Abstract
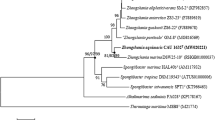
A gram-stain-negative, aerobic, rod-shaped bacterium strain CJK-A8-3T was isolated from a polyamine-enriched seawater sample collected from the Changjiang River estuary of China. The colonies were white and circular. Strain CJK-A8-3T grew optimally at 35 °C, pH 7.0 and 1.5% NaCl. Its polar lipids contained phosphatidylglycerol, phosphatidic acid, unidentified glycolipids, and a combination of phospholipids and glycolipids. The respiratory quinone was ubiquinone-10, and its main fatty acids were C16:0, 11-methyl C18:1ω7c and Summed Feature 8 (including C18:1ω7c/C18:1ω6c). The phylogenetic tree based on 16S rRNA genes placed strain CJK-A8-3T in a new linage within the genus Devosia. 16S rRNA gene sequence of strain CJK-A8-3T showed identities of 98.50% with Devosia beringensis S02T, 98.15% with D. oryziradicis, and 98.01% with D. submarina JCM 18935T. The genome size of strain CJK-A8-3T was 3.81 Mb with the DNA G + C content 63.9%, higher than those of the reference strains (60.4–63.8%). The genome contained genes functional in the metabolism of terrigenous aromatic compounds, alkylphosphonate and organic nitrogen, potentially beneficial for nutrient acquirement and environmental remediation. It also harbored genes functional in antibiotics resistance and balance of osmotic pressure, enhancing their adaptation to estuarine environments. Both genomic investigation and experimental verification showed that strain CJK-A8-3T could be versatile and efficient to use diverse organic nitrogen compounds as carbon and nitrogen sources. Based on phenotypic, chemotaxonomic, phylogenetic and genomic characteristics, strain CJK-A8-3T was identified as a novel Devosia species, named as Devosia aquimaris sp. nov. The type strain is CJK-A8-3T (= MCCC 1K06953T = KCTC 92162T).


Similar content being viewed by others
Data availability
The 16S rRNA gene sequence of strain CJK-A8-3T has been deposited in GenBank under the accession number MZ520614. Whole Genome Shotgun project of strain CJK-A8-3T has been deposited at GenBank under accession number JAIEZN000000000.
References
Bankevich A, Nurk S, Antipov D et al (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 23:455–477
Bourne Y, Henrissat B (2001) Glycoside hydrolases and glycosyltransferases: families and functional modules. Curr Opin Struct Biol 11(5):593–600
Brokaw AM, Eide BJ, Muradian M, Boster JM, Tischler AD (2017) Mycobacterium smegmatis PhoU proteins have overlapping functions in phosphate signaling and are essential. Front Microbiol 8:2523
Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH (2019) GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics (oxford, England) 36(6):1925–1927
Chen J, Xie JP (2011) Role and regulation of bacterial LuxR-like regulators. J Cell Biochem 112(10):2694–2702
Chen Y, Zhu S, Lin D et al (2019) Devosia naphthalenivorans sp. nov., isolated from East Pacific Ocean sediment. Int J Syst Evol Microbiol 69:1974–1979
Dong X, Cai M (2001) Determinative manual for routine bacteriology. Scientific Press, Beijing
Du J, Kook M, Akter S et al (2016) Devosia humi sp. nov., isolated from soil of a Korean pine (Pinus koraiensis) garden. Int J Syst Evol Microbiol 66:341–346
Du D, Xuan WK, Neuberger A et al (2018) Multidrug efflux pumps: structure, function and regulation. Nat Rev Microbiol 16(9):523–539
Fisch A, Bryskier A (2014) Phenicols. In: Bryskier A (ed) Antimicrobial Agents, pp 925–929
Galatis H, Martin K, Kampfer P, Glaeser SP (2013) Devosia epidermidihirudinis sp. nov., isolated from the surface of a medical leech. Antonie Van Leeuwenhoek 103:1165–1171
García-Estepa R, Argandoña M, Reina-Bueno M et al (2006) The ectD gene, which is involved in the synthesis of the compatible solute hydroxyectoine, is essential for thermoprotection of the halophilic bacterium Chromohalobacter salexigens. J Bacteriol 188(11):3774–3784
Gloster TM (2014) Advances in understanding glycosyltransferases from a structural perspective. Curr Opin Struct Biol 28:131–141
Guo LL, Wu D, Sun C et al (2020) Muricauda maritima sp. nov., Muricauda aequoris sp. nov. and Muricauda oceanensis sp. nov., three marine bacteria isolated from seawater. Int J Syst Evol Microbiol 70:6240–6250
