Abstract
It is standard cardiology practice for patients suffering from ventricular arrhythmias (the main cause of sudden cardiac death) belonging to high risk populations to be treated via the implantation of Subcutaneous Implantable cardioverter-defibrillators (S-ICDs). S-ICDs carry a risk of so-called T wave over sensing (TWOS), which can lead to inappropriate shocks that carry an inherent health risk. For this reason, according to current practice patients’ Electrocardiograms (ECGs) are manually screened by a cardiologist over 10 s to assess the T:R ratio—the ratio between the amplitudes of the T and R waves which is used as a marker for the likelihood of TWOS—with a plastic template. Unfortunately, the temporal variability of a patient’ T:R ratio can render such a screening procedure, which relies on an inevitably short ECG segment due to its manual nature, unreliable. In this paper, we propose and investigate a tool based on deep learning for the automatic prediction of the T:R ratios from multiple 10-second segments of ECG recordings capable of carrying out a 24-hour automated screening. Thanks to the significantly increased screening window, such a screening would provide far more reliable T:R ratio predictions than the currently utilized 10-second, template-based, manual screening is capable of. Our tool is the first, to the best of our knowledge, to fully automate such an otherwise manual and potentially inaccurate procedure. From a methodological perspective, we evaluate different deep learning model architectures for our tool, assess a range of stochastic-gradient-descent-based optimization methods for training their underlying deep-learning model, perform hyperparameter tuning, and create ensembles of the best performing models in order to identify which combination leads to the best performance. We find that the resulting model, which has been integrated into a prototypical tool for use by clinicians, is able to predict T:R ratios with very high accuracy. Thanks to this, our automated T:R ratio detection tool will enable clinicians to provide a completely automated assessment of whether a patient is eligible for S-ICD implantation which is more reliable than current practice thanks to adopting a significantly longer ECG screening window which better and more accurately captures the behavior of the patient’s T:R ratio than the current manual practice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
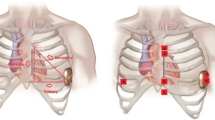
Sudden Cardiac Death (SCD) is one of the main causes of death worldwide. Most incidences of SCD can be attributed to ventricular arrhythmias (Adabag et al., 2010). For patients belonging to high risk populations, it is highly recommended that Implantable Cardioverter-Defibrillators (ICDs) be used for preventing ventricular arrhythmias from triggering an SCD (Kusumoto et al., 2018; Priori et al., 2015). S-ICDs are comprised of a can and of a subcutaneous leadFootnote 1 (shown in Fig. 1a), which can deliver a shock to treat episodes of ventricular arrhythmias. While S-ICDs have been proven to have equally effective sensing capabilities as other forms of ICD (Boersma et al., 2017), S-ICDs entirely avoid the classical complications other ICDs suffer from by utilizing a totally avascular approach (Knops et al., 2020). Unfortunately, the method that is currently used by S-ICDs to detect ventricular arrhythmias is known to be vulnerable to T Wave Over Sensing (TWOS) (van Rees et al., 2011). If the amplitude of the T and R waves are similar (Fig. 1b shows a diagram of the PQRST complex—the ECG (Electrocardiogram) of a single heartbeat—comprised of 5 main waveforms, i.e., the P, Q, R, S, and T waves), the T wave can be misinterpreted as a second R wave and this apparent doubling in heart rate can be incorrectly identified by the S-ICD as an episode of ventricular arrhythmia. Such an error can cause the S-ICD to deliver an inappropriate shock, leading to increased morbidity and mortality (van Rees et al., 2011).
a An S-ICD with the underlying human anatomy showing the can (pulse generator), the sensing electrodes (Pr and D) and the vectors between them. b Diagram of a single PQRST complex, comprised of the P, Q, R, S, and T waves. The figure also labels the QRS complex (a section of the PQRST complex featuring the Q, R, and S waves only). © Boston Scientific Corporation or its affiliates. Reproduced with permission
According to current practice, patients are screened before S-ICD implantation by analyzing short, non-invasive, three-lead, surface ECGs to assess their risk of TWOS. This ECG recording spans several PQRST complexes and, for its recording, the electrodes are placed in the locations that the sensing electrodes of an S-ICD would occupy. The T:R ratio (the ratio between the T wave’s and the R wave’s respective amplitudes) is used to predict a patient’s S-ICD implantation eligibility. Patients with high T:R ratios are deemed to have an increased risk of TWOS and, as such, are not eligible for S-ICD implantation. It is current practice to measure the T:R ratio by locating a plastic template (designed to fit PQRST complexes with acceptably low T:R ratios (below 1/3) completely within its boundary) over the PQRST complexes within a recording of the patient’s ECG. The patient passes the screening if the T:R ratio of at least one lead is suitably low. Despite this screening process, most of the inappropriate shocks that take place in patients implanted with S-ICDs are still caused by TWOS (Knops et al., 2020).
Crucially, the T:R ratio of a patient can vary over time due to frequent temporal variations in the R and T waves’ respective amplitudes due to multiple factors such as a variation in electrolyte levels (Madias et al., 2001; Madias, 2005; Fosbøl et al., 2008; Assanelli et al., 2013). In patients implanted with S-ICDs, these variations commonly go unnoticed as so-called silent TWOS episodes. As such, they carry a considerably high risk for a patient to be subjected to inappropriate shocks. The ECG recording currently used for the screening process has a short duration of roughly 10 s and, as such, it does not allow for taking the variability of the T:R ratio over an extended period into consideration. It follows that a patient whose T:R ratio is usually low could fail the screening and a patient whose T:R ratio is usually high could pass it.
In this paper, we outline a methodology for constructing models capable of accurately and reliably predicting the T:R ratio of multiple given 10-second, single-lead ECG recording. Thus, a three-lead, 24-hour ECG recording can then be broken into its constituent 10-second, single-lead ECG signals which can then be passed to our model to output a predicted T:R ratio for each ECG signal. This process would enable 24-hour automated screening, which, thanks to its significantly increased screening window, would provide far superior insight into the normal variations in the patient’s T:R ratio as well as a greater level of detail and reliability than the currently utilized 10-second screening is capable of.
The paper is organized as follows. In Sect. 2, we provide an overview of previous works that are related to ours. In Sect. 3, we first introduce our methodology and the preprocessing techniques we use to de-noise the ECG signals. We then detail the structure of the deep learning layers we use and the architectures of the deep learning models we propose for the T:R ratio prediction task. We then outline the way our models are evaluated, the way we build ensemble models, and the way hyperparameter tuning is carried out (which is key to the performances we achieve). In Sect. 4, we assess the accuracy of our models by selecting the best training algorithm, tune the corresponding hyperparameters, and create ensemble models. In Sect. 5, we demonstrate how our best-performing models can be incorporated into a clinical screening process for predicting the T:R ratio for a time much longer than what the current (manual) practice allows for.
2 Related works
To the best of our knowledge, the problem of predicting T:R ratios from ECG signals has not yet been explored in the literature. The only works on such a topic are the preliminary work we (the authors of this paper) published in (Dunn et al., 2021) (which has a strong clinical focus and lacks the more advanced machine-learning developments we outline here) and three other papers we published on the clinical aspects of applying our tool within a clinical environment (ElRefai et al., 2022a, b, c, 2023). In particular, due to our work being the first to automatize an otherwise manual procedure, it is not possible to compare it to any previously-established state of the art.
The developments from our preliminary work to this paper are significant. Firstly, we present a detailed methodology, providing details on how we construct and train our deep neural networks. Secondly, in this work we perform a much more extensive, reliable and detailed set of experiments to determine the best network architecture. Finally, we conduct numerous additional experiments to select the best optimisation algorithm to use for model training, and also tune the model’s hyperparameters (many details of these methods and algorithms are provided in our methodology Sect. 3). Beyond the methodological contributions, we find that the clinical tool resulting from the neural network models we developed is able to predict T:R ratios from ECG signals with a much greater accuracy than in our previous works.
