Abstract
Decision support systems are being developed to assist clinicians in complex decision-making processes by leveraging information from clinical knowledge and electronic health records (EHRs). One typical application is disease risk prediction, which can be challenging due to the complexity of modelling longitudinal EHR data, including unstructured medical notes. To address this challenge, we propose a deep state-space model (DSSM) that simulates the patient’s state transition process and formally integrates latent states with risk observations. A typical DSSM consists of three parts: a prior module that generates the distribution of the current latent state based on previous states; a posterior module that approximates the latent states using up-to-date medical notes; and a likelihood module that predicts disease risks using latent states. To efficiently and effectively encode raw medical notes, our posterior module uses an attentive encoder to better extract information from unstructured high-dimensional medical notes. Additionally, we couple a predictive clustering algorithm into our DSSM to learn clinically useful representations of patients’ latent states. The latent states are clustered into multiple groups, and the weighted average of the cluster centres is used for prediction. We demonstrate the effectiveness of our deep clustering-based state-space model using two real-world EHR datasets, showing that it not only generates better risk prediction results than other baseline methods but also clusters similar patient health states into groups.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
In the domain of personalized healthcare, understanding patients’ latent health states and making clinical decisions heavily rely on utilizing the information contained in electronic health records (EHRs). Patients’ health latent states can be implied by data of different types, including unstructured medical notes, laboratory testing results, clinical events, and other monitoring signals. To achieve personalized healthcare, it is crucial to identify each patient’s health latent states from a large volume of data, which requires intensive domain knowledge and labour resources. AI-based models can greatly assist the clinical decision process by modelling patient EHR data. In this paper, we focus on a novel approach to modelling longitudinal unstructured medical notes, which are collected from multiple hospital visits and used to predict disease risk over time.
To trace the trajectory of patients’ latent states, numerous studies have attempted to model latent variables. Both traditional machine learning techniques, such as L2-regularized logistic regression (Tang et al., 2020) and longitudinal K-means (Mullin et al., 2021), as well as deep learning methods, have been used to analyze longitudinal EHR data. Among the deep learning methods, recurrent neural networks (RNNs) have demonstrated their effectiveness in extracting longitudinal information from EHRs (Choi et al., 2016; Esteban et al., 2016; Lipton et al., 2015; Choi et al., 2016; Ma et al., 2017). However, RNNs have the limitation of being black-box models, making it difficult to have a probabilistic interpret the latent states of patients (Krishnan et al., 2017). To address this issue, several studies (Choi et al., 2016; Ma et al., 2017, 2020; Luo et al., 2020) have investigated the use of time-aware attention mechanisms for analyzing longitudinal EHRs. On the other hand, some researchers (Alaa & van der Schaar, 2019; Oezyurt et al., 2021; Alaa & van der Schaar, 2019) have integrated neural networks with the Hidden Markov Model (HMM) and State-Space Model (SSM) to parameterize state transitions and observations. Compared to RNNs and time-aware attention mechanisms, HMM and SSM with neural networks have the capacity to track changes in latent states through dynamic modelling. They are able to generate predictions and future observations from latent states through a generative model. In this paper, following the approach presented in prior works (Rangapuram et al., 2018; Li et al., 2021; Oezyurt et al., 2021), we couple the concept of state-space models with deep neural networks to introduce a novel framework known as the deep state-space model. This framework is designed specifically for modelling longitudinal patient data, and it incorporates the framework of variational autoencoders (VAEs) (Kingma & Welling, 2013) for learning the state transition and observation processes. Unlike most existing works, our model is one of the first attempts to apply deep state-space models to handle longitudinal unstructured medical notes.
In personalized healthcare, it is crucial to construct a decision-making model whose results are interpretable. Therefore, we also make efforts to provide interpretations of the latent states generated by our disease prediction model. The attention mechanism is frequently used to generate interpretable results, allowing the model to focus on specific parts of the input data when making predictions (Vaswani et al., 2017). RETAIN (Choi et al., 2016) and DIPOLE (Ma et al., 2017) utilized the self-attention mechanism to identify important input features from EHRs. Additionally, auxiliary medical knowledge can be integrated with EHR data using the cross-attention mechanism for disease risk prediction (Mullenbach et al., 2018; Niu et al., 2021a, b). There have also been attempts to couple HMM and SSM with attention mechanisms on latent states of patients in order to capture long-term disease dynamics and different disease states in the health trajectory (Oezyurt et al., 2021; Alaa & van der Schaar, 2019). However, merely understanding which important local features is insufficient for predicting disease progression and disease risk. Instead, we need a deeper understanding of the patient’s hidden states and the process of change in these states. Predictive clustering is a technique for providing cluster-level interpretations for latent states. It groups data samples into clusters in an unsupervised manner. Recently, the use of neural networks for learning latent representations from raw data has gained popularity in predictive clustering tasks (Lee & Van Der Schaar, 2020; Tzirakis et al., 2019; Ghosh et al., 2016). For example, ACTPC (Lee & Van Der Schaar, 2020) and CAMELOT (Aguiar et al., 2022) adopted the approach of learning discrete representations of patient health conditions to accurately describe the future outcome distribution.
In this paper, we will apply the predictive clustering algorithm to group patients’ latent states learnt from longitudinal unstructured medical notes. Specifically, each input medical note will be encoded as a continuous representation, and a similarity-based approach (Zhang et al., 2021) will be used to determine the probability of assigning the continuous representation to different clusters. The cluster assignment probability will be used as weights to obtain a weighted representation of cluster centre embeddings, which will then be used for disease risk prediction. Each latent state can be understood by interpreting the characteristics of its associated clusters. The latent states are updated by the deep state-state model. As illustrated in Fig. 1, our Deep State-space model with the Predictive Clustering for the Risk prediction of diseases, named DSPCR, consists of three modules: the prior module to learn the transition of patients’ latent states for generating the prior of current latent states based on the previous one, the posterior module to approximate the posterior distribution of latent states, and the likelihood module to generate predictions with the exploitation of the predictive clustering algorithm for the disease risk prediction. Our main contributions can be summarized as:
-
We develop a deep state-space model for disease risk prediction using longitudinal medical notes, where patient risks are treated as observations generated from a deep state-space transition process. Our deep state-space model, particularly designed for medical notes of the unstructured text data type, retains the characteristics of probabilistic models and exploits the representation power of deep neural networks.