Harwood CS, Parales RE (1996) The β-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol 50(1):553–590
Holmquist M (2000) Alpha beta-hydrolase fold enzymes structures, functions and mechanisms. Curr Protein Pept Sci 1(2):209–235
Hove Jensen B, Rosenkrantz TJ, Zechel DL, Willemoës M (2010) Accumulation of intermediates of the carbon-phosphorus lyase pathway for phosphonate degradation in phn mutants of Escherichia coli. J Bacteriol 192:370–374
Huang MM, Guo LL, Wu YH et al (2018) Pseudooceanicola lipolyticus sp. nov., a marine alphaproteobacterium, reclassification of Oceanicola flagellatus as Pseudooceanicola flagellatus comb. nov. and emended description of the genus Pseudooceanicola. Int J Syst Evol Microbiol 68:409–415
Jia YY, Sun C, Pan J et al (2014) Devosia pacifica sp. nov., isolated from deep-sea sediment. Int J Syst Evol Microbiol 64:2637–2641
Kampfer P, Busse HJ, Clermont D et al (2021) Devosia equisanguinis sp. nov., isolated from horse blood. Int J Syst Evol Microbiol 71:5090
Kempf B, Bremer E (1998) Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch Microbiol 170:319–330
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Komagata K, Susuki K (1987) Lipid and cell-wall systematics in bacterial systematics. Methods Microbiol 19:161–207
Kornelsen V, Kumar A (2021) Update on multidrug resistance efflux pumps in Acinetobacter spp. Antimicrob Agents Chemother 65(7):e00514-e521
Kumar S, Stecher G, Tamura K (2016) mega7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lee SD (2007) Devosia subaequoris sp. nov., isolated from beach sediment. Int J Syst Evol Microbiol 57:2212–2215
Lee I, Ouk Kim Y, Park SC, Chun J (2016) OrthoANI: an improved algo-rithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103
Leifson E (1963) Determination of carbohydrate metabolism of marine bacteria. J Bacteriol 85:1183–1184
Lenz KD, Klosterman KE, Mukundan H, Kubicek-Sutherland JZ (2021) Macrolides: from toxins to therapeutics. Toxins 13(5):347
Levasseur A, Drula E, Lombard V et al (2013) Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels 6(1):41
Ley Y, Cheng XY, Ying ZY, Zhou NY, Xu Y (2023) Characterization of two marine lignin-degrading consortia and the potential microbial lignin degradation network in nearshore regions. Microbiol Spectr 11(3):e04424-e4522
Li MT, Xu KQ, Watanabe M, Chen ZY (2007) Long-term variations in dissolved silicate, nitrogen, and phosphorus flux from the Yangtze River into the East China Sea and impacts on estuarine ecosystem. Estuar Coastal Shelf Sci 71:3–12
Lin D, Huang Y, Chen Y et al (2020) Devosia indica sp. nov., isolated from surface seawater in the Indian Ocean. Int J Syst Evol Microbiol 70:340–345
Lin SY, Tsai CF, Hameed A et al (2021) Description of Devosia faecipullorum sp. nov., harboring antibiotic- and toxic compound-resistance genes, isolated from poultry manure. Int J Syst Evol Microbiol 71:4901
Liu Q, Lu X, Tolar BB, Mou X, Hollibaugh JT (2015) Concentrations, turnover rates and fluxes of polyamines in coastal waters of the South Atlantic Bight. Biogeochemistry 123(1):117–133
Liu SM, Qi XH, Li X et al (2016) Nutrient dynamics from the Changjiang (Yangtze River) estuary to the East China Sea. J Mar Syst 154:15–27
Liu M, Liu H, Shi M et al (2021) Microbial production of ectoine and hydroxyectoine as high-value chemicals. Microb Cell Fact 20:76
Long MH, Mora JW (2023) Deoxygenation, acidification and warming in Waquoit Bay, USA, and a shift to pelagic dominance. Estuaries Coasts 46:941–958
Lu HM et al (2020) The evolution history of Fe–S cluster A-type assembly protein reveals multiple gene duplication events and essential protein motifs. Genome Biol Evol 12(3):160–173
Mahato NK, Gupta V, Singh P et al (2017) Microbial taxonomy in the era of OMICS: application of DNA sequences, computational tools and techniques. Antonie Van Leeuwenhoek 110:1357–1371
Martínez A, Ventouras LA, Wilson ST, Karl DM et al (2013) Metatranscriptomic and functional metagenomic analysis of methylphosphonate utilization by marine bacteria. Front Microbiol 4:340
Meier Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence- based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60
