Abstract
Due to the progress in image processing and Artificial Intelligence (AI), it is now possible to develop automated tool for the early detection and diagnosis of Alzheimer’s Disease (AD). Handcrafted techniques developed so far, lack generality, leading to the development of deep learning (DL) techniques, which can extract more relevant features. To cater for the limited labelled datasets and requirement in terms of high computational power, transfer learning models can be adopted as a baseline. In recent years, considerable research efforts have been devoted to developing machine learning-based techniques for AD detection and classification using medical imaging data. This survey paper comprehensively reviews the existing literature on various methodologies and approaches employed for AD detection and classification, with a focus on neuroimaging techniques such as structural MRI, PET, and fMRI. The main objective of this survey is to analyse the different transfer learning models that can be used for the deployment of deep convolution neural network for AD detection and classification. The phases involved in the development namely image capture, pre-processing, feature extraction and selection are also discussed in the view of shedding light on the different phases and challenges that need to be addressed. The research perspectives may provide research directions on the development of automated applications for AD detection and classification.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Healthcare management is one of the most pressing challenges for most governments, owing to the presence of various global phenomena, one of which is the ageing population. Medical research has a pivotal role in improving human lives since it allows for better diagnostic tools and medicines. Dementia, a disease affecting mainly the elderly, is an umbrella term for a neurological illness in which brain cell death causes memory loss and cognitive deterioration. Dementia is seventh in causing human death worldwide (Gauthier et al. 2021). The World Alzheimer Report 2021 (Gauthier et al. 2021) asserts that 55 million individuals are struggling with dementia currently. There are several types of dementia, the most common one being AD.
AD causes the death of neurons in the cortical region. The removal of debris found in neurons is the responsibility of two proteins known as beta-amyloid and tau. These proteins become harmful to the brain of a person who has been affected by AD. Abnormal tau increases, forming tangles inside neurons in time while beta-amyloid aggregates into plaques which steadily accumulate between neurons. Neurons lose their ability to communicate over time, which results in their death, and the hippocampus, responsible for learning and memory, starts to shrink. AD advances slowly before manifesting any symptoms (Association 2018). There exists a stage between AD and healthy known as mild cognitive impairment(MCI). MCI refers to patients in the initial stages of AD, whereby the patient shows cognitive loss but can still conduct daily tasks.
1.1 Diagnosis of AD
AD is diagnosed using elemental laboratory tests, quick cognitive tests, and structural imaging (H.Feldman et al. 2008). In addition to these assessments, functional imaging, neuropsychological testing, and cerebrospinal fluid (CSF) biomarker measurements are performed.
If a person is suspected of having AD, the first test that their doctor will recommend is a cognitive test. Cognitive tests are used to diagnose and assess the degree of memory and cognitive issues. The standard tests utilised to diagnose AD are the Mini-Mental State Examination (MMSE) and the Clinical Dementia Rating (CDR). The MMSE is divided into two sections, the first of which is entirely oral and includes attention, memory, and orientation. The second component evaluates the capacity to name items, execute written and verbal instructions, and reproduce a complex shape (F.Folstein et al. 1975). The exam is not timed, and the maximum possible score is 30. The CDR is built on a semi-structured conversation with the patient and a suitable observer. The cognitive capacities of the patient are graded on a five-point scale in six categories (memory, orientation, judgement, problem solving, communal affairs, and personal care). The global CDR is determined by clinical scoring standards (C.Morris 1993).
Laboratory testing is performed to rule out other comorbidities that could exacerbate the symptoms (Knopman et al. 2001). Vitamin B12 and thyroid functions are some of the recommended blood tests for routine examination. Knopman et al. (2001) stated that patients with B12 deficiency performed lower in cognitive tasks and patients with thyroid problems exhibited decreased mental status test scores and word fluency.
Neuropsychological testing can reveal the presence, nature, and severity of cognitive impairments (Belleville et al. 2014). Neuropsychological assessments assess a wide variety of cognitive processes, including attention and focus, speech and language ability, learning and memory, reasoning and problem solving (Rodney D. 1994). A clock drawing test (CDT) is a neuropsychological test used that uses a patient’s sketches of a clock to determine the onset of AD. The patient is instructed to draw numbers and hands to show the exact time. Irregularities in the drawings, such as erroneous number placement, omission of numerals, or inappropriate sequencing, indicate cognitive dysfunction (Harbi et al. 2017).