The tool we propose is based on deep learning and, in particular, on Convolutional Neural Networks (CNNs). In ECG analysis, CNNs have been used for classifying atrial fibrillation (Fan et al., 2018; Pourbabaee et al., 2018), heart attacks (Liu et al., 2017) and other arrhythmias (Kiranyaz et al., 2015; Zhang et al., 2020; Sangaiah et al., 2020; Lih et al., 2020), as well as for predicting blood pressure (Miao et al., 2020). Differently from these works where the multi-lead ECG is used as input for the CNN, in our model we include the additional step of transforming the 1-dimensional ECG signal of each lead into a 2-dimensional phase-space reconstruction (PSR) image which is then fed as input to the CNN. PSR images are typically used for computer vision tasks which learn to extract relevant features from them but, to the best of our knowledge, have never been used as input to a CNN. As a second element of novelty and differently from most of the literature where CNNs are used for classification (see Babu et al., 2016 for an exception), in this work we use them to perform a regression task.
In the literature, machine learning methods have been typically used to classify various Cardiovascular Diseases (CVDs) from ECG data (Vemishetty et al., 2019, 2016; Rocha et al., 2008; Roberts et al., 2001; Zhang et al., 2020; Pourbabaee et al., 2018; Kiranyaz et al., 2015; Fan et al., 2018; Lih et al., 2020). Machine learning has also been used for analyzing ECGs to detect heart attacks and seizures (Lee et al., 2014; Liu et al., 2017) as well as to predict patients’ blood pressure (Miao et al., 2020). Brain computer interfaces (BCI) have been created using brain ECG analysis for detecting which body part was being used to complete a task (Djemal et al., 2016; Chen et al., 2014). The creation of PSR images from ECG data is a commonly employed technique in ECG analysis. Manual approaches for extracting features from the PSR matrices (using box counting or calculating column and row statistics) have been used for, e.g., predicting CVD (Vemishetty et al., 2016, 2019; Rocha et al., 2008; Roberts et al., 2001), creating BCIs (Chen et al., 2014; Djemal et al., 2016) and detecting facial expressions Dawid (2019). Differently from such works, in this paper we input the entirety of the PSR matrix to a (deep learning) model which, during its training, learns to extract relevant features. We are not aware of another work where the entire PSR image is used in such a way.
3 Methodology
Predicting T:R ratios by locating T and R waves, measuring their peaks and calculating the T:R ratio (manually, with a plastic template) is vulnerable to TWOS when the T wave and R wave have similar characteristics. In this section, we propose a method for building deep learning models which are able to predict the T:R ratio of a single-lead ECG segments without explicitly detecting R and T peaks, which could then be used for 24-hour automated screening. A flowchat giving an overview of our methodology can be found in Fig. 2.
3.1 ECG signal preprocessing
Before using the ECG data as input to our deep learning models, we preprocess it to calculate the T:R ratios, filter out noise from the ECG signal and generate PSR images. This process, outlined in Fig. 3 (Lugovaya, 2005), is explained in this section.Footnote 2
The data used in this paper consists of 10-second three-lead ECG signals collected from a Holter recording. The vectors used by the leads in this recording are the same vectors that are used by the S-ICD. These three-lead ECG recordings are then broken into three single-lead ECG signals. We denote each 10-second segment with sampling frequency f by the vector \((x_1, \ldots , x_{10f})\). Each such segment is manually annotated with the positions of the T wave peaks at indexes \(\{T_1, \ldots , T_n\}\) and R wave peaks at indexes \(\{R_1, \ldots , R_n\}\). Using these annotations, we calculate the average T:R ratio (the dependent variable) as
This quantity is negative when the T wave has negative amplitude. While, in clinical use, positive and negative T:R ratios are considered as equivalent and, thus, only the magnitude of the T:R ratio is used, when analyzing the single lead ECG signals we note that positive or negative T waves are significantly different. Therefore, when building models for predicting T:R ratios of single-lead ECG signals, we retain the T:R ratio’s sign (i.e., its positivity or negativity).
The 10-second, single-lead segments of ECG signal are filtered to remove various forms of noise. Firstly, a one-dimensional Discrete Wavelet Transformation (DWT) is used to correct the baseline drift. We then create a 9 level decomposition of the ECG signal using the Daubechies 8 (db8) wavelet. The level 9 coefficients are used to reconstruct the unstable baseline of the ECG signal which is then subtracted from the original signal. The resulting signal has a stable baseline of 0. Secondly, the 50Hz power-line noise is suppressed using bandstop filtering. Finally, any remaining high frequency noise is suppressed using a lowpass filter. Figure 4a show an ECG segment with a significant baseline drift before any filtering. Figure 4b shows the same segment after filtering has been applied.
TWOS is particularly likely to occur in PQRST complexes with small R waves. To address this, the standard algorithm employed within S-ICDs will search the QRS complex (shown in Fig. 1a) for negative Q or S waves with greater amplitude than that of the R wave. Provided that the amplitude of the largest wave in the QRS complex is significantly larger than that of the T wave, the S-ICD will not deliver a shock. For this reason, we are not strictly interested in predicting the T:R ratio but, rather, the ratio between the amplitudes of the wave of greatest amplitude in the QRS complex and the T wave. To account for this in our analysis, after filtering (including baseline drift correction), we search for the peak of greatest magnitude within a narrow region surrounding each R peak label. For simplicity, in the rare case that the largest peak found is not the R peak, these new peaks are assigned as the new R peak despite, in fact, being a Q or an S wave. As R waves are always positive, to ensure that all signals handed to our models have a positive wave labeled as the R wave, when the R peak label is moved to a Q or an S wave, the whole ECG signal is flipped, making these waves positive. Figure 4c gives an example of an ECG segment (which has had filtering applied) with a very small R wave and a large negative wave in the QRS complex. Figure 4d shows the same segment with its reassigned R peaks after negative QRS flipping.
a The first 5 s of an unfiltered 10-second ECG segment showing significant baseline drift with R peaks in red and T peaks in blue. b The signal shown in Fig. 4a post-filtering. c The first 5 s of a 10second ECG segment with very small R wave but a large negative wave in the QRS complex with R peaks in red and T peaks in blue. d The signal shown in Fig. 4c after flipping and relabeling. e A \(32 \times 32\) pixel PSR image created from 10 seconds of single-lead ECG signal. f The PSR image shown in Fig. 4e darkened for readability
The one-dimensional time series \(x_1, x_2, \ldots , x_n\) representing the filtered and potentially flipped single ECG signal of a single lead is then transformed to a two-dimensional Phase Space Reconstruction (PSR) image. While high-dimension PSR transformations have been used in the field of BCI (Chen et al., 2014; Djemal et al., 2016), two-dimensional PSR transformations are more commonly used in the literature on ECG analysis (Rocha et al., 2008; Krishnan et al., 2007; Vemishetty et al., 2019; Roberts et al., 2001). The transformed time-series is given as
where \(q = \max \left\{ |x_i |: i = 1, \ldots , n\right\} \) and each scaled point \(\frac{1}{q}x_i\) in the time-series signal is mapped to a phase space vector comprised of the original scaled point and the scaled point \(\tau \) readings before it \(\left[ (\frac{1}{q}x_{i-\tau }, \frac{1}{q}x_{i})\right] \). The phase space plot (the plot of all phase space vectors in \(\mathbb {R}^2\), ranging from \(-1\) to \(+1\) on each axis) is divided into \(N^2\) squares which we denote by \(g_{ij}\) for all \(i,j\in \{1,\ldots ,N\}\), of size \(\frac{2}{N} \times \frac{2}{N}\), with \(N \in \mathbb {Z}^+\). C is the phase space matrix with dimension \(N\times N\) and is constructed in such a way that, for each \(i,j \in N\), the entry \(c_{ij}\) is equal to the number of phase space vectors in B that fall within the square area \(g_{ij}\). We construct P by normalizing C such that, for each \(i,j \in N\), the element \(p_{ij}\) corresponds to the proportion of all of the phase points which fall within \(g_{ij}\). We calculate P as follows:
At this point, the typical approach in the literature is to extract features from the PSR images to be used as inputs to numerical models that are used for classifying ECG signals into categories (Vemishetty et al., 2019; Krishnan et al., 2007; Rocha et al., 2008). Differently, in this paper we consider these matrices as \(N \times N\) (with \(N = 32\)) pixel images and rely on deep learning architectures typically used for computer vision to autonomously perform feature selection. Figure 4e gives an example of one of the PSR images. Figure 4f shows the same PSR image darkened for readability.
