-of-care lung ultrasound for the assessment of pneumonia: a narrative review in the covid-19 era. J Med Ultrason 48:31–43
Buonsenso D, Chiaretti A, Curatola A, Morello R, Giacalone M, Parri N (2021) Pediatrician performed point-of-care ultrasound for the detection of ingested foreign bodies: case series and review of the literature. J Ultrasound 24:107–114
Jahanshir A, Moghari SM, Ahmadi A, Moghadam PZ, Bahreini M (2020) Value of point-of-care ultrasonography compared with computed tomography scan in detecting potential life-threatening conditions in blunt chest trauma patients. Ultrasound J 12:1–10
Kumar R, Kumbharkar P, Vanam S, Sharma S (2023) Medical images classification using deep learning: a survey. Multimed Tools Appl 1–46
Mehta T, Mehendale N (2021) Classification of x-ray images into covid-19, pneumonia, and tb using cgan and fine-tuned deep transfer learning models. Res Biomed Eng 37:803–813
Bhosale RD, Yadav D (2023) Customized convolutional neural network for pulmonary multi-disease classification using chest x-ray images. Multimed Tools Appl 1–35
Oh Y, Park S, Ye JC (2020) Deep learning covid-19 features on cxr using limited training data sets. IEEE Trans Med Imaging 39(8):2688–2700
Hussain E, Hasan M, Rahman MA, Lee I, Tamanna T, Parvez MZ (2021) Corodet: A deep learning based classification for covid-19 detection using chest x-ray images. Chaos Solit Fractals 142:110495
Khan AI, Shah JL, Bhat MM (2020) Coronet: A deep neural network for detection and diagnosis of covid-19 from chest x-ray images. Comput Methods Programs Biomed 196:105581
Karthik R, Menaka R, Hariharan M (2021) Learning distinctive filters for covid-19 detection from chest x-ray using shuffled residual cnn. Appl Soft Comput 99:106744
Ibrahim DM, Elshennawy NM, Sarhan AM (2021) Deep-chest: Multi-classification deep learning model for diagnosing covid-19, pneumonia, and lung cancer chest diseases. Comput Biol Med 132:104348
Ibrahim AU, Ozsoz M, Serte S, Al-Turjman F, Yakoi PS (2021) Pneumonia classification using deep learning from chest x-ray images during covid-19. Cogn Comput 1–13
Srivastava G, Chauhan A, Jangid M, Chaurasia S (2022) Covixnet: A novel and efficient deep learning model for detection of covid-19 using chest x-ray images. Biomed Signal Process Control 78:103848
Toraman S, Alakus TB, Turkoglu I (2020) Convolutional capsnet: A novel artificial neural network approach to detect covid-19 disease from x-ray images using capsule networks. Chaos Solit Fractal 140:110122
Gupta H, Bansal N, Garg S, Mallik H, Prabha A, Yadav J (2023) A hybrid convolutional neural network model to detect covid-19 and pneumonia using chest x-ray images. Int J Imaging Syst Technol 33(1):39–52
Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR (2020) Automated detection of covid-19 cases using deep neural networks with x-ray images. Comput Biol Med 121:103792
Shuja J, Alanazi E, Alasmary W, Alashaikh A (2021) Covid-19 open source data sets: a comprehensive survey. Appl Intell 51:1296–1325
Chowdhury ME, Rahman T, Khandakar A, Mazhar R, Kadir MA, Mahbub ZB, Islam KR, Khan MS, Iqbal A, Al Emadi N et al (2020) Can ai help in screening viral and covid-19 pneumonia? Ieee Access 8:132665–132676
Rahman T, Khandakar A, Qiblawey Y, Tahir A, Kiranyaz S, Kashem SBA, Islam MT, Al Maadeed S, Zughaier SM, Khan MS et al (2021) Exploring the effect of image enhancement techniques on covid-19 detection using chest x-ray images. Comput Biol Med 132:104319
Kermany D, Zhang K, Goldbaum M (2018) Labeled optical coherence tomography (oct) and chest x-ray images for classification (2018). Mendeley Data, v2. https://doi.org/10.17632/rscbjbr9sj. https://nihcc.app.box.com/v/ChestXray-NIHCC
Rahman T, Khandakar A, Kadir MA, Islam KR, Islam KF, Mazhar R, Hamid T, Islam MT, Kashem S, Mahbub ZB et al (2020) Reliable tuberculosis detection using chest x-ray with deep learning, segmentation and visualization. IEEE Access 8:191586–191601
Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM (2017) Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2097–2106
Alzubaidi L, Zhang J, Humaidi AJ, Al-Dujaili A, Duan Y, Al-Shamma O, Santamaría J, Fadhel MA, Al-Amidie M, Farhan L (2021) Review of deep learning: concepts, cnn architectures, challenges, applications, future directions. J Big Data 8:1–74
Garbin C, Zhu X, Marques O (2020) Dropout vs batch normalization: an empirical study of their impact to deep learning. Multimed Tools Appl 79:12777–12815
Wang Q, Ma Y, Zhao K, Tian Y (2020) A comprehensive survey of loss functions in machine learning. Ann Data Sci 1–26
Lin T-Y, Goyal P, Girshick R, He K, Dollár P (2017) Focal loss for dense object detection. In: Proceedings of the IEEE International conference on computer vision, pp 2980–2988
Nvidia Developer (2023) Jetson AGX Xavier Developer Kit. Last Accessed 20 May 2023. https://developer.nvidia.com/embedded/jetson-agx-xavier-developer-kit
Mittal S (2019) A survey on optimized implementation of deep learning models on the nvidia jetson platform. J Syst Archit 97:428–442
Murtaza G, Shuib L, Abdul Wahab AW, Mujtaba G, Mujtaba G, Nweke HF, Al-garadi MA, Zulfiqar F, Raza G, Azmi NA (2020) Deep learning-based breast cancer classification through medical imaging modalities: state of the art and research challenges. Artif Intell Rev 53:1655–1720
Khaire UM, Dhanalakshmi R (2020) High-dimensional microarray dataset classification using an improved adam optimizer (iadam). J Ambient Intell Humaniz Comput 11(11):5187–5204
Xu Z, Lv Z, Li J, Sun H, Sheng Z (2022) A novel perspective on travel demand prediction considering natural environmental and socioeconomic factors. IEEE Intell Transp Syst Mag 15(1):136–159
Funding
Open access funding provided by HEAL-Link Greece.
Author information
Authors and Affiliations
Contributions
Methodology, T.S.; investigation, T.S.; software, T.S.; formal analysis, T.S.; conceptualization, T.S.; resources, T.S.; project administration, T.S.; visualization, T.S.; validation, T.S.; writing—original draft preparation, T.S.; writing—review and editing, T.S.; supervision, M.D. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Ethical and informed consent for data used
Not Applicable.
Competing interests
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sanida, T., Dasygenis, M. A novel lightweight CNN for chest X-ray-based lung disease identification on heterogeneous embedded system. Appl Intell 54, 4756–4780 (2024). https://doi.org/10.1007/s10489-024-05420-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-024-05420-2