Metcalf WW, Wanner BL (1993) Evidence for a fourteen-gene, phnC to phnP locus for phosphonate metabolism in Escherichia coli. Gene 129:27–32
Minnikin DE, O’Donnell AG, Goodfellow M et al (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Mohd Nor MN, Sabaratnam V, Tan GYA (2017) Devosia elaeis sp. nov., isolated from oil palm rhizospheric soil. Int J Syst Evol Microbiol 67:851–855
Moore L, Moore E, Murray R, Stackebrandt E, Starr M (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Nakagawa Y, Sakane T, Yokota A (1996) Transfer of “Pseudomonas riboflavin” (Foster 1944), a gram-negative, motile rod with long-chain 3-hydroxy fatty acids, to Devosia riboflavina gen. nov., sp. nov., nom. rev. Int J Syst Bacteriol 46:16–22
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32(1):268–274
Nishibori N, Fujihara S, Nishijima T (2006) Changes in intracellular polyamine concentration during growth of Heterosigma akashiwo (Raphidophyceae). Fish Sci 72:350–355
Pang Y, Lu W, Chen M, Yan Y et al (2022) Devosia salina sp. nov., isolated from South China Sea sediment. Int J Syst Evol Microbiol 72:5258
Park S, Jung YT, Kim S, Yoon JH (2016) Devosia confluentis sp. nov., isolated from the junction between the ocean and a freshwater lake, and reclassification of two Vasilyevaea species as Devosia enhydra comb. nov. and Devosia mishustinii comb. nov. Int J Syst Evol Microbiol 66:3935–3941
Parks DH, Imelfort M, Skennerton CT et al (2015) CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055
Parte AC, Carbasse JS, Meier-Kolthoff JP, Reimer LC, GöKER M (2020) List of Prokaryotic names with Standing in nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol 70:5607–5612
Peters P, Galinski EA, Trüper HG (1990) The biosynthesis of ectoine. FEMS Microbiol Lett 71:157–162
Reasoner DJ, Geldreich EE et al (1985) A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol 49:1–7
Reitzer L (2003) Nitrogen assimilation and global regulation in Escherichia coli. Annu Rev Microbiol 57:155–176
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131
Rivas R, Willems A, Subba-Rao NS et al (2003) Description of Devosia neptuniae sp. nov. that nodulates and fixes nitrogen in symbiosis with Neptunia natans, an aquatic legume from India. Syst Appl Microbiol 26:47–53
Roberts MC, Schwarz S (2016) Tetracycline and phenicol resistance genes and mechanisms: importance for agriculture, the environment, and humans. J Environ Qual 45(2):576–592
Romanenko LA, Tanaka N, Svetashev VI (2013) Devosia submarina sp. nov., isolated from deep-sea surface sediments. Int J Syst Evol Microbiol 63:3079–3085
Saitou N, Nei M (1987) The neighbor- joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, MIDI technical note 101. USFCC Newsl 20:1–6
Schwarz G, Mendel RR (2006) Molybdenum cofactor biosynthesis and molybdenum enzymes. Annu Rev Plant Biol 57:623–647
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069
Shi W, Zhang B, Jiang Y et al (2021) Structural basis of copper-efflux-regulator-dependent transcription activation. iScience 24(5):10244
Siegele DA (2005) Universal stress proteins in Escherichia coli. J Bacteriol 187(18):6253–6254
Talwar C, Nagar S, Kumar R et al (2020) Defining the environmental adaptations of genus Devosia: insights into its expansive short peptide transport system and positively selected genes. Sci Rep 10:1151
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position- specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tyler B (1978) Regulation of the assimilation of nitrogen compounds. Ann Rev Biochem 47:1127–1162
Vanparys B, Heylen K, Lebbe L, De Vos P (2005) Devosia limi sp. nov., isolated from a nitrifying inoculum. Int J Syst Evol Microbiol 55:1997–2000
Vermassen A, Leroy S, Talon R et al (2019) Cell wall hydrolases in bacteria: insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Front Microbiol 10:331
Wang W-X, Rainbow PS (2020) Environmental pollution of the Pearl River Estuary, China. Status and Impact of Contaminants in a Rapidly Developing Region. In book series: Estuaries of the World (EOTW). Springer Berlin, Heidelberg
Westby CA, Enderlin CS, Steinberg NA, Joseph CM, Meeks JC (1987) Assimilation of 13NH4+ by Azospirillum brasilense grown under nitrogen limitation and excess. J Bacteriol 169:4211–4214
