Abstract
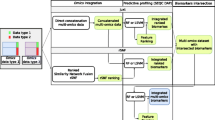
Numerous research has been conducted to define the molecular and clinical aspects of various tumors from a multi-omics point of view. However, there are significant obstacles in integrating multi-omics via Machine Learning (ML) for biomarker identification and cancer subtype classification. In this research, iMVAN, an integrated Multimodal Variational Autoencoder and Network fusion, is presented for biomarker discovery and classification of cancer subtypes. First, MVAE is used on multi-omics data consisting of Copy Number Variation (CNV), mRNA, and Reverse Protein Phase Array (rppa) to discover the biomarkers associated with distinct cancer subtypes. Then, multi-omics integration is accomplished by fusing similarity networks. Ultimately, the MVAE latent data and network fusion are given to a Simplified Graph Convolutional Network (SGC) for categorizing cancer subtypes. The suggested study extracts the top 100 features, which are then submitted to the KEGG analysis and survival analysis test. The survival study identifies nine biomarkers, including AGT, CDH1, CALML5, ERBB2, CCND1, FZD6, BRAF, AR, and MSH6, as poor prognostic markers. In addition, the cancer subtypes are classified, and the performance is assessed. The experimental findings demonstrate that the iMVAN performed well, with an accuracy of 87%.






Similar content being viewed by others
Code Availability
The code for the iMVAN is available at the following link: https://github.com/Arwin94/iMVAN
References
Amina B, Lynda AK, Sonia S, Adel B, Jelloul BH, Miloud M, Tewfik S et al (2021) Fibroblast growth factor receptor 1 protein (fgfr1) as potential prognostic and predictive marker in patients with luminal b breast cancers overexpressing human epidermal receptor 2 protein (her2). Indian J Pathol Microbiol 64(2):254
Asperti A, Trentin M (2020) Balancing reconstruction error and kullback-leibler divergence in variational autoencoders. IEEE Access 8:199,440–199,448
Berrar D (2019) Cross-validation
Bi K, He MX, Bakouny Z, Kanodia A, Napolitano S, Wu J, Grimaldi G, Braun DA, Cuoco MS, Mayorga A et al (2021) Tumor and immune reprogramming during immunotherapy in advanced renal cell carcinoma. Cancer Cell 39(5):649–661
Bouchalova K, Kharaishvili G, Bouchal J, Vrbkova J, Megova M, Hlobilkova A (2014) Triple negative breast cancer-bcl2 in prognosis and prediction. review. Current drug targets 15(12):1166–1175
Çevik A, Weber GW, Eyüboğlu BM, Oğuz KK, Initiative ADN (2017) Voxel-mars: a method for early detection of alzheimer’s disease by classification of structural brain mri. Ann Oper Res 258:31–57
Chaudhary KR (2022) Knnimputer — way to impute missing values. https://www.analyticsvidhya.com/blog/2020/07/-knnimputer-a-robust-way-to-impute-missing-values-using-scikit-learn/
Chen W, Chen Y, Zhang K, Yang W, Li X, Zhao J, Liu K, Dong Z, Lu J (2021) Agt serves as a potential biomarker and drives tumor progression in colorectal carcinoma. Int Immunopharmacol 101(108):225
Cheng LH, Hsu TC, Lin C (2021) Integrating ensemble systems biology feature selection and bimodal deep neural network for breast cancer prognosis prediction. Scientific Reports 11(1):1–10. https://doi.org/10.1038/s41598-021-92864-y
Chierici M, Bussola N, Marcolini A, Francescatto M, Zandonà A, Trastulla L, Agostinelli C, Jurman G, Furlanello C (2020) Integrative Network Fusion: A Multi-Omics Approach in Molecular Profiling. Frontiers in Oncology 10(June). https://doi.org/10.3389/fonc.2020.01065