-
To understand the patients’ latent states learned from the large volume of unstructured raw medical notes, we proposed a deep state-space-based predictive clustering algorithm.
-
To demonstrate the performance of the proposed model, we use two publicly available EHR datasets for both quantitative and qualitative evaluations.
2 Related work
2.1 Disease risk prediction with deep learning methods
In recent years, there has been an increasing interest in the utilization of deep learning methods for predicting disease risk. By harnessing the robust feature extraction capabilities inherent in deep neural networks such as Convolutional Neural Networks (CNNs) (Razavian & Sontag, 2015; Che et al., 2017), RNNs (Xu et al., 2018; Ma et al., 2017; Choi et al., 2016), and BERT (Alsentzer et al., 2019), alongside the advantages offered by parallel processing with GPU/TPU, deep learning methods exhibit substantial potential for enhancing the accuracy and efficiency of risk prediction. These techniques can be classified into two categories based on the type of EHR data: continuous numeric data and unstructured medical notes. For continuous numeric data, several deep learning models have been developed, including RETAIN (Choi et al., 2016), DIPOLE (Ma et al., 2017), RAIM (Xu et al., 2018), and ConCare (Ma et al., 2020), which used RNNs to extract features from laboratory test results or clinical codes. GRAM (Choi et al., 2017) and KAME (Ma et al., 2018), on the other hand, used a knowledge graph to learn embeddings that improve accuracy and interpretation with both sufficient and insufficient EHR data. For unstructured medical notes, models such as CAML (Mullenbach et al., 2018), LEAM (Wang et al., 2018), and LERP (Niu et al., 2021b) utilized the cross-attention mechanism between medical notes and additional clinical information to extract valuable medical phrases for prediction. MNN (Qiao et al., 2019) tried the attention mechanism to guide feature extraction from medical notes using latent information contained in medical codes.
2.2 Modelling longitudinal EHR data
Longitudinal EHR data stores patient health information collected during multiple hospital visits. To model the longitudinal information of EHR data, RNNs, HMM, and AttDMM (Oezyurt et al., 2021) are often used to describe the variations of latent states over several hospital visits of a patient, the structure of which is shown in Fig. 2a, b, and c, respectively.
Typical neural network-based sequential models for modelling longitudinal data. a Recurrent Neural Networks (RNNs). b Deep Hidden Markov Model (HMM). c Attentive Deep Markov Model (AttDMM). \(\Diamond \) denotes a deterministic representation, \(\bigcirc \) denotes probabilistic states, \(\blacksquare \) denotes the neural networks, and \(\triangledown \) denotes the observations and outcomes
For example, GAMENET (Shang et al., 2019), an RNN-based model with an attention mechanism was developed for disease diagnosis and drug recommendation with consecutive hospital visits. CausalHMM (Li et al., 2021) proposed a causal hidden Markov model that learns separate latent representations through supervised tasks such as medical image reconstruction and risk prediction. AttDMM, such as the one proposed in Oezyurt et al. (2021), had been utilized to model longitudinal EHR data, including by tracing patients’ latent states and predicting disease risk from laboratory test results. In addition, ACTPC (Lee & Van Der Schaar, 2020) used a deep predictive clustering of time-series data samples to understand disease progression.
2.3 Predictive clustering-based prediction models
In the previous works for disease risk prediction, the attention mechanism was frequently applied to identify important information and provide interpretation. For example, RETAIN (Choi et al., 2016) and DIPOLE (Ma et al., 2017) used the time-aware attention mechanism to identify the important hospital visits for patients; CAML (Mullenbach et al., 2018) and LDAM (Niu et al., 2021a) adopted the label-dependent attention mechanism to improve the prediction and interpretation. However, there have been relatively few attempts to use unsupervised clustering methods to provide interpretability to predict disease risk, especially for longitudinal medical notes. This is because traditional unsupervised clustering models, such as K-means, hierarchical clustering, and other unsupervised attempts (Zhang et al., 2019; Giannoula et al., 2018), are commonly struggling to meet our prediction requirements. Recently, there have been some attempts to apply an unsupervised clustering model, Predictive Clustering, on structured numeric EHRs to help make predictions over time. Predictive clustering is an unsupervised method but can be used as a visualization to show the latent states of patients by grouping data samples under the guidance of supervised classes. For example, in Lee and Van Der Schaar (2020), a predictive clustering model called ACTPC was proposed as a way to group patients’ latent states into different clusters based on the embedding of their cluster centre, which is guided by a supervised task. In Aguiar et al. (2022), the CAMELOT was developed, which is based on ACTPC but replaces the undifferentiated selector network and is capable of end-to-end training. In this paper, we focus on integrating the predictive clustering algorithm into our disease risk prediction model using unstructured medical notes.
3 Method
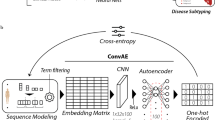
Figure 3 illustrates the workflow of the disease prediction process. The process involves several key steps which are: collecting unstructured medical notes, pre-processing medical notes by removing non-alphabetic characters and stop-words, encoding data to get the latent states, generating the cluster distribution of latent states, and making disease risk prediction. This workflow allows for the efficient and effective prediction of clinical disease risks using unstructured medical notes from EHRs. The following part of this section will focus on introducing our DSPCR model for disease risk prediction using longitudinal medical notes.
3.1 The overview of our model
Suppose each patient n is characterized by a sequence of observations: \(\varvec{x}^{n} = \{\varvec{x}^{n}_1,\ldots ,\varvec{x}^{n}_t,\ldots ,\) \( \varvec{x}^{n}_{T_n}\}\), where each element \(\varvec{x}^{n}_t\) represents the medical notes containing \(N_t^{n}\) words collected at the t-th hospital visit, and \(T_n\) denotes the total number of visits. Let \(\varvec{y}^{n}=\{\varvec{y}^{n}_1,\ldots ,\varvec{y}^{n}_t,\ldots ,\varvec{y}^{n}_{T_n}\}\) indicate the presence of different disease risks observed during multiple visits, where each vector \(\varvec{y}^{n}_t\) contains 1 and 0 values. In the task of predicting the disease risks of patient n, \(\varvec{x}^{n}\) will be used to get the predicted value of \(\varvec{y}^{n}\). All notations to be used in the following subsections are listed in Table 1.