Xu L, Zhang Y, Read N et al (2017) Devosia nitraria sp. nov., a novel species isolated from the roots of Nitraria sibirica in China. Antonie Van Leeuwenhoek 110:1475–1483
Yoo SH, Weon HY, Kim BY et al (2006) Devosia soli sp. nov., isolated from greenhouse soil in Korea. Int J Syst Evol Microbiol 56:2689–2692
Yoon JH, Kang SJ, Park S, Oh TK (2007) Devosia insulae sp. nov., isolated from soil, and emended description of the genus Devosia. Int J Syst Evol Microbiol 57:1310–1314
Zhang DC, Redzic M, Liu HC et al (2012) Devosia psychrophila sp. nov. and Devosia glacialis sp. nov., from alpine glacier cryoconite, and an emended description of the genus Devosia. Int J Syst Evol Microbiol 62:710–715
Zhang H, Yohe T, Huang L et al (2018) dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46(W1):W95–W101
Zhang YX, Yu Y, Luo W et al (2021) Devosia beringensis sp. nov., isolated from surface sediment of the Bering Sea. Int J Syst Evol Microbiol 71:4995
Acknowledgements
We thank Dr. Huirong Li from Polar Research Institute of China for providing Devosia strain D. beringensis S02T. We also appreciated Dr. Aharon Oren for the help with the consultation of species name. Samples were collected onboard of R/V Zheyuke II implementing the open research cruise NORC2019-03-03 supported by NSFC Shiptime Sharing Project. The work was supported by the National Natural Science Foundation of China (grants No. 42176038), Science Foundation of Donghai Laboratory (No. DH-2022KF0211), Scientific Research Fund of the Second Institute of Oceanography, MNR (No. JG1521) and the Project of State Key Laboratory of Satellite Ocean Environment Dynamics, Second Institute of Oceanography (No. SOEDZZ2204).
Funding
The funding was provided by National Natural Science Foundation of China (Grant No 42176038), Science Foundation of Donghai Laboratory (Grant No DH-2022KF0211), the Scientific Research Fund of the Second Institute of Oceanography, MNR (Grant No. JG1521), Project of State Key Laboratory of Satellite Ocean Environment Dynamics,Second Institute of Oceanography (Grant No SOEDZZ2204).
Author information
Authors and Affiliations
Contributions
QL designed research. YRQ, MYL, YHW and CHH performed isolation, deposition and polyphasic taxonomy. YRQ and QL conducted the growth experiment on nitrogen substrates. MYL, YRQ and QL performed the genome analysis. MYL, YRQ and QL drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10482_2023_1924_MOESM2_ESM.tif
Supplemental Fig. 1 Neighbor-joining phylogenetic tree based on 16S rRNA gene sequences showing the phylogenetic relationship of strain CJK-A8-3Tand other isolated Devosia strains. Caulobacter vibrioides CB51T was used as an outgroup. Bootstrap values (>50.0%) based on 1000 replications were shown at branch nodes. Bar, 0.01 substitutions per nucleotide position (TIF 11381 KB)
10482_2023_1924_MOESM3_ESM.tif
Supplemental Fig. 2 Polar lipid diagram of strain CJK-A8-3T, a, total lipid; b, amino lipid; c, glycolipid; d, phospholipid. These abbreviations indicate phosphatidic acid (PA), phosphatidylglycerol (PG), a combination of phospholipids and glycolipids (PGL) and two unidentified glycolipids (GL1, GL2) (TIF 1061 KB)
10482_2023_1924_MOESM4_ESM.tif
Supplemental Fig. 3 Growth curves of strain CJK-A8-3T in modified marine agar 2216 (MA) media individually added with different concentrations (50, 200, 500 and 1000 nM) of six nitrogen substrates: (a) putrescine, (b) spermidine, (c) spermine, (d) glutamic acids, (e) urea and (f) ammonium chloride. Error bars represent the standard errors of three replicates of each treatment. Asterisks indicate the significant differences (One-Way ANOVA; p<0.05) between cell densities at the stationary phase cultured in modified MA with nitrogen compounds and those in modified MA without extra substrate addition (control) (TIF 939 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lai, M., Qian, Y., Wu, YH. et al. Devosia aquimaris sp. nov., isolated from seawater of the Changjiang River estuary of China. Antonie van Leeuwenhoek 117, 29 (2024). https://doi.org/10.1007/s10482-023-01924-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10482-023-01924-y