Corda G, Sala G, Lattanzio R, Iezzi M, Sallese M, Fragassi G, Lamolinara A, Mirza H, Barcaroli D, Ermler S et al (2017) Functional and prognostic significance of the genomic amplification of frizzled 6 (fzd6) in breast cancer. The Journal of pathology 241(3):350–361
De Santo I, McCartney A, Migliaccio I, Di Leo A, Malorni L (2019) The emerging role of esr1 mutations in luminal breast cancer as a prognostic and predictive biomarker of response to endocrine therapy. Cancers 11(12):1894
Delgado FM, Gómez-Vela F (2019) Computational methods for gene regulatory networks reconstruction and analysis: A review. Artificial intelligence in medicine 95:133–145
Dhillon A, Singh A, Bhalla VK (2023) A systematic review on biomarker identification for cancer diagnosis and prognosis in multi-omics: from computational needs to machine learning and deep learning. Archives of Computational Methods in Engineering 30(2):917–949
Eicher T, Kinnebrew G, Patt A, Spencer K, Ying K, Ma Q, Machiraju R, Mathé EA (2020) Metabolomics and multi-omics integration: a survey of computational methods and resources. Metabolites 10(5):202
Gokgoz N, Öktem H (2021) Modeling of tumor-immune system interaction with stochastic hybrid systems with memory: A piecewise linear approach. Advances in the Theory of Nonlinear Analysis and its Application 5(1):25–38
Gu T, Zhao X (2019) Integrating multi-platform genomic datasets for kidney renal clear cell carcinoma subtyping using stacked denoising autoencoders. Scientific Reports 9(1):1–11. https://doi.org/10.1038/s41598-019-53048-x
Guo H, Wang S, Ju M, Yan P, Sun W, Li Z, Wu S, Lin R, Xian S, Yang D et al (2021) Identification of stemness-related genes for cervical squamous cell carcinoma and endocervical adenocarcinoma by integrated bioinformatics analysis. Frontiers in Cell and Developmental Biology 9(642):724
Hamilton W, Ying Z, Leskovec J (2017) Inductive representation learning on large graphs. Advances in neural information processing systems 30
Hira MT, Razzaque MA, Angione C, Scrivens J, Sawan S, Sarkar M (2021) Integrated multi-omics analysis of ovarian cancer using variational autoencoders. Scientific Reports 11(1). https://doi.org/10.1038/s41598-021-85285-4
Jung I, Kim M, Rhee S, Lim S, Kim S (2021) Monti: A multi-omics non-negative tensor decomposition framework for gene-level integrative analysis. Front Genet 1635
Kaur P, Singh A, Chana I (2021) Computational techniques and tools for omics data analysis: state-of-the-art, challenges, and future directions. Archives of Computational Methods in Engineering 28(7):4595–4631
Kaur P, Singh A, Chana I (2022) Bsense: a parallel bayesian hyperparameter optimized stacked ensemble model for breast cancer survival prediction. J Comput Sci 60(101):570
Kurozumi S, Alsaleem M, Monteiro CJ, Bhardwaj K, Joosten SE, Fujii T, Shirabe K, Green AR, Ellis IO, Rakha EA et al (2020) Targetable erbb2 mutation status is an independent marker of adverse prognosis in estrogen receptor positive, erbb2 non-amplified primary lobular breast carcinoma: a retrospective in silico analysis of public datasets. Breast Cancer Res 22:1–11
Kuter S, Bolat K, Akyurek Z (2022) A machine learning-based accuracy enhancement on eumetsat h-saf h35 effective snow-covered area product. Remote Sens Environ 272(112):947
Lánczky A, Győrffy B (2021) Web-based survival analysis tool tailored for medical research (KMplot): Development and implementation. Journal of Medical Internet Research 23(7):1–7. https://doi.org/10.2196/27633
Li S, Jiang L, Tang J, Gao N, Guo F (2020) Kernel Fusion Method for Detecting Cancer Subtypes via Selecting Relevant Expression Data. Front Genet 11(September):1–10. https://doi.org/10.3389/fgene.2020.00979