3.2 Neural network layers
Here, we briefly overview the 5 types of layer that we use within the architectures of our neural networks (presented in the next subsection).
A dense layer consists of a number of neurons, each of which is connected to all the neurons in the previous layer. For each neuron of index i, the neuron’s outputs \(z_i\) is given by
where b is a vector containing a bias for each neuron in the layer, \(w_i\) is a vector of weights for the ith neuron containing a weight associated to each element in the input and \(\odot \) is the Hadamard product.
Next, we consider convolutional layers. In a standard convolutional layer, 3-dimensional filters, also referred to as kernels, are applied to the input at regular intervals (the size of these intervals is referred to as the stride) to transform it into a 3-dimensional output comprised of a number of 2-dimensional feature maps. Let k be the index of a feature map. The output for a neuron in row i and column j of the feature map k in a convolutional layer with 3-dimensional input x is given by
where \(s_h\) and \(s_w\) are, respectively, the vertical and horizontal strides, \(f_h\), \(f_w\) and \(f_{n'}\) are the height, width and depth of this layer’s filters (where \(f_{n'}\) is equal to the number of feature maps in the previous layer), \(b_k\) is the bias applied to the feature map k and \(w_k\) is the matrix of weights defining the 3-dimensional filter used to generate the k-th feature map. Hence, \(w_{uvk'k}\) is the weight in row u and column v of the two dimensional slice of the filter which connects to feature map \(k'\) of the input. For a detailed introduction to CNNs, we refer the reader to (Wu, 2017).
We also consider batchnorm layers. Batch normalization, or batchnorm, is a tool for reparameterizing neural networks to significantly speed up training and reduce generalization error, allowing us to forgo the use of dropout (the process by which neurons are randomly turned on and off during training) (Goodfellow et al., 2016). For regularizing CNNs, batchnorm has been shown to perform better than dropout (Garbin et al., 2020). It can be applied to any layer and normalizes the distribution across the minibatch for each output in a layer, thus ensuring that throughout training the inputs to the following layer always have the same distribution (Garbin et al., 2020). Batchnorm is applied within its own layer. The minibatch outputs \(z = (z^{(1)}, \dots , z^{(m^B)})\) of a batch normalization layer with minibatch inputs \(X = (X^{(1)}, \dots , X^{(m^B)})\) are given by
where \(m^B\) is the size of the minibatch and \(X^{(n)}\) is the matrix containing the output from the previous layer for the n-th data point in the minibatch. The outputs \(z^{(n)}\), the minibatch mean \(\mu _B\), the standard deviation \(\sigma _B\) and the scaling and shifting parameters \(\gamma \) and \(\beta \) are all matrices with the same shape as the input \(X^{(n)}\). The learnable parameters in this layer are the vectors \(\gamma \) and \(\beta \). The Hadamard product \(\odot \) and Hadamard division \(\oslash \) (also referred to as element-wise product and division, respectively), are used in these calculations. We apply the activation function in its own layer, after batchnorm is performed (as was done in the paper by Ioffe & Szegedy, 2015 where batchnorm is introduced).
In our activation layers, we use the generally recommended activation function rectified linear unit or ReLU (Nair & Hinton, 2010; Glorot et al., 2011) given by
This function is applied element wise to the output of the layer preceding it.
The last layer we consider is a skip connection layer. Addition skip connections as utilized in ResNet (He et al., 2016) are used within our deep CNNs, whereby the output of one layer is added to the input of a layer located significantly deeper into the network. Models with skip connections have a smoother loss surface (Balduzzi et al., 2017; Li et al., 2017), which makes optimizing the model parameters easier and thus speeds up training.
3.3 Model architectures
We now discuss the architectures of the deep learning models we use to predict T:R ratios from \(32 \times 32\) pixel PSR images.
Each model consists of N feature-extraction blocks feeding into a regression block. Figure 5 shows this general structure. Following preliminary experiments (Dunn et al., 2021) where a wide range of model architectures were accessed, we found two model architectures to provide a good level of accuracy: the MLP5 and Complex CNN5 models. Their names refer to the fact that, in these models, the regression block is preceded by 5 Multi Layer Perception (MLP) feature extraction blocks or by 5 Complex Convolutional Neural Network (Complex CNN) feature extraction blocks, respectively.
Let us first consider the MLP5 model shown in Fig. 6. Each of the 5 MLP feature extraction blocks has a dense layer of 1024 neurons, a batchnorm layer and, then, an activation layer. These layers extract abstract features from the PSR images and pass them on to the regression block. Before being used as input into the first 1-dimensional (1D) dense layer, the \(32 \times 32\) pixel PSR images are flattened into a 1D vector. The regression block derives the T:R ratio from the features’ output from the final feature extraction block. It also utilizes two dense layers containing 256 and 64 neurons, each followed by batchnorm and activation layers before a final fully connected layer with a single neuron which generates the predicted T:R ratio. Further details on the structure of the MLP5 feature extraction blocks and the regression block are given in Tables 1 and 2 in the Appendix.
A diagram of the MLP5 model, consisting of 5 MLP feature extraction blocks and the regression block. The batchnorm and activation layers are not represented. The neurons in the flattened input, feature extraction blocks and regression block are labeled i, e and r, respectively. Their superscript denotes which layer they belong to, while the subscript refers to their position in that layer
Let us now focus on the Complex CNN5 model, in which five Complex CNN feature extraction blocks are used to extract features from the PSR image which are then flattened and passed to the regression block. The convolutional layers used within this model aim to exploit the multi-dimensional nature of the PSR image inputs. As convolutional layers take three-dimensional (3D) inputs, the original PSR images are considered to have shape \(32 \times 32 \times 1\). Figure 7 shows the first feature extraction block of the Complex CNN5 model. The Complex CNN feature extraction block contains two convolutional layers (each followed by batchnorm and activation layers) with relatively small kernels which feed into a convolutional layer with stride 2 and a larger kernel (followed by batchnorm and activation layers). The first two convolutional layers extract features from the PSR images, while the third reduces the size of the feature maps and increases their number. While pooling layers are typically used to decrease the size of the feature maps, using a convolutional layer to do this allows an additional opportunity to extract more complex features from the outputs of the previous layers. Addition skip connections are also included, which add the input of the block to the output of the first two convolutional layers, speeding up training.Footnote 3 Each subsequent Complex CNN feature extraction block has twice as many feature maps as its predecessor had and the height and width of these feature maps are half the size of those in the previous Complex CNN feature extraction block. The extracted features’ output by the 5th block are flattened and then input into the regression block. Because of this halving in height and width of the feature maps, the kernel size of the convolutional layers must also decrease. Details on the exact structure of the Complex CNN feature extraction blocks can be found in Table 3 in the Appendix.
3.4 Model evaluation
For each model architecture, we perform 10 rounds of 10-Fold CV. For each round of 10-fold CV, the data is shuffled before being split into 10 equally sized folds. Then, at each iteration of the 10-fold CV one fold is reserved as the testing set. A randomly sampled 20% of the remaining 9 folds is used to form the validation set which is used to prevent overfitting while training. The other 80% of the non-testing data is used as training set. Figure 8 illustrates our 10-fold CV process as well as the training of an ensemble model (see the next subsection for the introduction of ensemble models).
Part A gives an overview of how training and testing sets are iteratively sampled within a single 10-fold CV. Part B illustrates how within each round of a single 10-fold CV, having reserved a testing set, 5 sub-models are trained with different training and validation sets and their predictions for the testing set are averaged giving the ensemble model’s prediction. If we consider an ensemble model with only one sub-model, this becomes exactly the same as the 10-fold CV process for training non-ensemble models, which we described in Sect. 3.4
In this paper, our models are trained for at most 1000 epochs, employing early stopping (Prechelt, 1998) for regularization in model training. It is often observed during training that both the training and validation errors initially decrease and, at some point, the validation error begins to increase while the training error continues to decrease. The error on the validation set serves as a proxy for the generalization error which is used to determine when overfitting has begun. Therefore, in order to achieve the lowest possible validation error, we reset the model parameters to their values before overfitting began and the validation error started increasing. This means that we continue training our model until the validation error has not improved for a given number of epochs (referred to as the patience of the optimization algorithm) instead of continuing to train until we reach a local minimum of the training error (Bengio et al., 2017). In all of our experiments, we use a patience of 200 epochs.