Our DSPCR model adopts the sequential Bayesian updating approach using the up-to-date prior from the state transition model and the likelihood determined by the latest observation to update the current latent state, by computing its posterior distribution according to the Bayes’ rule. In our work, we adopt this approach to infer the patient’s latent state \(\varvec{z}_t^{n}\) at each hospital visit t. Figure 4 gives an overview of our model: a prior module generates the prior distribution of latent state \(\varvec{z}_t^{n}\) from previous latent states; the posterior module approximates the posterior distribution of \(\varvec{z}_t^{n}\) by encoding the information contained in \(\varvec{x}_{t}^{n}\); and the likelihood module adopts the predictive clustering algorithm to generate the observation \(\varvec{y}_{t}^{n}\).
To infer the parameters and latent states of DSPCR, our optimization objective consists of two components: the evidence lower bound on the log data likelihood (ELBO) (Krishnan et al., 2017), and the clustering loss. The ELBO term measures the divergence between the prior and posterior distributions of latent states and also examines the expected likelihood of generating observations. Here, we adopt a Gaussian variational approximation approach such that the distribution of latent states follows the Gaussian distribution, where the mean and standard deviation are approximated by \(\varvec{x}_{t}^{n}\). The clustering loss is to constrain the latent space such that latent states \(\varvec{z}_t^n\) for all n and t can fall into different clusters.
The overview of the proposed DSPCR model. It contains three components: the prior module, the posterior module, and the likelihood module. Key variables are described as follows: \(\varvec{x}_{t}^{n}\) is medical notes of patient n at time t; \(\varvec{z}_{t-1}^{n}\) is the latent state; \(\varvec{Z}_{t-1}^{n} =[\varvec{z}_1^{n},..,\varvec{z}_{t-1}^{n}]\) contains latent states of all past visits; \({\varvec{\mu }^{n}_{t}}\) and \({\varvec{\sigma }^{n}_{t}}\) denote the mean and standard deviation of latent states, where the subscript (p) and (q) indicate the prior and the posterior; \(\hat{\varvec{z}}_{t}^{n}\) is the sampled vector of latent states; \(\varvec{c}_{1:K}\) contains the embeddings of K cluster centers; \(\varvec{o}_{t}^{n}\) and its normalized version \(\varvec{s}_{t}^{n}\) indicates the similarity between \(\hat{\varvec{z}}_{t}^{n}\) and \(\varvec{c}_{k}\) for all \(k\in \{1,\ldots ,K\}\); \(\varvec{u}_t^n\) is the weighted average of \(\varvec{c}_{1:K}\), where the weight is given by \(\varvec{s}_{t}^{n}\); and \(\varvec{\hat{y}}_{t}^{n}\) is the predicted risk vector
3.2 Attentive encoder for the posterior approximation
In this subsection, we focus on describing the posterior module of our DSPCR model as shown in Fig. 5. The variational approximation of the posterior is \( q_\phi (\varvec{z}_{t}^{n} \mid \varvec{Z}_{t-1}^{n},\varvec{x}_{t}^{n})\), where \({\varvec{\mu }^{n}_{t}}^{(q)}\) and \({\varvec{\sigma }^{n}_{t}}^{(q)}\) denote the mean and standard deviation of the posterior respectively, and \(\varvec{Z}_{t-1}^{n} =[\varvec{z}_1^{n},..,\varvec{z}_{t-1}^{n}]\) contains the latent states of all past visits. Specifically, the posterior is parameterized by the attentive encoder network and fully connected networks using \(\varvec{Z}_{t-1}^{n}\) and \(\varvec{x}_{t}\).
For the embedding step, Clinical-BERT (Alsentzer et al., 2019) and the self-attention mechanism is used to embed medical notes \(\varvec{x}_{t}^{n}\) into latent representations. Clinical-BERT is a language understanding model which has been trained on a large clinical corpus with the aim of facilitating various downstream disease-prediction tasks (Johnson et al., 2016). The embedded data is denoted as \(\varvec{E}_{t}^{n} \in {\mathbb {R}}^{D \times N_{t}^{n}}\), where D is the embedding size. For the integrating step, we adapt the self-attention mechanism to assist in capturing the information contained in consecutive words. Firstly, a scaled-dot similarity matrix \(\varvec{G}_{t}^{n} \in {\mathbb {R}}^{ N_{t}^{n} \times N_{t}^{n}}\) is used to represent the similarity between each token from \(\varvec{E}_{t}^{n}\) as follows:
where \(f_1\) and \(f_2\) are two fully connected networks, and \((.)^T\) is the matrix transpose operator. A max-pooling layer together with the SoftMax activation is then adopted to generate an attentive embedding vector of medical notes \(\varvec{e}_{t}^{n} \in {\mathbb {R}}^{D}\):
and
where \(f_3\) is a fully connected network, \(\varvec{g}_{t}^{n} \in {\mathbb {R}}^{N_{t}^{n}}\) is the self-attention score vector, \(\varvec{E}_{t,i}^{n}\) is the i-th column of \(\varvec{E}_{t}^{n}\), \(g_{t,i}^{n}\) is the i-th element of \(\varvec{g}_{t}^{n}\). With \(\varvec{e}_{t}^{n}\) containing the weighted information from \(\varvec{x}_{t}^{n}\), the next step is to combine \(\varvec{e}_{t}^{n}\) with \(\varvec{Z}_{t-1}^{n}\) to generate \(\varvec{v}_{t}^{n}\) as:
where \(f_4\) is a fully connected network, g is the forget gate adopted from the long short-term memory (LSTM) (Hochreiter & Schmidhuber, 1997), BiGRU is the bidirectional Gated Recurrent Unit (GRU) network (Chung et al., 2014), and \(\oplus \) is the concatenation operator. The weighted representation \(\varvec{v}_{t}^{n}\) is then fed into two fully connected networks \(f_5\) and \(f_6\) for generating \({\varvec{\mu }^{n}_{t}}^{(q)}\) and \({\varvec{\sigma }^{n}_{t}}^{(q)}\), respectively. In practice, we can get a sampled state vector from:
where \(\epsilon \in {\mathcal {N}}\ (0,\textbf{I})\) is the random noise.