Li Y, Wu T, Peng Z, Tian X, Dai Q, Chen M, Zhu J, Xia S, Sun A, Yang W et al (2022) Ets1 is a prognostic biomarker of triple-negative breast cancer and promotes the triple-negative breast cancer progression through the yap signaling. American Journal of Cancer Research 12(11):5074
Liu P, Li F, Lin J, Li L, Wang L (2019) Cdh1 as a therapeutic target for breast cancer treatment. Scientific reports 9(1):1–13
Liu X, Lei F, Xia G, Zhang Y, Wei W (2022) Adjmix: simplifying and attending graph convolutional networks. Complex & Intelligent Systems, pp 1–10
Lu M, Zhan X (2018) The crucial role of multiomic approach in cancer research and clinically relevant outcomes. EPMA J 9(1):77–102
Matissek KJ, Onozato ML, Sun S, Zheng Z, Schultz A, Lee J, Patel K, Jerevall PL, Saladi SV, Macleay A et al (2018) Expressed gene fusions as frequent drivers of poor outcomes in hormone receptor-positive breast cancerfrequent expressed gene fusions in hr+ breast cancer. Cancer discovery 8(3):336–353
Pavanelli AC, Mangone FR, Yoganathan P, Bessa SA, Nonogaki S, de Toledo Osório CA, de Andrade VP, Soares IC, de Mello ES, Mulligan LM et al (2022) Comprehensive immunohistochemical analysis of ret, bcar1, and bcar3 expression in patients with luminal a and b breast cancer subtypes. Breast Cancer Res Treat 192(1):43–52
Rajpal S, Agarwal M, Kumar V, Gupta A, Kumar N (2021) Triphasic DeepBRCA-A Deep Learning-Based Framework for Identification of Biomarkers for Breast Cancer Stratification. IEEE Access 9:103,347–103,364. https://doi.org/10.1109/ACCESS.2021.3093616
Ramadan A, Hashim M, Abouzid A, Swellam M (2021) Clinical impact of pten methylation status as a prognostic marker for breast cancer. Journal of Genetic Engineering and Biotechnology 19(1):1–11
Dn Ren, Chen J, Li Z, Yan H, Yin Y, Wo D, Zhang J, Ao L, Chen B, Ito TK et al (2015) Lrp5/6 directly bind to frizzled and prevent frizzled-regulated tumour metastasis. Nat Commun 6(1):1–13
Roberts ME, Jackson SA, Susswein LR, Zeinomar N, Ma X, Marshall ML, Stettner AR, Milewski B, Xu Z, Solomon BD et al (2018) Msh6 and pms2 germ-line pathogenic variants implicated in lynch syndrome are associated with breast cancer. Genetics in Medicine 20(10):1167–1174
Rocca J (2022) Understanding Variational Autoencoders (VAEs). https://towardsdatascience.com/understanding-variational-autoencoders-vaes-f70510919f73
Rodriguez-Ruiz ME, Buqué A, Hensler M, Chen J, Bloy N, Petroni G, Sato A, Yamazaki T, Fucikova J, Galluzzi L (2019) Apoptotic caspases inhibit abscopal responses to radiation and identify a new prognostic biomarker for breast cancer patients. Oncoimmunology 8(11):e1655,964
Sarkar JP, Saha I, Sarkar A, Maulik U (2021) Machine learning integrated ensemble of feature selection methods followed by survival analysis for predicting breast cancer subtype specific miRNA biomarkers. Comput Biol Med 131(January):104,244. https://doi.org/10.1016/j.compbiomed.2021.104244
Savku E, Azevedo N, Weber G (2017) Optimal control of stochastic hybrid models in the framework of regime switches. In: Modeling, Dynamics, Optimization and Bioeconomics II: DGS III, Porto, Portugal, February 2014, and Bioeconomy VII, Berkeley, USA, March 2014-Selected Contributions 3, Springer, pp 371–387
Sun D, Li A, Tang B, Wang M (2018) Integrating genomic data and pathological images to effectively predict breast cancer clinical outcome. Comput Methods Prog Biomed 161:45–53
Taylan P, Yerlikaya-Özkurt F, Bilgic Ucak B, Weber GW (2021) A new outlier detection method based on convex optimization: application to diagnosis of parkinson’s disease. J Appl Stat 48(13–15):2421–2440