While, during the training of our models, we look to minimize the Mean Squared Error (MSE), when we come to evaluate their performance in predicting T:R ratios of the testing PSR images there are additional metrics which we are interested in. In particular, we record the training time for each model as, should two models give comparable accuracy, the one with a shorter training time is preferable as it would be less computationally expensive to retrain it if and when new data became available. Similarly, we record the number of epochs the model trained for before early stopping took place. The average prediction time, i.e., the average amount of time taken for the model to predict each testing PSR image’s T:R ratio, is also recorded. A lower average prediction time indicates that predicting T:R ratios in real time would be less computationally expensive and, therefore, the model would be more suitable for incorporation into a device with limited computational resources. It is worth noting here that we only compare the average prediction time of models with different architectures as, after training, the average prediction time for models with the same architecture should be consistent. We assess the accuracy of the models’ predictions of the T:R ratios of the testing PSR images using Root Mean Squared Error (RMSE) and Mean Absolute Error (MAE), defined by
where, for each \(i \in \{1, \dots , n\}\), \(y_i\) is the true T:R ratio, \(\hat{y}_i\) is the predicted T:R ratio, and n is the number of PSR images. RMSE, while similar to its squared counterpart the Mean Squared Error (MSE), does not see such a broad distribution of errors and, thus, lends itself to a graphical comparison of errors over multiple rounds of CV. MAE is the most intuitive of these errors and, therefore, it is important when discussing the impact of the prediction of a model.
3.5 Ensemble models
As a final inclusion to improve the performance of our models, we construct and evaluate ensemble models.
Within each round of CV after the testing set has been excluded, the remaining data is split into 5 non-overlapping folds. 5 sub-models are then trained, each using one of these 5 folds as a validation set and the remaining 4 as a training set. The predictions of the ensemble models for the testing set are calculated by taking the average of the predictions of the sub-models. This is illustrated in Fig. 8.
By creating an ensemble model, we ensure that each data point which is not used for testing will be used for training by all but one of the sub-models. In contrast, when training a single model, the data points in the validation set are never used for training.
Having determined the best model architecture, the best training algorithm and the best hyperparameter selection, we will construct and evaluate an ensemble model at each iteration of the 10 rounds of 10-fold CV.
4 Experiments
In this section, we compare the use of various optimization techniques to fine tune our models for accurately predicting T:R ratios of S-ICD implantation candidates.
4.1 Data sets
To train and evaluate our models, we rely on the Southampton General Hospital ECG (SGH-ECG) data set, which was collected by our partners at the Southampton General Hospital for our preliminary study (Dunn et al., 2021). To construct the SGH-ECG data set, we randomly sampled 10-second ECG segments from 24-hour ECG recordings. A total of 390 segments (sampled at 500 Hz with annotated R and T peaks) were collected. These 24-hour recordings were originally collected for the clinical research study HEART TWO.Footnote 4 The dataset comprises a total of 18 patients with an equal number of males and females of a mean age of 53.16 years. 4 of them (22.22%) displayed a structurally-normal heart, 3 (16.67%) an adult congenital heart disease, 3 (16.67%) a hypertrophic cardiomyopathy, and 8 (44.44%) a congestive hart failure. Each of the 10-second ECG segments was used to generate a \(32 \times 32\) pixel PSR image as well as its corresponding T:R ratio using the preprocessing techniques detailed in Sect. 3.1.
Due to the relatively small size of our data set, at each round of CV we combine the training data with 310 \(32\times 32\) pixel PSR images with associated T:R ratios derived from the open source ECG Identification (ECG-ID) Database (Lugovaya, 2005), containing 10-second ECG segments with a sampling rate of 500Hz, with R and T peak annotations. In this dataset, 44 and 46 of the participants were, respectively, male and female and the patients’ age ranged from 13 to 75. We do not add this data to the testing and validation sets as we are only interested in evaluating the performance of our models for patients with specific heart conditions which the participants of the ECG-ID study did not have. We remark that, as the SGH-ECG dataset contains patients with structurally normal hearts, there is no additional value to be gained in assessing our models on the ECG-ID dataset. Additionally, we have a higher level of confidence in the quality of the SGH-ECG dataset and, as such, this is the only data set we will use in the remainder of the paper to measure the performance of the models we consider.
4.2 Training algorithms
In all of our experiments, training is carried out via minibatch Stochastic Gradient Descent (minibatch SGD), a variant of gradient descent that makes use of the fact that the average gradient on a minibatch of m randomly sampled data points is an unbiased estimator of the gradient on all the data points. Let X and Y be the sets of observations and labels and, for all \(i = 1, \dots , |X |\), let \((x_i,y_i)\) be an observation-label pair. The formulas by which minibatch SGD calculates the unbiased estimate \({\hat{g}}\) of the gradient g and uses it to update the model parameters \(\theta \) at each step are \(\hat{g}:= \frac{1}{m} \sum _{x_i \in \bar{X}}^{} \nabla _\theta L(f(x_i;\theta ),y_i)\) and \(\theta := \theta - \epsilon \hat{g}\), where \(\bar{X} \subseteq X\) is the set of m data points in the minibatch and \(f(x_i)\) is the output of the model with \(x_i\) given as input. Training is carried out for a number of epochs. An epoch is comprised of a number of descent steps such that the gradient is calculated on each data point in the training set at least once.
A popular inclusion to the minibatch SGD algorithm is momentum (a technique designed to leverage the fact that, when the gradient is small but consistent across multiple descent steps, we can afford to take larger steps in that direction, thus achieving an accelerating learning Bishop, 1995). In cases where the gradient is small, SGD can exhibit a slow convergence. When implementing the classical momentum method (Polyak, 1964), at each descent step an exponentially decaying history of the previous descent steps is added to the product of the gradient of the loss function and the learning rate. In this paper, we implement Nesterov momentum, a variant of momentum wherein, rather than calculating the gradient using the current value of the model parameters \(\theta \), the gradient is calculated using the parameters after the momentum has been applied (Sutskever et al., 2013).
We consider 3 SGD variants with an adaptive learning rate in which the learning rate is scaled for each individual model parameter: AdaGrad (where the learning rates corresponding to each model parameter are scaled by the inverse of the sum of the squared partial derivatives over all training iterations Duchi et al., 2011), RMSprop (an adaptation of AdaGrad designed to converge more effectively in non-convex optimization problems—such as the training of multi-layer neural networks—by diminishing the learning rate proportionally to the inverse of an exponentially decaying window of squared gradients) and ADAM (Kingma & Ba 2014) (in which an exponentially decaying history is kept of both the first and second order moments of the gradient). The formulas by which ADAM updates the model parameters \(\theta \) at each iteration are \(s:= \rho _1 r + (1-\rho _1)g\), \(\hat{s}:= \frac{s}{1-{p_1}^t}\), \(r:= \rho _2 r + (1-\rho _2)g\odot g\), \(\hat{r}:= \frac{r}{1-{p_2}^t}\), \(\Delta \theta := -\epsilon \frac{\hat{s}}{\delta + \sqrt{\hat{r}}}\), and \(\theta := \theta + \Delta \theta \), where t is the number of descent steps that the algorithm has been running for, \(\rho _1\) and \(\rho _2\) are additional hyperparameters and \(\odot \) is the Hadamard product.
The principal requirement when initializing the model parameters of a neural network (thereby choosing a starting point for the optimization method) for training via SGD variants is to break symmetry (Goodfellow et al., 2016). To achieve this, we initialize the weights randomly using the Glorot normalized initialization method (Glorot & Bengio, 2010) with 0 mean and a variance of \(\text {Var}\left[ W\right] = \frac{2}{n_{\text {in}}+n_{\text {out}}}\), where W is the matrix of weights in a given layer, \(n_\text {in}\) is the number of nodes in the previous (input) layer and \(n_\text {out}\) is the number of nodes in this layer’s output. Differently from the original work, where a uniform distribution is used, in our work we sample the weights from a Normal distribution as this is the default implementation in TensorFlow, the Python package we use to train and evaluate our neural networks.
