Abstract
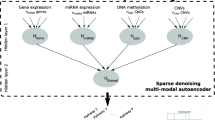
The integration of multi-omics data is suitable for early detection and is also significant to a wide variety of cancer detection and treatment fields. Accurate prediction of survival in cancer patients remains a challenge due to the ever-increasing heterogeneity and complexity of cancer. The latest developments in high-throughput sequencing technologies have rapidly produced multi-omics data of the same cancer sample. Recently, many studies have shown to extract biologically relevant latent features to learn the complexity of cancer by taking advantage of deep learning. In this paper, we propose a Shared representation learning method by employing the Autoencoder structure for Multi-Omics (SAMO) data, which is inspired by the recent success of variational autoencoders to extract biologically relevant features. Variational autoencoders are a deep neural network approach capable of generating meaningful latent spaces. We address the problem of losing information when integrating multiple data sources. We formulate a distributed latent space jointly learned by separated variational autoencoders on each data source in an unsupervised manner. Firstly, we pre-trained the variational autoencoders separately, which produce shared latent representations. Secondly, we fine-tuned only the encoders and latent representations with a supervised classifier for the prediction task. Here, we used a lung cancer multi-omics data combined illumina human methylation 27 K and gene expression RNA seq. datasets from The Cancer Genome Atlas (TCGA) data portal.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Dai, X., Li, T., Bai, Z., Yang, Y., Liu, X., Zhan, J., Shi, B.: Breast cancer intrinsic subtype classification, clinical use and future trends. Am. J. Cancer Res. 5(10), 2929 (2015)
Liang, M., Li, Z., Chen, T., Zeng, J.: Integrative data analysis of multi-platform cancer data with a multimodal deep learning approach. IEEE/ACM Trans. Comput. Biol. Bioinf. 12(4), 928–937 (2014)
Prat, A., Pineda, E., Adamo, B., Galván, P., Fernández, A., Gaba, L., Díez, M., Viladot, M., Arance, A., Muñoz, M.: Clinical implications of the intrinsic molecular subtypes of breast cancer. The Breast 24, S26–S35 (2015)
Blanco-Calvo, M., Concha, Á., Figueroa, A., Garrido, F., Valladares-Ayerbes, M.: Colorectal cancer classification and cell heterogeneity: a systems oncology approach. Int. J. Mol. Sci. 16(6), 13610–13632 (2015)
Deng, S.P., Zhu, L., Huang, D.S.: Predicting hub genes associated with cervical cancer through gene co-expression networks. IEEE/ACM Trans. Comput. Biol. Bioinf. 13(1), 27–35 (2015)
Hanahan, D., Weinberg, R.A.: Hallmarks of cancer: the next generation. Cell 144(5), 646–674 (2011)
Chaudhary, K., Poirion, O.B., Lu, L., Garmire, L.X.: Deep learning–based multi-omics integration robustly predicts survival in liver cancer. Clin. Cancer Res. 24(6), 1248–1259 (2018)
Vasaikar, S.V., Straub, P., Wang, J., Zhang, B.: LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 46(D1), D956–D963 (2018)
Zhang, W., Feng, H., Wu, H., Zheng, X.: Accounting for tumor purity improves cancer subtype classification from DNA methylation data. Bioinformatics 33(17), 2651–2657 (2017)
Lee, S., Lim, S., Lee, T., Sung, I., Kim, S.: Cancer subtype classification and modeling by pathway attention and propagation. Bioinformatics (2020)
Wang, Z., Wang, Y.: Exploring DNA methylation data of lung cancer samples with variational autoencoders. In: 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp. 1286–1289. IEEE (2018)
Zhang, X., Zhang, J., Sun, K., Yang, X., Dai, C., Guo, Y.: Integrated multi-omics analysis using variational autoencoders: application to pan-cancer classification. In: 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp. 765–769. IEEE (2019)
Wang, Z., Wang, Y.: Extracting a biologically latent space of lung cancer epigenetics with variational autoencoders. BMC Bioinformatics 20(18), 1–7 (2019)
Ng, A.: Sparse autoencoder. CS294A Lecture Notes, 72(2011), 1–19 (2011)
Kusner, M.J., Paige, B., Hernández-Lobato, J.M.: Grammar variational autoencoder. arXiv preprint arXiv:1703.01925 (2017)
Le, L., Patterson, A., White, M.: Supervised autoencoders: improving generalization performance with unsupervised regularizers. In: Advances in Neural Information Processing Systems, pp. 107–117 (2018)
Zhang, Y., Lee, K., Lee, H.: Augmenting supervised neural networks with unsupervised objectives for large-scale image classification. In: International Conference on Machine Learning, pp. 612–621 (2016)
Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (No. 2020R1A2B5B02001717) and (2019K2A9A2A06020672).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Ryu, K.H., Park, K.H., Namsrai, OE., Pham, VH., Batbaatar, E. (2021). Shared Representation with Multi-omics Distributed Latent Spaces for Cancer Subtype Classification. In: Pan, JS., Li, J., Ryu, K.H., Meng, Z., Klasnja-Milicevic, A. (eds) Advances in Intelligent Information Hiding and Multimedia Signal Processing. Smart Innovation, Systems and Technologies, vol 212. Springer, Singapore. https://doi.org/10.1007/978-981-33-6757-9_52
Download citation
DOI: https://doi.org/10.1007/978-981-33-6757-9_52
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-33-6756-2
Online ISBN: 978-981-33-6757-9
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)