3.3 State transition network for the prior generation
In the framework of sequential Bayesian inference, the state transition network is used to generate the prior distribution of the current latent state from the previous one. Here, we represent the prior distribution for patient n at time t as:
where the mean and standard deviation of the prior \({\varvec{\mu }^{n}_{t}}^{(p)}\) and \({\varvec{\sigma }^{n}_{t}}^{(p)}\) are parameterized by a GRU network (Chung et al., 2014) and two fully-connected layers \(f_7\) and \(f_8\). Here, \(f_7\) is used to generate the mean vector while \(f_8\) is used to derive the standard deviation vector of the latent states as follows:
3.4 Predictive clustering for the likelihood estimation
In our likelihood module, we integrate predictive clustering into our deep state-space model. All latent states are clustered into K groups, whose center embeddings are denoted as \(\varvec{c}_{1:K}=[\varvec{c}_{1},\ldots ,\varvec{c}_{k},\ldots ,\varvec{c}_{K}]\). Each sampled latent state \(\hat{\varvec{z}}_{t}^{n}\) is approximated as a weighted average of \(\varvec{c}_{1:K}\), where the weight \(\varvec{s}_t^n\) is determined by the similarity between the latent states to each cluster embedding. The weighted average of centre embeddings \(\varvec{u}_t^n\) is used to predict disease risks.
The first step is to detect clusters of latent states and also derive embeddings of cluster centres. Following the approach developed in Van der Maaten and Hinton (2008); Zhang et al. (2021), the probability of assigning \(\hat{\varvec{z}}_{t}^{n}\) to the k-th cluster is calculated by measuring the similarity between \(\hat{\varvec{z}}_{t}^{n}\) and \(\varvec{c}_k\) based on the Student’s t-distribution as follows:
where \(\alpha \) is the degree of freedom of the Student’s t-distribution, and \(\hat{\varvec{z}}_{t}^{n}\) is the latent representation of \(\varvec{x}_{t}^{n}\) generated by the posterior module using Eq. (5). A SoftMax layer is then used to normalize \(\varvec{o}_{t}^{n}=[o_{t}^{n1};\ldots ;o_{t}^{nK}]\) as:
With \(\varvec{s}_{t}^{n}\), we can obtain the weighted average of cluster centre embedding as:
where \(\varvec{c}_{1:K} \in {\mathbb {R}}^{K \times D}\). To learn \(\varvec{c}_{1:k}\), we utilize an auxiliary probability \(w_{t}^{nk}\) as discussed in Xie et al. (2016):
where \(f_k = {\textstyle \sum _{n=1}^{N}} o_{t}^{nk}\) is the soft cluster frequency with the batch size of N. To make the cluster assignment probability close to the auxiliary probability, we will minimize the KL divergence between them, which is defined as:
The clustering-oriented loss averaged across N samples is:
In the likelihood module, a fully connected network \(f_9\) is used to get the predictive values of risks:
The log-likelihood of observing each element of \(\varvec{y}_{t}^{n}\) with the given latent state \(\varvec{z}_{t}^{n}\) can be then defined as:
With the adoption of the Bayesian variational inference, the evidence lower bound (ELBO) related loss is defined as:
where KL(.) measures the Kullback–Leibler divergence between two distributions. \(p_\theta (\varvec{y}_{t}^{n}\mid \varvec{z}_{t}^{n})\) is the likelihood of observing \(\varvec{y}_{t}^{n}\) given the latent state \(\varvec{z}_{t}^{n}\). When \(t>1\), \(q_\phi (\varvec{z}_{t}^{n}\mid \varvec{Z}_{t-1}^{n},\varvec{x}_{t}^{n})\), and \(p_\theta (\varvec{z}_{t}^{n}\mid \varvec{z}_{t-1}^{n})\) are the posterior and the prior of \(\varvec{z}_{t}^{n}\), respectively. For \(\varvec{z}_{1}^{n}\), its prior and posterior are represented as \(p_\theta (\varvec{z}_1)\) and \(q_\phi (\varvec{z}_{1}^{n} \mid \varvec{x}_{1}^{n})\) respectively. \(\phi \) and \(\theta \) represent the parameters of neural networks for the distribution approximation.
The training procedure to optimize DSPCR by minimizing the losses defined in Eqs. (14) and (17) is shown in Algorithm 1.
4 Experiments
4.1 Experimental dataset
Our model and the comparative baselines were trained and evaluated on two publicly available datasets, which are MIMIC-IIIFootnote 1 and N2C2-2014Footnote 2 datasets as summarized in Table 2.
4.1.1 The MIMIC-III dataset
MIMIC-III (Medical Information Mart for Intensive Care III) (Johnson et al., 2016) is a large, publicly accessible dataset that comprises de-identified health data for patients hospitalized at the Beth Israel Deaconess Medical Center’s intensive care unit (ICU) in Boston, Massachusetts. It contains 53,423 EHRs collected from 38,597 patients. The average length of stay in ICUs of patients in MIMIC-III is 4.9 days. We choose upper-level categories of disease risks as defined in Harutyunyan et al. (2019) (acute disease risk, mixed disease risk, and chronic disease risk) to evaluate the performance of the proposed model in the task of risk prediction. The data processing tool in Harutyunyan et al. (2019) is used to extract EHR data and risk indicators from the MIMIC-III data. The stop-words and non-alphabet characters are removed from the medical notes. To check the effect of using data from multiple visits, we extract a longitudinal subset of MIMIC-III, which contains 9,759 EHRs from patients with two or more hospital visits. The average number of visits in the subset data is 2.61. The same data-splitting strategy as in Harutyunyan et al. (2019) was adopted to get the training and test datasets at the ratio of 4:1 for performance evaluation.