In this paper, we consider the choice of SGD-based algorithm used to train the model parameters as a hyperparameter of the model itself which should be selected optimally. We then look to tune the hyperparameters of these SGD-based algorithms, namely, batch size and global learning rate.
Running the 10 rounds of 10-fold CV is computationally very expensive. In our experiments, having selected the best performing model architecture we are only able to run the 10 rounds of 10-fold CV 16 times, meaning that we can only assess 16 different hyperparmeter combinations. Due to this restriction, we opted for tuning the hyperparameters sequentially, first evaluating SGD, SGD with Nesterov Momentum, AdaGrad, RMSProp, RMSprop with momentum and ADAM to determine the SGD-based algorithm which gives the best accuracy with default hyperparameter selections. We then tune its batch size and finally tune its global learning rate while using the batch size found in the previous step. We should note that this approach has the drawback of not fully considering the interaction of the hyperparameters with each other. Ideally, we would instead use an algorithm such as TPE or Gaussian optimisation (Bergstra et al., 2011). However, these algorithms are typically initialized using random search to evaluate tens or hundreds of hyperparameter combinations to generate an initial image of the hyperparameter space (Bergstra et al., 2011; Ozaki et al., 2020). As we do not have the computational resources to evaluate this many hyperparameter combinations, we would not be able to properly initialize these algorithms. As for grid search and random search, which are widely popular alternatives for hyperparameter tuning due to their simplicity, with only 16 evaluation points these methods would also be ineffective.Footnote 5
4.3 Experiment 1: architecture comparison
Firstly, we compare the two model architectures proposed in Sect. 3.3, i.e., Complex CNN5 and MLP5.
In this experiment, we use ADAM as the minibatch SGD variant for training, as “...Adam works well in practice and compares favorably to other stochastic optimization methods” (Kingma & Ba, 2014). We use the generally accepted default selections of batch size 32 (Benigo 2012; Master & Luschi, 2018) and learning rate 0.001 as suggested in the original paper (Kingma & Ba, 2014). These experiments are conducted using Tensorflow in Python 3.10.
Figure 9 comprises 5 violin plots of the distributions of 5 performance metrics (detailed in Sect. 3.4) over the 100 total rounds of CV for the MLP5 architecture (shown in blue) and the Complex CNN5 model (shown in red). The MLP5 model completes its training in, on average, 493 s. This is significantly quicker than the time of the Complex CNN5, whose average training time is 1262 s despite taking roughly the same number of epochs to complete training.
The MLP5 models are found to be able to predict the testing PSR images’ T:R ratios much quicker than the Complex CNN5 models, with an average prediction time per PSR image of 0.0075 s compared to an average of 0.0119 s for the Complex CNN5 one. This shorter prediction time is valuable, as it indicates that the MLP5 models would require fewer computational resources to make predictions of the T:R ratio in real time than the Complex CNN5 models would.
In our analysis, our primary concern is accuracy. Here, we consider RMSE and MAE as defined in Eq. (2). The MLP5 models achieve an average RMSE of 0.105 and an average MAE of 0.0567 compared to the Complex CNN5 model’s average RMSE of 0.111 and MAE of 0.0624. For these reasons, we conclude that the MLP5 architecture leads to better performance as well as to shorter training and prediction times.
We must note that, while the way our models will be delivered to clinicians (and also updated and maintained) is yet to be determined from a software engineering perspective, the shorter prediction and training time of MLP5 is preferable as it may enable to model to be run on less powerful computing devices and also to be updated/retrained more regularly when new data becomes available.
In the subsequent experiments, we will look to select the best SGD variant for training these MLP5 models. We will not consider the prediction times in the subsequent analysis, as they should be the same for all models with the same architecture.
4.4 Experiment 2: optimizer comparison
In the previous experiment, we assumed ADAM was the preferred minibatch SGD variant. In this experiment, we test such an assumption by evaluating 6 minibatch SGD variants: SGD, SGD with Nesterov Momentum, AdaGrad, RMSProp, RMSprop with momentum and ADAM. At this stage, we still use the default batch size and learning rate of, respectively, 32 and 0.001. We perform 10 rounds of 10-fold CV, training in each fold an MLP5 model with each of the 6 minibatch SGD variants for a total of 100 models trained using each minibatch SGD variant and evaluated on a distinct testing set.
Figure 10 shows the distribution of the RMSEs across the 100 models for each minibatch SGD variant (left) as well as an expanded version of the same graph (right). The trend shown in these results is that the prediction error is lower for SGD variants which are better suited to the non-convex setting (such is the case when training a deep neural network for T:R ratio prediction from PSR images). MLP5 models trained using ADAM exhibit an average RMSE of 0.105 compared to those trained using basic minibatch SGD, which have an average RMSE of 0.121. We observe that the training time and the number of epochs before early stopping is generally greater for more complex versions of minibatch SGD such as for ADAM and RMSprop. This can be seen in Fig. 17 in Section B of the Appendix.
With this experiment, we have confirmed that ADAM does indeed give the best accuracy (with a default hyperparameter selection) than a number of popular minibatch SGD algorithms and, hence, is the optimization algorithm we will use going forward.
4.5 Experiment 3: tuning batch size
Batch size is a hyperparameter of all minibatch SGD based algorithms. In previous experiments, we used the generally recommended batch size of 32 (Benigo 2012, Master & Luschi, 2018). In this experiment, we evaluate batch sizes of \(2^n\) for \(n\in \{3, 4, 5, 6, 7, 8\}\) (while still using a learning rate of 0.001) by performing 10 rounds of 10-fold CV, in each fold training an MLP5 model with each of the 6 different batch sizes for a total of 100 models trained with each batch size and evaluated on a distinct testing set.
Figure 11 shows the distribution of the RMSEs across the 100 models trained with each batch size (left) as well as an expanded version of the same graph (right). As can be seen, increasing the batch size steadily decreases the RMSE up to a batch size of 128, which achieves an average RMSE of 0.1029, compared to the previously used batch size of 32 which leads to an average RMSE of 0.1053.
Figure 18 in Section B of the Appendix shows that, while the number of epochs before earlystopping increases as the batch size increases, the training time decreases. In one epoch, minibatches are propagated forward though the network and their errors are used to calculate gradients and update the parameters. This is repeated until all data has been seen at least once. This means that a large batch size results in the parameters being updated far fewer times per epoch. This is the reason why, in Fig. 18, we see that the models with a large batch size typically take more epochs to train as each epoch represents fewer descent steps. However, despite training for more epochs, the large reduction in the number of times the model parameters are updated results in a drastic reduction in training time for models trained with larger batch sizes.
Using a batch size of 128 leads to the best average RMSE of 0.1029 while also resulting in a significant reduction in average training time compared to using a batch size 32 and, as such, 128 is the batch size we will use going forward.
4.6 Experiment 4: tuning learning rate
ADAM is considered robust to the choice of hyperparameters \(\rho _1\) and \(\rho _2\) and, as such, we only tune the global learning rate \(\epsilon \) by searching a logarithmic range \(10^{-n}\) for \(n \in \{1, 2, 3, 4, 5, 6\}\) while using the best-performing batch size of 128 found in the previous experiment (Bengio et al., 2017). We evaluate these learning rates by carrying out 10 rounds of 10-fold CV, in each fold training an MLP5 model with each of the different learning rates for a total of 100 models trained with each learning rate and evaluated on a distinct testing set.
Figure 11 shows the distribution of the RMSEs across the 100 models trained with each learning rate (left) as well as an expanded version of the same graph (right). Figure 11 does not show the results for models trained with a learning rate of \(10^{-5}\) and \(10^{-6}\). However, the trend demonstrated in Fig. 11 (that, for learning rates less than 0.01, the average RMSE increases as the learning rate decreases) continues for these values. We observe that using a learning rate of 0.01 gives an average RMSE of 0.1020 as opposed to the RMSE of 0.1029 given by the previously used default of 0.001 (Fig. 12).
Figure 19 in Section B of the Appendix shows that, as we would expect, the number of epochs increases for smaller learning rates and, as the batch size is fixed at 128 in this experiment, the training time is proportional to the number of epochs and so it also increases as the learning rate becomes smaller.
We conclude from this experiment that an MLP5 model trained using ADAM with a batch size of 128 and a learning rate of 0.01 offers improved accuracy while taking less time to train.