Temoçin BZ, Weber GW (2014) Optimal control of stochastic hybrid system with jumps: a numerical approximation. J Comput Appl Math 259:443–451
Tomozumi Imamichi (2022) DAVID Bioinformatics Resources. https://david.ncifcrf.gov/
Valla M, Klæstad E, Ytterhus B, Bofin AM (2022) Ccnd1 amplification in breast cancer-associations with proliferation, histopathological grade, molecular subtype and prognosis. J Mammary Gland Biol Neoplasia 27(1):67–77
Vasaikar SV, Straub P, Wang J, Zhang B (2018) LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 46(D1):D956–D963. https://doi.org/10.1093/nar/gkx1090
Wang B, Mezlini AM, Demir F, Fiume M, Tu Z, Brudno M, Haibe-Kains B, Goldenberg A (2014) Similarity network fusion for aggregating data types on a genomic scale. Nature Methods 11(3):333–337
Wang T, Shao W, Huang Z, Tang H, Zhang J, Ding Z, Huang K (2021) MOGONET integrates multi-omics data using graph convolutional networks allowing patient classification and biomarker identification. Nature Communications 12(1):1–13. https://doi.org/10.1038/s41467-021-23774-w
Weber GW, Yasar O (2004) Discrete tomography: A modern inverse problem reconsidered by optimization. J Comp Tech 9:115–121
Weber GW, Kropat E, Alparslan Gök SZ (2008) Semi-infinite and conic optimization in modern human life and financial sciences under uncertainty. In: ISI Proceedings of 20th Mini-EURO conference, Continuous Optimization and Knowledge-Based Technologies, Neringa, Lithuania, pp 180–185
Weber GW, Uğur Ö, Taylan P, Tezel A (2009) On optimization, dynamics and uncertainty: a tutorial for gene-environment networks. Discret Appl Math 157(10):2494–2513
Wu F, Souza A, Zhang T, Fifty C, Yu T, Weinberger KQ (2019) Simplifying graph convolutional networks. In International Conference on Machine Learning 2019 2019-May 24 (pp. 6861–6871). PMLR
Xing X, Yang F, Li H, Zhang J, Zhao Y, Gao M, Huang J, Yao J (2021) An Interpretable Multi-Level Enhanced Graph Attention Network for Disease Diagnosis with Gene Expression Data. Proceedings - 2021 IEEE International Conference on Bioinformatics and Biomedicine, BIBM 2021 pp 556–561
Yıldırım MH (2015) Electricity market modeling using stochastic and robust optimization
Yu Z, Huang F, Zhao X, Xiao W, Zhang W (2021) Predicting drug–disease associations through layer attention graph convolutional network. Brief Bioinform 22(4):bbaa243
Zhang C, Chen Y, Zeng T, Zhang C, Chen L (2022) Deep latent space fusion for adaptive representation of heterogeneous multi-omics data. Brief Bioinform 23(2):1–15
Zhang L, Fang C, Xu X, Li A, Cai Q, Long X (2015) Androgen receptor, egfr, and brca1 as biomarkers in triple-negative breast cancer: a meta-analysis. BioMed research international 2015
Acknowledgements
We are thankful to Dr. Vikas Sharma, an Assistant Professor in School of Mathematics, TIET, Patiala, for his thorough examination of the mathematical concepts presented in this work. His valuable suggestions and recommended changes have significantly enhanced the overall quality and rigor of the mathematical analysis.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical Approval
This article does not contain any study on human participants or animals performed by any of the authors
Competing interests
Authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dhillon, A., Singh, A. & Bhalla, V.K. iMVAN: integrative multimodal variational autoencoder and network fusion for biomarker identification and cancer subtype classification. Appl Intell 53, 26672–26689 (2023). https://doi.org/10.1007/s10489-023-04936-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-023-04936-3