4.1.2 The N2C2-2014 dataset
N2C2-2014 (Kumar et al., 2015) is a collection of EHRs and associated annotations for use in natural language processing (NLP) research, which consists of de-identified EHRs from two different hospitals, which contains 1,304 medical notes from 296 individuals in the N2C2-2014, with an average of 4.42 visits per patient. We also remove all stop-words and non-alphabet characters from medical notes. We select four more disease-related disease risk factors, i.e., hyperlipidemia, hypertension, coronary artery disease (CAD), and diabetes as our prediction targets. Performance evaluation makes use of the 4:1 splitting strategy between the training and test datasets.
4.2 Baseline methods
To properly evaluate the proposed methods, we compare our model DSPCR to different baseline models from two distinct categories: Class 1 methods are entirely supervised models for disease risk prediction, whereas Class 2 methods integrate unsupervised predictive clustering in supervised prediction tasks.
The Class 1 baseline methods are listed as follows:
-
SVM and XGBOOST: Support Vector Machines (SVM) and eXtreme Gradient Boosting (XGBOOST) are two popular machine learning algorithms that are used for classification tasks. The word2vec is used to encode medical notes.
-
CAML: Convolutional Attention for Multi-Label classification (CAML) (Mullenbach et al., 2018) is a state-of-the-art disease classification method that provides interpreted classification results based on convolutional neural networks between medical notes and label embeddings by using a cross-attention mechanism.
-
\(\mathcal {B}\)+CAML: For a fair comparison, we use Clinical-BERT (Alsentzer et al., 2019) to replace the encoder layer of CAML.
-
\(\mathcal {B}\)+CAML+ConCare: We incorporate the time-aware attention mechanism from ConCare (Ma et al., 2020) into \(\mathcal {B}\)+CAML to model longitudinal patient hospitalization information.
-
RETAIN: REverse Time AttentIoN model (RETAIN) (Choi et al., 2016) is an RNNs-based interpretable disease risk prediction model by using a reverse time-aware attention mechanism.
-
\(\mathcal {B}\)+RETAIN: For a fair comparison, we use Clinical-BERT (Alsentzer et al., 2019) to replace the encoder layer of RETAIN.
-
DIPOLE: An efficient and accurate DIagnosis Prediction mOdEL (DIPOLE) (Ma et al., 2017) apply a Bi-directional RNNs (Schuster & Paliwal, 1997) with the dual time-aware attention mechanism to replace the reverse time-aware attention mechanism of simple RNNs, resulting in a method that can focus on both future and past information.
-
\(\mathcal {B}\)+DIPOLE: For a fair comparison, we use Clinical-BERT (Alsentzer et al., 2019) to replace the encoder layer of DIPOLE.
The Class 2 baseline methods are listed as follows:
-
Deep K-means: K-means is a well-known unsupervised clustering algorithm. To be able to handle complex medical notes and predict disease risks, we use a deep neural network version of K-means with the Clinical-BERT and fully connected layers for medical node encoding. The K-means model will be trained using all medical note data to discover clusters and hence get cluster centre embeddings by calculating the mean embedding vector of all instances in each cluster. The embedding of the centre to which each instance belongs will then be used for risk prediction.
-
CAMELOT: ACTPC (Lee & Van Der Schaar, 2020) and CAMELOT (Aguiar et al., 2022) are two state-of-the-art predictive clustering algorithms for disease risk prediction, with CAMELOT demonstrating improved predictive performance and training methodologies. Both ACTPC and CAMELOT are not capable of processing unstructured medical data. Instead of concentrating on modelling numerical time-series health monitoring signals, we revised CAMELOT by using Clinical-BERT and the self-attention mechanism to encode medical nodes instead of an RNN-based encoder.
We trained all models with PyTorch on NVIDIA TESLA V100S GPU. The learning rate is set to \(1e^{-5}\) for Clinical-BERT-related models and \(1e^{-3}\) for others, the embedding size of medical notes data is 768, the size of the latent state is 384, the drop-out rate is set to 0.3, and all models are optimized by ADAM. All competing models were trained five times with a fixed set of five different seeds and the results are presented in terms of average indicator performance. The source code of our model can be accessed via.Footnote 3
4.3 The performance of disease risk prediction
4.3.1 Evaluation metrics for disease prediction
We typically employ accuracy, precision, recall, F1 scores, and ROC-AUC score to evaluate the predictive performance of all comparative models.
Precision: Precision is defined as the ratio of correctly predicted positive samples to all predicted positive samples:
Recall: Recall is defined as the ratio of correctly predicted positive samples to all original true positive samples:
F1 score: The F1 score is the harmonic mean of precision and recall:
Accuracy: Accuracy represents the ratio of correctly predicted samples to the total number of samples:
where FN, TN, FP, and TP refer to false negatives, true negatives, false positives, and true positives, respectively.
ROC-AUC: ROC-AUC score measures the area under the receiver operating characteristic curve.
To provide a more thorough perspective of the evaluation results’ performance, we compute micro and macro precision, average, recall, and F1 scores, and display their overall and individual performance in terms of disease risk.
where j indicates the class index and L is the number of classes.
4.3.2 Comparison with purely supervised baselines
From Table 3, we find that deep neural network-based models, especially for Clinical-BERT-related models, have superior predictive power than classical machine learning methods such as SVM and XGBOOST. Although they lead in recall metrics, they do not pay attention to precision metrics. Moreover, we have observed that comparative models utilizing longitudinal data, such as the time-aware attention-based \(\mathcal {B}\)+CAML+ConCare, as well as RNNs-based RETAIN and DIPOLE, exhibit notable improvements in terms of micro and macro F1 scores. These findings strongly imply that incorporating historical information with a time-aware attention mechanism and RNNs models from past hospital visits can significantly enhance disease risk prediction. Among all the baseline models, our DSPCR model consistently achieves higher F1 scores for both datasets. Furthermore, we conducted a comprehensive evaluation of the ROC-AUC score performance. Given that \(\mathcal {B}\)+DIPOLE achieved the highest micro and macro F1 scores among all baseline models, we proceeded to compare our model DSPCR with \(\mathcal {B}\)+DIPOLE separately for different disease risks on both the MIMIC-III dataset and N2C2-2014 dataset. From the analysis of Figs. 6 and 7, it is evident that our model DSPCR consistently achieves superior evaluation performance in terms of the ROC-AUC score on the two datasets. These findings serve as compelling evidence of the efficacy of our deep state-space model in accurately modelling longitudinal data for predictive tasks. Additionally, to eliminate the effect of disease selection on the N2C2-2014 dataset, we performed a similar evaluation of all comparative models for the subset disease risks, as presented in the Appendix. As shown in Table 6, we can obtain consistent findings as before.
