Abstract
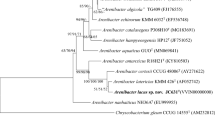
In this study, we reported a Gram-stain-negative, ovoid to rod-shaped, atrichous, and facultative anaerobe bacteria strain named YMD61T, which was isolated from the intertidal sediment of Yangma island, China. Growth of strain YMD61T occurred at 10.0–45.0 °C (optimum, 30.0 °C), pH 7.0–10.0 (optimum, 8.0) and with 0–3.0% (w/v) NaCl (optimum, 2.0%). Phylogenetic tree analysis based on 16 S rRNA gene or genomic sequence indicated that strain YMD61T belonged to the genus Fuscovulum and was closely related to Fuscovulum blasticum ATCC 33,485T (96.6% sequence similarity). Genomic analysis indicated that strain YMD61T contains a circular chromosome of 3,895,730 bp with DNA G + C content of 63.3%. The genomic functional analysis indicated that strain YMD61T is a novel sulfur-metabolizing bacteria, which is capable of fixing carbon through an autotrophic pathway by integrating the processes of photosynthesis and sulfur oxidation. The predominant respiratory quinone of YMD61T was ubiquinone-10 (Q-10). The polar lipids of YMD61T contained phosphatidylglycerol, phosphatidylethanolamine, phosphatidylcholine, five unidentified lipids, unidentified aminolipid and unidentified aminophospholipid. The major fatty acids of strain YMD61T contained C18:1ω7c 11-methyl and summed feature 8 (C18:1 ω 7c or/and C18:1 ω 6c). Phylogenetic, physiological, biochemical and morphological analyses suggested that strain YMD61T represents a novel species of the genus Fuscovulum, and the name Fuscovulum ytuae sp. nov. is proposed. The type strain is YMD61T (= MCCC 1K08483T = KCTC 43,537T).


Similar content being viewed by others
Data availability
No datasets were generated or analysed during the current study.
Code availability
The GenBank accession number of the 16S rRNA gene sequence of strain YMD61T is OP740932. The GenBank accession number of complete genome sequence of strain YMD61T is CP124535.
Abbreviations
- FAME:
-
Fatty acid methyl esters
- ANI:
-
Average nucleotide identity
- GGDC:
-
Genome-to-genome distance calculator
- DSMZ:
-
Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH
- DDH:
-
DNA–DNA hybridizations.
References
Brinkhoff T, Giebel HA, Simon M (2008) Diversity, ecology, and genomics of the Roseobacter clade: a short overview. Arch Microbiol 189:531–539
Buchan A, González JM, Moran MA (2005) Overview of the marine Roseobacter lineage. Appl Environ Microb 71:5665–5677
Buchan A, Gonzalez JM, Moran MA (2005a) Overview of the marine Roseobacter lineage. Appl Environ Microbiol 71:5665–5677
Cao W, Wong MH (2007) Current status of coastal zone issues and management in China: a review. Environ Int 33:985–992
Chen L, Xiong Z, Sun L et al (2012) VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res 40(D1):D641–D645
Collins M (1985) Isoprenoid quinone analysis in bacterial classification and identification. Chemical methods in Bacterial Systematics. Academic, London, pp 267–287
Eckersley K, Dow CS (1980) Rhodopseudomonas blastica sp. nov., a member of the Rhodospirillaceae. J Gen Microbiol 119:465–473
Gilman IS, Edwards EJ (2020) Crassulacean acid metabolism. Curr Biol 20(2):R57–R62
Grabarczyk DB, Berks BC (2017) Intermediates in the Sox sulfur oxidation pathway are bound to a sulfane conjugate of the carrier protein SoxYZ. PLoS ONE 12(3):e0173395
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Hiraishi A, Ueda Y (1994) Intrageneric structure of the genus Rhodobacter:transfer of Rhodobacter sulfidophilus and related marine species to the genus Rhodovulum gen. Nov. Int J Syst Bacteriol 44:15–23
Hugler M, Sievert SM (2011) Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Ann Rev Mar Scie 3:261–289
Jia B, Raphenya AR, Alcock B et al (2017) CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45(Database issue):D566–D573
Kim OS, Cho YJ, Lee K et al (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62(3):716–721
Kim M, Oh HS, Park SC et al (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351
Kumar S, Stecher G, Li M et al (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Lee I, Ouk Kim Y, Park SC et al (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103
Liu B, Pop M (2009) ARDB-antibiotic resistance genes database. Nucleic Acids Res 37(suppl 1):D443–D447