Figure 13 shows the average training and validation error at each epoch of each of the 100 models trained using ADAM with a batch size of 128 and a learning rate of 0.01 in each of the 100 rounds of CV performed in this experiment. As can be seen, while the validation error is higher than the training error during training, both decrease rapidly before stabilizing and reduce slowly over the rest of the training. For the purposes of this plot where early stopping occurs, the errors are kept constant for the remainder of the 1000 epochs (Fig. 14).
4.7 Experiment 5: creating ensemble models
In the previous experiments, we found that the a tuned MLP5 model trained using ADAM with a batch size of 128 and a global learning rate of 0.01 gave the best accuracy. In this experiment, we assess the benefits of ensemble models by performing 10 rounds of 10-fold CV and, in each fold, training 5 MLP5 submodels, for a total of 100 ensemble models each containing 5 tuned MLP5 submodels. We compare each of these MLP5 ensemble models to a tuned MLP5 model and to a ’default’ MLP5 model trained using ADAM with a batch size of 32 and a learning rate of 0.001 (the naive hyperparameter selections we initially considered to be good defaults).
Figure 11 shows the distribution of the RMSEs of the 100 MLP5 ensemble models on their respective testing sets, compared with those of the tuned MLP5 models and the default MLP5 models (left) as well as to an expanded version of the same graph (right). As can be seen, the default MLP5 models achieve an average RMSE of 0.1052 while the tuned MLP5 models achieve an average of 0.1024 and the ensemble MLP5 models an average of 0.0928. This represents a reduction in average RMSE of 12% for the ensemble MLP5 models but of only 2% for the tuned MLP5 models. If, however, we examine the upper quartile of these distributions, we see that both the ensemble MLP5 models and the tuned MLP5 models offer a more significant improvement over the base MLP5 models. The ensemble MLP5 models’ RMSE distribution has an upper quartile of 0.1002 compared to the tuned MLP5 models’ 0.1147 and to the base MLP5 models’ 0.1259, which is a reduction of 9% for the tuned models and of 20% for the ensemble models. We do not examine the number of epochs or the training duration for this experiment as the number of epochs used to train each submodel is expected to be the same as for the tuned MLP5 models and, likewise, the training time for the whole ensemble MLP5 model is expected to be simply 5 times that of a single tuned MLP5 model.
To this point, we have focused on RMSE because it makes the violin plots we have used more easily interpretable than MSE, and it provides a stronger penalization to large prediction errors than MAE. However, in order to put the performance of our model in context, we will once again examine MAE, as it as very easy to interpret as the average absolute error across each prediction. The default MLP5 models achieve an average MAE of 0.0567 compared to our best performing model, the ensemble MLP5 model, which achieves an average MAE of 0.0461. This represents a substantial reduction in average absolute prediction error (MAE) of 19%.
5 Discussion
In the previous section, we showed that, through optimization algorithm selection, hyperparameter tuning and creating ensemble models, we were able to reduce the average MAE across the 100 CV rounds to 0.0461, a reduction of 19%, and the mean and upper quartile of the distribution of RMSEs over the 100 CV rounds by 12% and 20%, respectively.
In this section, we discuss the clinical impact of our models’ ability to predict the T:R ratios of 10-second, single-lead, ECG segments. As described in Sect. 3.1, from a clinical perspective the sign of the T:R ratio is irrelevant. Differently, our model is expected to output both positive and negative predicted T:R ratios depending on the sign of the T wave. For use in clinical analysis, we can simply compute the absolute value of the model.
Figure 15 shows the predicted absolute T:R ratios for each PSR image (notice that, during each round of 10-fold CV, each PSR image is used for testing exactly once) generated using the SGH-ECG dataset when it was reserved for testing in 10-fold CV, plotted against their true absolute T:R ratios. The green line represents a perfect labeling. As can be seen, the absolute value of the outputs of the ensemble MLP5 model predicts the true absolute T:R ratios with very low error.
Having established that this model can be used to accurately and autonomously predict T:R ratios from 10-second ECG segments, we can now use it to efficiently perform screening to assess the normal variations in the patients’ T:R ratios across multiple leads over a 24-hour period. 8640 non-overlapping 10-second, single-lead ECG segments are created by breaking down a 24-hour, three-lead ECG recording. The 10-second segments are transformed following the process laid out in Sect. 3.1. This results in 8640 chronologically ordered PSR images. For each lead, a time series of T:R ratios can be generated by inputting the PSR images into the model. As previously mentioned, for clinical use we consider the absolute values of the predicted T:R ratio. These time series of T:R ratios corresponding to each lead of the ECG recording can enable clinicians to easily analyze the normal behaviors of the patient’s T:R ratio over a much longer screening window than the 10-second screening process used in current practice.
We used such a method to create Fig. 16, which shows the variation of the T:R ratio on each lead over a 24-hour period. For readability, we have smoothed the time series of T:R ratios by plotting a moving average with a window of half an hour. For more details on the clinical applications of this tool, we refer the reader to our other works in this area (ElRefai et al., 2022a, b, c) where we determine what the T:R ratio cut-off used in the screening should be and evaluate the variation of the T:R ratio of patients with various heart conditions.
6 Conclusions
TWOS is the leading cause of inappropriate shocks for S-ICD implanted patients. Due to the fact that the T:R ratio can vary significantly over time, the current screening process of using a 10 s ECG recording to estimate a patient’s typical T:R ratio does not reliably capture the normal behavior of the latter. In this paper, we have developed models which enable autonomous T:R ratio prediction from PSR images of 10-second, single-lead ECG signals with a high degree of accuracy. These models can be incorporated into a tool enabling clinicians to perform 24-hour, automated screenings to assess patients’ normal T:R ratio variations with greater detail and reliability.
Following 100 total rounds of CV, we have determined that the MLP5 model is preferable to the Complex CNN5 model as it has a shorter training time, can make predictions faster and, most importantly, it predicts T:R ratios with higher accuracy (with an average RMSE of 0.105 compared to the Complex CNN5’s average RMSE of 0.111).
Creating ensemble models comprised of MLP5 models, trained using the best-performing SGD based optimization method and with a tuned batch size and global learning rate, we were able to greatly reduce the prediction error compared to the models which used default selections for these hyperparameters. We were able to reduce the average MAE across the 100 CV rounds to 0.0461, a reduction of 19%, and the mean and upper quartile of the distribution of RMSEs over the 100 CV rounds by 12% and 20% respectively.
Currently, we are using our proposed screening tool to assess the temporal variations of the T:R ratio in patients belonging to patient groups who are likely to be implanted an S-ICDs. We are also investigating whether the increased detail provided by our tool can allow the cut-off threshold for the screening to be increased above the current value of 1/3, allowing patients who had, under the previous screening process, been determined to be at too high a risk of TWOS to be safely implanted with S-ICDs. Finally, we hope to determine in future research if fluctuations in a patient’s T:R ratios can provide indication that a cardiac episode is impending.
Notes
Using a single lead, the S-ICD is able to record the ECG signals on 3 vectors—see Fig. 1a. When using a Holter ECG recording to assess the patients’ S-ICD implantation eligibility, numerous leads are used. We refer to a single lead of a Holter recording as the ECG recording for a single vector.
The preprocessing techniques we use in this paper to derive PSR images from 10-second single-lead ECG segments were first illustrated in our preliminary work on the clinical impact of this type of tool (Dunn et al., 2021). For the sake of completeness, we briefly overview these methods here.
In preliminary experiments (Dunn et al., 2021) a more basic CNN feature extraction block with a single convolutional layer, no skip connection and a pooling layer for dimensionality reduction was evaluated and found to predict T:R ratios with far inferior accuracy.
This study was performed with favorable opinion from the REC (17/SC/0623) and with R &D (RHMCAR0528) approval. This study was conducted in accordance with the Research Governance Framework for Health and Social Care (2005), Good Clinical Practice and their relevant updates.
In our preliminary experiments (Dunn et al., 2021) we assessed the use of image augmentation schemes. However, we did not find that they significantly improved the models’ performance and, hence, we do not evaluate these methods here.
References
Adabag, A. S., Luepker, R. V., Roger, V. L., & Gersh, B. J. (2010). Sudden cardiac death: Epidemiology and risk factors. Nature Reviews Cardiology, 7(4), 216–225.