4.3.3 Comparison with clustering-based baselines
From Table 3, the deep K-means model and CAMELOT exhibit similar predictive performance on the MIMIC-III dataset and the latter show better performance on the N2C2-2014 dataset with higher F1 scores. Clustering-based baseline models tend to have no significant improvement in predictive accuracy compared to Class 1 methods due to their limited ability to handle unstructured data. However, they remain unique in providing insight into the underlying status of patients at the clustering level. Remarkably, our DSPCR model outperforms both deep K-means models and CAMELOT with higher values of all metrics for both datasets and also supports clustering-level evidence for patents’ latent state. This observation implies that our model is the state-of-the-art predictive clustering method in the disease risk prediction task utilizing unstructured medical note data.
4.3.4 Model complexity analysis
Figure 8 illustrates the computation time of our model, DSCPR, alongside several baseline models. All comparative models were executed with the same batch size and number of epochs on the Tesla V100S GPU and Xeon Gold 6226 CPU. It is evident from Fig. 8 that SVM, XGBOOST, and CAML exhibit the shortest computation times compared to the other models being compared. However, it is noteworthy that these models demonstrate the poorest evaluation performance on the MIMIC-III dataset and N2C2-2014 dataset. On the other hand, the \(\mathcal {B}\)+CAML+ConCare model shows the highest computation time while maintaining a relatively satisfactory evaluation performance. In addition, our model, DSPCR, and the baseline model \(\mathcal {B}\)+DIPOLE exhibit similar computational performance on the MIMIC-III dataset and N2C2-2014 data in Fig. 8. However, our model outperforms \(\mathcal {B}\)+DIPOLE in terms of evaluation metrics such as micro/macro F1 scores and accuracy.
4.4 The performance of clustering latent health states
Our model can not only predict disease risks but can also group latent states into different clusters. Here, we would like to demonstrate the performance of clustering patients’ latent health states in both quantitative and qualitative ways.
4.4.1 Quantitative evaluation
We adopt standard clustering evaluation metrics, including Silhouette score (SIL) (Rousseeuw, 1987), Davies-Bouldin Index (DBI) (Davies & Bouldin, 1979), and Variance Ratio Criterion (VRC) (Caliński & Harabasz, 1974) to evaluate the performance of clustering results in the absence of cluster labels.
-
Silhouette score (SIL) reflects the consistency of the clustering results by measuring the degree of dispersion between clusters. The SIL score ranges in \([-1, +1]\): if the score is close to 1, it means that the sample has a reasonable clustering result; if it is close to -1, it is more appropriate if the sample was clustered in its neighbouring cluster; if it is close to 0, then it indicates that the sample is on the boundary of two clusters (Rousseeuw, 1987).
-
Davies-Bouldin Index (DBI) measures the ratio between the intra-cluster dispersion and inter-cluster separation. A lower DBI value implies better clustering results (Davies & Bouldin, 1979).
-
Variance Ratio Criterion (VRC) measures a ratio of the sum of inter-cluster dispersion and the sum of intra-cluster dispersion for all clusters. A larger VRC value indicates better clustering results (Caliński & Harabasz, 1974).
Table 4 shows the performance of clustering on both datasets. We can find that our DSPCR model outperforms all comparative models with the best SIL, DBI, and VRC values. This finding suggests that we can produce state-of-the-art clustering results while retaining the best predictive performance.
4.4.2 Case studies
To visualize the cluster-level evidence provided by our model DSPCR for the latent states of patients, we show the cluster assignment and chief complaints of six randomly selected patients with three hospital visits from the test dataset in Fig. 9. In this section, we aim to investigate the effectiveness of the predictive clustering module of our model DSPCR: 1) whether it can accurately track the latent states of patients across hospital visits 2) whether patients with similar chief complaints are assigned the same cluster-ID, otherwise assigned to different cluster-ID. From the cluster assignment indicated by green blocks, the answer to the first question is obvious: the detected latent health states of patients vary with different hospital visits. For the second question, we can also obtain the answer from Fig. 9. Patient 1 is assigned to cluster 1 for all three hospital visits; patient 2 is assigned to cluster 3, cluster 4, and cluster 1 on the 1st, 2nd, and 3rd visit, respectively; patient 3 first stays in cluster 3 and then remains in cluster 4 for the rest two visits; patients 4 stays in cluster 4 for the first two visits and then moves to cluster 5; patient 5 experiences a state transition in a similar way to patient 4, but the first two visits were assigned to cluster 2; patient 6 remains in cluster 3 for all three hospital visits. As a result, we can conclude that patients with the same chief complaint are assigned the same clustering ID.
4.5 Ablation study
We conduct ablation studies to investigate the impacts of 1) adopting the state-space modelling approach to incorporate historical information via the prior module and 2) utilizing predictive clustering in the likelihood module for prediction. DSPCR-B is the ablated version of DSPCR in which both the state transition network and the predictive clustering modules are removed. DSPCR-C keeps the predictive clustering network of DSPCR while the state transition network is discarded. From Table 5, we can find that DSPCR-B and DSPCR-C have similar risk prediction performances. Considering the interpretability of latent states brought by DSPCR-C, we would say that DSPCR-C is better than DSPCR-B whose results lack interpretability. After comparing DSPCR with its ablated version DSPCR-C, we can find that the complete version can obtain higher F1 values, especially for the N2C2-2014 dataset. This observation reflects the impacts of adopting the state-space modelling approach.