Liu J, Bao Y, Zhang X et al (2021) Rhodobacter kunshanensis sp. nov., a novel bacterium isolated from activated sludge. Curr Microbiol 78(10):3791–3797
Martin MF (2020) Older than genes: the acetyl coa pathway and origins. Front Microbiol 4:11817
Martin U, Rashmi P, Arathi R (2015) The Pathogen-host interactions database (PHI-base): additions and future developments. Nucleic Acids Res 43(Database issue):D645–D655
Medema MH, Blin K, Cimermancic P et al (2011) antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 39(suppl 2):W339–W346
Meier-Kolthoff JP, Auch AF, Klenk HP et al (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60
Minnikin DE, Odonnell AG, Goodfellow M et al (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Meth 2:233–241
Na SI, Kim YO, Yoon SH et al (2018) UBCG: up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol 56:281–285
Pan K, Wang WX (2012) Trace metal contamination in estuarine and coastal environments in China. Sci Total Environ 421:3–16
Pester M, Knorr KH, Friedrich MW et al (2012) Sulfate-reducing microorganisms in wetlands–fameless actors in carbon cycling and climate change. Front Microbiol 3:72–91
Petersen TN, Brunak S, von Heijne G et al (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8(10):785–786
Rabus R, Venceslau SS, Woehlbrand L et al (2015) Pereira A post-genomic view of the ecophysiology, catabolism and biotechnological relevance of sulphate-reducing prokaryotes. Adv Microb Physiol 66:55–321
Rzhetsky A, Nei M (1992) A simple method for estimating and testing minimum-evolution trees. Mol Biol 9:945–967
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. USFCC News Lett 20:1–6
Selje N, Simon M, Brinkhoff T (2004) A newly discovered Roseobacter Cluster in temperate and polar oceans. Nature 427:445–448
Suresh G, Lodha TD, Indu B et al (2019) Taxogenomics resolves conflict in the genus Rhodobacter: a two and half decades pending thought to reclassify the genus Rhodobacter. Front Microbiol 10:2480
Suresh G, Lodha T, Indu B et al (2020) Taxogenomics resolves conflict in the genus Rhodobacter: a two and half decades pending thought to reclassify the genus Rhodobacter. Front Microbiol 11:1111
Tatusova T, Dicuccio M, Badretdin A et al (2016) NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res 44:6614–6624
Wayne LG, Moore WEC, Stackebrandt E et al (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Evol Microbiol 37:463–464
Wilms R, Sass H, Kopke B et al (2006) Specific bacterial, archaeal and eukaryotic communities in tidal-flat sediments along a vertical profile of several meters. Appl Environ Microbiol 72:2756–2764
Wu S, Li R, Xie S et al (2019) Shi depth-related change of sulfate-reducing bacteria community in mangrove sediments: the influence of heavy metal contamination. Mar Pollut Bull 140:443–450
Xian WD, Liu ZT, Li MM (2020) Rhodobacter flagellatus sp. nov., a thermophilic bacterium isolated from a hot spring. Int J Syst Evol Microbiol 70(3):1541–1546
Xu P, Li WJ, Tang SK et al (2005) Naxibacter alkalitolerans gen. nov., Sp nov., a novel member of the family ‘Oxalobacteraceae’ isolated from China. Int J Syst Evol Microbiol 55:1149–1153
Xu XD, Zhang J, Sun QL et al (2021) Description of Psychrosphaera ytuae sp. nov., isolated from the deep-sea cold seep sediment of South China Sea. Int J Syst Evol Microbiol 71(8):004983
Acknowledgements
This work was supported by the Natural Science Foundation of Shandong Province (ZR2020MC198), the Development Plan of Youth Innovation Team in Colleges and Universities of Shandong Province (2022KJ269) and Yantai University Doctoral Start-up Foundation (HX20B34).
Author information
Authors and Affiliations
Contributions
BZZ: investigation, writing original draft. XDX: isolated the bacterium. DDZ: investigation. XPL: funding acquisition, phylogenetic and genomic characterisation. XDJ: supervision and writing-reviewing and editing. JZ: conceptualization, funding acquisition, funding acquisition.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, Bz., Xu, Xd., Zhou, Dd. et al. Description of Fuscovulum ytuae sp. nov, a facultative autotroph isolated from the intertidalite of Yangma island, China. Antonie van Leeuwenhoek 117, 52 (2024). https://doi.org/10.1007/s10482-024-01947-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10482-024-01947-z