Assanelli, D., Di Castelnuovo, A., Rago, L., Badilini, F., Vinetti, G., Gianfagna, F., Salvetti, M., Zito, F., Donati, M. B., De Gaetano, G., & Iacoviello, L. (2013). T-wave axis deviation and left ventricular hypertrophy interaction in diabetes and hypertension. Journal of Electrocardiology, 46(6), 487–491.
Babu, G. S., Zhao, P., Li, & X.-L. (2016). Deep convolutional neural network based regression approach for estimation of remaining useful life. In International Conference on Database Systems for Advanced Applications (pp. 214–228). Springer
Balduzzi, D., Frean, M., Leary, L., Lewis, J., Ma, K. W. D., & McWilliams, B. (2017). The shattered gradients problem: If resnets are the answer, then what is the question?. In International Conference on Machine Learning (pp. 342–350). PMLR.
Bengio, Y. (2012). Practical recommendations for gradient-based training of deep architectures. In Neural networks: Tricks of the trade (pp. 437–478). Springer.
Bengio, Y., Goodfellow, I., & Courville, A. (2017). Deep learning (Vol. 1). MIT Press.
Bergstra, J., Bardenet, R., Bengio, Y., & Kégl, B. (2011). Algorithms for hyper-parameter optimization. In: J. Shawe-Taylor, R. Zemel, P. Bartlett, F. Pereira, K. Q. Weinberger, K.Q. (Eds.), Advances in Neural Information Processing Systems (Vol. 24, pp. 1–9).
Bishop, C. M. (1995). Neural networks for pattern recognition. Oxford University Press.
Boersma, L., Barr, C., Knops, R., Theuns, D., Eckardt, L., Neuzil, P., Scholten, M., Hood, M., Kuschyk, J., Jones, P., & Duffy, E. (2017). Implant and midterm outcomes of the subcutaneous implantable cardioverter-defibrillator registry: The effortless study. Journal of the American College of Cardiology, 70(7), 830–841.
Chen, M., Fang, Y., & Zheng, X. (2014). Phase space reconstruction for improving the classification of single trial EEG. Biomedical Signal Processing and Control, 11, 10–16.
Dawid, A. (2019). PSR-based research of feature extraction from one-second EEG signals: A neural network study. SN Applied Sciences, 1(12), 1–12.
Djemal, R., Bazyed, A. G., Belwafi, K., Gannouni, S., & Kaaniche, W. (2016). Three-class EEG-based motor imagery classification using phase-space reconstruction technique. Brain Sciences, 6(3), 36.
Duchi, J., Hazan, E., & Singer, Y. (2011). Adaptive subgradient methods for online learning and stochastic optimization. Journal of Machine Learning Research, 12(7), 2121–2159.
Dunn, A. J., ElRefai, M. H., Roberts, P. R., Coniglio, S., Wiles, B. M., & Zemkoho, A. B. (2021). Deep learning methods for screening patients’ S-ICD implantation eligibility. Artificial Intelligence in Medicine, 119, 102139.
ElRefai, M., Abouelasaad, M., Conibear, I., Wiles, B. M., Dunn, A. J., Coniglio, S., Zemkoho, A. B., & Roberts, P. R. (2022). The use of artificial intelligence and deep learning methods in subcutaneous implantable cardioverter defibrillator screening to optimise selection in special patient populations. Europace, 24, 053–448.
ElRefai, M., Abouelasaad, M., Dunn, A. J., Coniglio, S., Zemkoho, A. B., Wiles, B. M., & Roberts, P. R. (2022). Eligibility for subcutaneous implantable cardiac defibrillator utilising artificial intelligence and deep learning methods for prolonged screening: Where is the cut-off? Europace, 24, 053–447.
ElRefai, M., Abouelasaad, M., Wiles, B. M., Dunn, A. J., Coniglio, S., Zemkoho, A. B., Morgan, J., & Roberts, P. R. (2023). Correlation analysis of deep learning methods in S-ICD screening. Annals of Noninvasive Electrocardiology. https://doi.org/10.1111/anec.13056
ElRefai, M., Abouelasaad, M., Wiles, B. M., Dunn, A. J., Coniglio, S., Zemkoho, A. B., & Roberts, P. R. (2022). Deep learning-based insights on T: R ratio behaviour during prolonged screening for S-ICD eligibility. Journal of Interventional Cardiac Electrophysiology. https://doi.org/10.1007/s10840-022-01245-6
Fan, X., Yao, Q., Cai, Y., Miao, F., Sun, F., & Li, Y. (2018). Multiscaled fusion of deep convolutional neural networks for screening atrial fibrillation from single lead short ECG recordings. IEEE Journal of Biomedical and Health Informatics, 22(6), 1744–1753.
Fosbøl, E. L., Seibæk, M., Brendorp, B., Torp-Pedersen, C., Køber, L., Investigations, D. (2008). Prognostic importance of change in QRS duration over time associated with left ventricular dysfunction in patients with congestive heart failure: The DIAMOND study. Journal of Cardiac Failure, 14(10), 850–855.
Garbin, C., Zhu, X., & Marques, O. (2020). Dropout vs. batch normalization: An empirical study of their impact to deep learning. Multimedia Tools and Applications, 79, 1–39.
Glorot, X., & Bengio, Y. (2010). Understanding the difficulty of training deep feedforward neural networks. In Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics. JMLR Workshop and Conference Proceedings (pp. 249–256).
Glorot, X., Bordes, A., & Bengio, Y. (2011). Deep sparse rectifier neural networks. In Proceedings of the Fourteenth International Conference on Artificial Intelligence and Statistics. JMLR Workshop and Conference Proceedings (pp. 315–323).
Goodfellow, I., Bengio, Y., & Courville, A. (2016). Deep learning. MIT Press.
He, K., Zhang, X., Ren, S., & Sun, J. (2016). Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (pp. 770–778).
Ioffe, S., & Szegedy, C. (2015). Batch normalization: Accelerating deep network training by reducing internal covariate shift. In International Conference on Machine Learning (pp. 448–456). PMLR.
Kingma, D. P., & Ba, J (2014). Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980.
Kiranyaz, S., Ince, T., & Gabbouj, M. (2015). Real-time patient-specific ECG classification by 1-d convolutional neural networks. IEEE Transactions on Biomedical Engineering, 63(3), 664–675.
Knops, R. E., Olde Nordkamp, L. R., Delnoy, P.-P.H., Boersma, L. V., Kuschyk, J., El-Chami, M. F., Bonnemeier, H., Behr, E. R., Brouwer, T. F., Kääb, S., & Mittal, S. (2002). Subcutaneous or transvenous defibrillator therapy. New England Journal of Medicine, 383(6), 526–536.
Krishnan, S. M., Dutt, D. N., Chan, Y., & Anantharaman, V (2007). Phase space analysis for cardiovascular signals. In Advances in Cardiac Signal Processing (pp. 339–354). Springer.
Kusumoto, F. M., Bailey, K. R., Chaouki, A. S., Deshmukh, A. J., Gautam, S., Kim, R. J., Kramer, D. B., Lambrakos, L. K., Nasser, N. H., & Sorajja, D. (2018). Systematic review for the 2017 AHA/ACC/HRS guideline for management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: A report of the American college of cardiology/American heart association task force on clinical practice guidelines and the heart rhythm society. Circulation, 138(13), 392–414.
Lee, S.-H., Lim, J. S., Kim, J.-K., Yang, J., & Lee, Y. (2014). Classification of normal and epileptic seizure EEG signals using wavelet transform, phase-space reconstruction, and euclidean distance. Computer Methods and Programs in Biomedicine, 116(1), 10–25.
Li, H., Xu, Z., Taylor, G., Studer, C., & Goldstein, T. (2017). Visualizing the loss landscape of neural nets. arXiv preprint arXiv:1712.09913.
Lih, O. S., Jahmunah, V., San, T. R., Ciaccio, E. J., Yamakawa, T., Tanabe, M., Kobayashi, M., Faust, O., & Acharya, U. R. (2020). Comprehensive electrocardiographic diagnosis based on deep learning. Artificial Intelligence in Medicine, 103, 101789.