4.6 Sensitivity analysis
In the above experiments, we set the number of clusters to 8 and 16 for MIMIC-III and N2C2-2014 respectively. The way we set these numbers follows the strategy adopted in Lee and Van Der Schaar (2020), where the number of clusters is set as \(2^L\) and L is the number of disease risks. Here, we investigate how the number of clusters would affect the performance. The micro and macro F1 scores for different numbers of clusters during the training process are shown in Fig. 10. We can see that for the MIMIC-III dataset, all models have experienced sharp decreases in performance evaluation metrics in the first 20 epochs, followed by consistent increases in the following epochs. For the N2C2 dataset, the F1 scores remain largely stable after the first few epochs.
A noteworthy finding from Fig. 10 is that the red line is generally above the other two lines after 20 epochs, suggesting that 8 and 16 are appropriate choices of the cluster number for MIMIC-III and N2C2-2014 respectively.
5 Conclusion
In this paper, a novel deep state-space modelling with the predictive clustering model is proposed to predict disease risks using longitudinal unstructured medical notes. The deep state-space model, which both inherits the representation power of deep neural networks and retains the structured representations of probabilistic models, has been successfully applied to model longitudinal medical notes generated from multiple hospital visits. To encode raw medical notes from their original vocabulary space into latent representations, the clinical language model together with the attention mechanism is utilized. Notably, we adopt the predictive clustering approach to represent patient latent states from different hospital visits. Our work would help to move towards interpretable AI for clinical decision-making by providing cluster-level evidence for the prediction. When applied to real-world EHR datasets, our model demonstrated its strong predictive power and ability to group different patient states. The proposed model will greatly assist clinicians in the disease risk prediction task by uncovering the information hidden in longitudinal medical notes.
References
Aguiar, H., Santos, M., Watkinson, P., & Zhu, T. (2022). Learning of cluster-based feature importance for electronic health record time-series. In International conference on machine learning (pp. 161–179). PMLR.
Alaa, A., & van der Schaar, M. (2019). Attentive state-space modeling of disease progression.
Alsentzer, E., Murphy, J.R., Boag, W., Weng, W.-H., Jin, D., Naumann, T., & McDermott, M. (2019). Publicly available clinical bert embeddings. arXiv preprint arXiv:1904.03323
Caliński, T., & Harabasz, J. (1974). A dendrite method for cluster analysis. Communications in Statistics-Theory and Methods, 3(1), 1–27.
Che, Z., Cheng, Y., Sun, Z., & Liu, Y. (2017). Exploiting convolutional neural network for risk prediction with medical feature embedding. arXiv preprint arXiv:1701.07474
Choi, E., Bahadori, M. T., Schuetz, A., Stewart, W. F., & Sun, J. (2016). Doctor ai: Predicting clinical events via recurrent neural networks. In Machine learning for healthcare conference (pp. 301–318). PMLR
Choi, E., Bahadori, M.T., Song, L., Stewart, W.F., & Sun, J. (2017). Gram: graph-based attention model for healthcare representation learning. In Proceedings of the 23rd ACM SIGKDD international conference on knowledge discovery and data mining (pp. 787–795).
Choi, E., Bahadori, M. T., Sun, J., Kulas, J., Schuetz, A., & Stewart, W. (2016). Retain: An interpretable predictive model for healthcare using reverse time attention mechanism. Advances in neural information processing systems, Vol. 29.
Chung, J., Gulcehre, C., Cho, K., & Bengio, Y. (2014). Empirical evaluation of gated recurrent neural networks on sequence modeling. arXiv preprint arXiv:1412.3555
Davies, D. L., & Bouldin, D. W. (1979). A cluster separation measure. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2, 224–227.
Esteban, C., Staeck, O., Baier, S., Yang, Y., & Tresp, V. (2016). Predicting clinical events by combining static and dynamic information using recurrent neural networks. In 2016 IEEE international conference on healthcare informatics (ICHI) (pp. 93–101). IEEE.
Ghosh, S., Cheng, Y., & Sun, Z. (2016). Deep state space models for computational phenotyping. In 2016 IEEE international conference on healthcare informatics (ICHI) (pp. 399–402). IEEE.
Giannoula, A., Gutierrez-Sacristán, A., Bravo, Á., Sanz, F., & Furlong, L. I. (2018). Identifying temporal patterns in patient disease trajectories using dynamic time warping: A population-based study. Scientific Reports, 8(1), 1–14.
Harutyunyan, H., Khachatrian, H., Kale, D. C., Ver Steeg, G., & Galstyan, A. (2019). Multitask learning and benchmarking with clinical time series data. Scientific Data, 6(1), 96.
Hochreiter, S., & Schmidhuber, J. (1997). Long short-term memory. Neural Computation, 9(8), 1735–1780.
Johnson, A. E., Pollard, T. J., Shen, L., Li-Wei, H. L., Feng, M., Ghassemi, M., Moody, B., Szolovits, P., Celi, L. A., & Mark, R. G. (2016). Mimic-iii, a freely accessible critical care database. Scientific Data, 3(1), 1–9.
Kingma, D.P., & Welling, M. (2013). Auto-encoding variational bayes. arXiv preprint arXiv:1312.6114
Krishnan, R., Shalit, U., & Sontag, D. (2017). Structured inference networks for nonlinear state space models. In Proceedings of the AAAI conference on artificial intelligence, Vol. 31.
Kumar, V., Stubbs, A., Shaw, S., & Uzuner, Ö. (2015). Creation of a new longitudinal corpus of clinical narratives. Journal of Biomedical Informatics, 58, 6–10.
Lee, C., & Van Der Schaar, M. (2020). Temporal phenotyping using deep predictive clustering of disease progression. In International conference on machine learning (pp. 5767–5777). PMLR.
Li, J., Wu, B., Sun, X., & Wang, Y. (2021). Causal hidden Markov model for time series disease forecasting. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition (pp. 12105–12114).
Lipton, Z. C., Kale, D. C., Elkan, C., & Wetzel, R. (2015). Learning to diagnose with lstm recurrent neural networks. arXiv preprint arXiv:1511.03677
Luo, J., Ye, M., Xiao, C., & Ma, F. (2020). Hitanet: Hierarchical time-aware attention networks for risk prediction on electronic health records. In Proceedings of the 26th ACM SIGKDD international conference on knowledge discovery & data mining (pp. 647–656).
Ma, F., Chitta, R., Zhou, J., You, Q., Sun, T., & Gao, J. (2017). Dipole: Diagnosis prediction in healthcare via attention-based bidirectional recurrent neural networks. In Proceedings of the 23rd ACM SIGKDD international conference on knowledge discovery and data mining (pp. 1903–1911).
Ma, F., You, Q., Xiao, H., Chitta, R., Zhou, J., & Gao, J. (2018). Kame: Knowledge-based attention model for diagnosis prediction in healthcare. In Proceedings of the 27th ACM international conference on information and knowledge management (pp. 743–752).
Ma, L., Zhang, C., Wang, Y., Ruan, W., Wang, J., Tang, W., Ma, X., Gao, X., & Gao, J. (2020). Concare: Personalized clinical feature embedding via capturing the healthcare context. In Proceedings of the AAAI conference on artificial intelligence (Vol. 34, pp. 833–840).
Mullenbach, J., Wiegreffe, S., Duke, J., Sun, J., & Eisenstein, J. (2018). Explainable prediction of medical codes from clinical text. arXiv preprint arXiv:1802.05695
Mullin, S., Zola, J., Lee, R., Hu, J., MacKenzie, B., Brickman, A., Anaya, G., Sinha, S., Li, A., & Elkin, P. L. (2021). Longitudinal k-means approaches to clustering and analyzing ehr opioid use trajectories for clinical subtypes. Journal of Biomedical Informatics, 122, 103889.
Niu, S., Qin, Y., Song, Y., Guo, Y., & Yang, X. (2021). Label dependent attention model for disease risk prediction using multimodal electronic health records. In Proceedings of the IEEE conference on data mining (pp. 455–464).
Niu, S., Song, Y., Qin, Y., Guo, Y., & Yang, X. (2021). Label-dependent and event-guided interpretable disease risk prediction using ehrs. In Proceedings of the IEEE international conference on bioinformatics and biomedicine (BIBM)
Oezyurt, Y., Kraus, M., Hatt, T., & Feuerriegel, S. (2021). Attdmm: An attentive deep markov model for risk scoring in intensive care units. arXiv preprint arXiv:2102.04702
Qiao, Z., Wu, X., Ge, S., & Fan, W. (2019). Mnn: Multimodal attentional neural networks for diagnosis prediction. Extraction, 1, 1.
Rangapuram, S. S., Seeger, M. W., Gasthaus, J., Stella, L., Wang, Y., & Januschowski, T. (2018). Deep state space models for time series forecasting. Advances in Neural Information Processing Systems, 31, 7785–7794.
Razavian, N., & Sontag, D. (2015). Temporal convolutional neural networks for diagnosis from lab tests. arXiv preprint arXiv:1511.07938
Rousseeuw, P. J. (1987). Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. Journal of Computational and Applied Mathematics, 20, 53–65.
Schuster, M., & Paliwal, K. K. (1997). Bidirectional recurrent neural networks. IEEE Transactions on Signal Processing, 45(11), 2673–2681.
Shang, J., Xiao, C., Ma, T., Li, H., & Sun, J. (2019). Gamenet: Graph augmented memory networks for recommending medication combination. In Proceedings of the AAAI conference on artificial intelligence (Vol. 33, pp. 1126–1133).
Tang, S., Chappell, G. T., Mazzoli, A., Tewari, M., Choi, S. W., & Wiens, J. (2020). Predicting acute graft-versus-host disease using machine learning and longitudinal vital sign data from electronic health records. JCO Clinical Cancer Informatics, 4, 128–135.
Tzirakis, P., Nicolaou, M.A., Schuller, B., & Zafeiriou, S. (2019). Time-series clustering with jointly learning deep representations, clusters and temporal boundaries. In 2019 14th IEEE international conference on automatic face & gesture recognition (FG 2019) (pp. 1–5). IEEE.
Van der Maaten, L., & Hinton, G. (2008). Visualizing data using t-sne. Journal of Machine Learning Research, 9(11).
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A.N., Kaiser, Ł., & Polosukhin, I. (2017). Attention is all you need. In Advances in neural information processing systems (pp. 5998–6008).
Wang, G., Li, C., Wang, W., Zhang, Y., Shen, D., Zhang, X., Henao, R., & Carin, L. (2018). Joint embedding of words and labels for text classification. In Proceedings of the 56th annual meeting of the association for computational linguistics (volume 1: Long papers) (pp. 2321–2331). Association for Computational Linguistics, Melbourne, Australia. https://aclanthology.org/P18-1216
Xie, J., Girshick, R., & Farhadi, A. (2016). Unsupervised deep embedding for clustering analysis. In International conference on machine learning (pp. 478–487). PMLR.
Xu, Y., Biswal, S., Deshpande, S. R., Maher, K. O., & Sun, J. (2018). Raim: Recurrent attentive and intensive model of multimodal patient monitoring data. In Proceedings of the 24th ACM SIGKDD international conference on knowledge discovery & data mining (pp. 2565–2573).
Zhang, D., Nan, F., Wei, X., Li, S., Zhu, H., McKeown, K., Nallapati, R., Arnold, A., & Xiang, B. (2021). Supporting clustering with contrastive learning. arXiv preprint arXiv:2103.12953
Zhang, X., Chou, J., Liang, J., Xiao, C., Zhao, Y., Sarva, H., Henchcliffe, C., & Wang, F. (2019). Data-driven subtyping of Parkinson’s disease using longitudinal clinical records: A cohort study. Scientific Reports, 9(1), 1–12.
Acknowledgements
This work is supported by the National Key Research and Development Program of China (No. 2021ZD0113303), and the National Natural Science Foundation of China (Nos. 62022052, 62276159).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no competing interests to declare that are relevant to the content of this article. This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix A: Subset evaluation on N2C2-2014 dataset
Appendix A: Subset evaluation on N2C2-2014 dataset
See Table 6.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Niu, S., Ma, J., Yin, Q. et al. A deep clustering-based state-space model for improved disease risk prediction in personalized healthcare. Ann Oper Res (2024). https://doi.org/10.1007/s10479-023-05817-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10479-023-05817-1