Liu, W., Zhang, M., Zhang, Y., Liao, Y., Huang, Q., Chang, S., Wang, H., & He, J. (2017). Real-time multilead convolutional neural network for myocardial infarction detection. IEEE Journal of Biomedical and Health Informatics, 22(5), 1434–1444.
Lugovaya, T. S. (2005). Biometric human identification based on electrocardiogram. Master’s thesis, Faculty of Computing Technologies and Informatics, Electrotechnical University LETI, Saint-Petersburg, Russian Federation.
Madias, J. E. (2005). QTc interval in patients with changing edematous states: Implications on interpreting repeat QTc interval measurements in patients with anasarca of varying etiology and those undergoing hemodialysis. Pacing and Clinical Electrophysiology, 28(1), 54–61.
Madias, J. E., Bazaz, R., Agarwal, H., Win, M., & Medepalli, L. (2001). Anasarca-mediated attenuation of the amplitude of electrocardiogram complexes: A description of a heretofore unrecognized phenomenon. Journal of the American College of Cardiology, 38(3), 756–764.
Masters, D., & Luschi, C. (2018). Revisiting small batch training for deep neural networks. arXiv preprint arXiv:1804.07612.
Miao, F., Wen, B., Hu, Z., Fortino, G., Wang, X.-P., Liu, Z.-D., Tang, M., & Li, Y. (2020). Continuous blood pressure measurement from one-channel electrocardiogram signal using deep-learning techniques. Artificial Intelligence in Medicine, 108, 101919.
Nair, V., & Hinton, G. E. (2010) Rectified linear units improve restricted Boltzmann machines. In ICML.
Ozaki, Y., Tanigaki, Y., Watanabe, S., & Onishi, M. (2020). Multiobjective tree-structured parzen estimator for computationally expensive optimization problems. In Proceedings of the 2020 Genetic and Evolutionary Computation Conference (pp. 533–541).
Polyak, B. T. (1964). Some methods of speeding up the convergence of iteration methods. USSR Computational Mathematics and Mathematical Physics, 4(5), 1–17.
Pourbabaee, B., Roshtkhari, M. J., & Khorasani, K. (2018). Deep convolutional neural networks and learning ECG features for screening paroxysmal atrial fibrillation patients. IEEE Transactions on Systems, Man, and Cybernetics: Systems, 48(12), 2095–2104.
Prechelt, L. (1998). Early stopping-but when?. In Neural networks: Tricks of the trade (pp. 55–69). Springer.
Priori, S. G., Blomström-Lundqvist, C., Mazzanti, A., Blom, N., Borggrefe, M., Camm, J., Elliott, P. M., Fitzsimons, D., Hatala, R., Hindricks, G., Kirchhof, P., Kjeldsen, K., Kuck, K. H., Hernandez-Madrid, A., Nikolaou, N., Norekval, T. M., Spaulding, C., & Van Veldhuisen, D. J. (2016). 2015 ESC Guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac Death. The Task Force for the Management of Patients with Ventricular Arrhythmias and the Prevention of Sudden Cardiac Death of the European Society of Cardiology. Giornale Italiano di Cardiologia, 17(2), 108–170.
Roberts, F. M., Povinelli, R. J., & Ropella, K. M. (2001). Identification of ECG arrhythmias using phase space reconstruction. In European Conference on Principles of Data Mining and Knowledge Discovery (pp. 411–423). Springer.
Rocha, T., Paredes, S., De Carvalho, P., Henriques, J., & Antunes, M. (2008). Phase space reconstruction approach for ventricular arrhythmias characterization. In 2008 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (pp. 5470–5473). IEEE.
Sangaiah, A. K., Arumugam, M., & Bian, G.-B. (2020). An intelligent learning approach for improving ECG signal classification and arrhythmia analysis. Artificial Intelligence in Medicine, 103, 101788.
Sutskever, I., Martens, J., Dahl, G., & Hinton, G. (2013). On the importance of initialization and momentum in deep learning. In International Conference on Machine Learning (pp. 1139–1147). PMLR.
van Rees, J. B., Borleffs, C. J. W., de Bie, M. K., Stijnen, T., van Erven, L., Bax, J. J., & Schalij, M. J. (2011). Inappropriate implantable cardioverter-defibrillator shocks: Incidence, predictors, and impact on mortality. Journal of the American College of Cardiology, 57(5), 556–562.
Vemishetty, N., Acharyya, A., Das, S., Ayyagari, S., Jana, S., Maharatna, K., & Puddu, P. E. (2016). Classification methodology of CVD with localized feature analysis using phase space reconstruction targeting personalized remote health monitoring. In 2016 Computing in Cardiology Conference (CinC) (pp. 437–440). IEEE.
Vemishetty, N., Gunukula, R. L., Acharyya, A., Puddu, P. E., Das, S., & Maharatna, K. (2019). Phase space reconstruction based CVD classifier using localized features. Scientific Reports, 9(1), 1–18.
Wu, J. (2017). Introduction to convolutional neural networks. National Key Lab for Novel Software Technology. Nanjing University, China.
Zhang, J., Liu, A., Gao, M., Chen, X., Zhang, X., & Chen, X. (2020). ECG-based multi-class arrhythmia detection using spatio-temporal attention-based convolutional recurrent neural network. Artificial Intelligence in Medicine, 106, 101856.
Funding
Open access funding provided by Università degli studi di Bergamo within the CRUI-CARE Agreement. The work of Anthony J. Dunn is jointly funded by Decision Analysis Services Ltd andf EPSRC through the studentship with Reference EP/R513325/1. The work of Alain B. Zemkoho is supported by the EPSRC grant EP/V049038/1. The work of Stefano Coniglio and Alain B. Zemkoho is supported by The Alan Turing Institute under the EPSRC grants EP/N510129/1 and EP/W037211/1.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Mohamed ElRefai has received unrestricted grant from Boston Scientific. Paul R. Roberts has received honoraria from Boston Scientific and Medtronic and Research funding from Boston Scientific. Benedict M. Wiles has received unrestricted research grant and consultancy fees from Boston Scientific. Anthony J. Dunn, Stefano Coniglio and Alain B. Zemkoho have no financial or proprietary interests in any material discussed in this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendix A: Model architectures
In this section, we provide details on the regression and feature-extraction blocks used in our models. Details on the feature extraction block that is used to construct the MLP5 and Complex CNN5 models are reported in Table 1. Details on the feature extraction blocks used for the MLP5 models are reported in Table 2. The structures of the 5 Complex CNN feature extraction blocks used within the Complex CNN5 model are given by Table 3, where n denotes which of the 5 feature extraction blocks we are referring to.
Appendix B: Experiment graphs
In this section, we provide additional figures from the experiments reported in Sect. 4. Figure 17 shows the training time and number of epochs before early stopping for the various SGD variants evaluated in Sect. 4.4, Fig. 18 shows the training time and number of epochs before early stopping for the various batch sizes evaluated in Sect. 4.5, and Fig. 19 shows the training time and number of epochs before early stopping for the various global learning rates evaluated in Sect. 4.6.
Left: Violin plots showing the distribution of the training time of the 100 MLP5 models trained with each SGD variant evaluated in the 10 rounds of 10-fold CV. Right: Violin plots showing the distribution of the number of epochs required to train the 100 MLP5 models trained with each SGD variant evaluated in the 10 rounds of 10-fold CV
Left: Violin plots showing the distribution of the training time of the 100 MLP5 models trained with each batch size evaluated in the 10 rounds of 10-fold CV. Right: Violin plots showing the distribution of the number of epochs required to train the 100 MLP5 models trained with each batch size evaluated in the 10 rounds of 10-fold CV
Left: Violin plots showing the distribution of the training time of the 100 MLP5 models trained with each global learning rate evaluated in the 10 rounds of 10-fold CV. Right: Violin plots showing the distribution of the number of epochs required to train the 100 MLP5 models trained with each global learning rate evaluated in the 10 rounds of 10-fold CV
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dunn, A.J., Coniglio, S., ElRefai, M. et al. Deep learning and hyperparameter optimization for assessing one’s eligibility for a subcutaneous implantable cardioverter-defibrillator. Ann Oper Res 328, 309–335 (2023). https://doi.org/10.1007/s10479-023-05326-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10479-023-05326-1






