Biomarkers supply detailed measures of abnormal changes in the brain, which may be used to identify people who are forming the fundamental pathology of AD (Craig-Schapiro et al. 2009). Biomarkers may aid in early diagnosis, which is especially difficult due to the lack of identifiable signs and symptoms associated with AD (Craig-Schapiro et al. 2009). CSF is a valuable source of biomarkers because it provides a thorough picture of the brain’s biochemical and metabolic properties (Sharma and Singh 2016). The CSF biomarkers amyloid-beta, total-tau (T-tau), and phosphorylated tau (P-tau) allow researchers to understand the functional changes that accompany AD (Blennow and Zetterberg 2018). Several investigations have found a connection between P-tau levels in CSF and neocortical tangle counts (Buerger et al. 2006). CSF T-tau levels were shown to be considerably greater in AD patients, a conclusion that has since been replicated in hundreds of articles utilising different research methods (Olsson et al. 2016). However, Stomrud et al. (2007) discovered that CSF beta-amyloid, not T-tau or P-tau, anticipates future cognitive deterioration in the elderly.
Lumbar punctures are utilised in patients to recover CSF fluids, which are intrusive and unpleasant, making diagnosis difficult and irreproducible (Sharma and Singh 2016). However, neuroimaging biomarkers, able to detect brain atrophy and amyloid plaque deposition, are less invasive (Yu et al. 2021). Rathore et al. (2017) stated that neuroimaging enabled the evaluation of diseased brain alterations linked with AD in vivo.
Manual segmentation of brain scans entails tracing structures slice by slice, which is time-consuming, expensive, and imprecise owing to human error (Akkus et al. 2017). Computer-aided systems are preferred to address the problems faced during manual brain scan segmentation. These are designed to assist radiologists in understanding brain images and detecting brain disorders (Lazli et al. 2020).
1.2 Motivation of this study
In the new global economy, caring for AD patients has become a central issue due to the substantial costs. In 2018, $227 billion were estimated for the total payments for all individuals with AD or other dementia (Association 2018). According to the World Alzheimer Report 2021 (Gauthier et al. 2021), less than 25% of persons with dementia get diagnosed, and in low-income nations, it is less than 10%. Furthermore, the document reveals that 30% of people are misdiagnosed. As a result, effective diagnostic methods are required to assist clinicians in interpreting the findings of the tests used to diagnose AD.
Research is being complemented with the development of artificial intelligence (AI) in the medical field. This field is maturing, with a plethora of well-known methods and algorithms. Machine learning (ML) models are being utilised to predict AD. Feature extraction, feature dimension reduction and classification are the general steps used to develop automated applications. The structures must be manually outlined by defining regions of interest (ROI) (Falahati et al. 2014). These methods, however, demand specialised knowledge and numerous optimisation phases, which can be time-consuming (Jo et al. 2019). Furthermore, typical ML algorithms are not generalisable (Akkus et al. 2017).
DL models, on the other hand, take raw data as input. As a result, the DL models automatically recognise the highly notable characteristics in the supplied data set (Oh et al. 2019). DL algorithms only took off as they became more accessible due to the rise in processing power of Graphics Processing Units (GPUs). The most often used DL model is Convolutional Neural Networks (CNN). Transfer learning (TL) is when proven pre-trained CNNs are utilised in the initialisation step, and they are then re-trained for new tasks by fine-tuning their last layers. TL is especially effective in cases of insufficient data, which is prevalent in the medical field. TL has shown excellent performance in classifying the 1000 classes of ImageNet. For this study, it was of interest to investigate whether TL models can be adopted in the development of AD detection or classification.
2 Methodology
Several computer-aided systems have been utilised to detect AD. The aim of this study is to review existing TL based classification models for the detection of AD. The following steps were pursued to conduct the review:
-
1.
The research questions (RQs) were first defined.
-
2.
The search strategy was applied.
-
3.
The inclusion and exclusion criteria were defined to choose relevant articles.
-
4.
Finally, data extraction was performed in order to answer the RQs.
2.1 Research questions
The research questions for this review have been established and elaborated as follows:
-
RQ1: What are the neuroimaging techniques used to diagnose AD? There exists several neuroimaging techniques. This question answers which neuroimaging techniques are adept for the diagnosis of AD and in which areas of neuroimaging are research still going on.
-
RQ2: What datasets are available to train the TL models? The foundation of ML, DL and TL is the availability of large volumes of data to train the models. However, in medical imaging, it is difficult to obtain labelled data. It is therefore imperative to identify the datasets that are available to train the models.
-
RQ3: What techniques are used to augment the data given that a large volume of data is usually required for the training of Artificial Intelligence (AI) models ? In medical imaging, as mentioned above, it is difficult to obtain labelled data. Moreover, often times, there is a class imbalance, whereby there is a over-representation of the healthy class and the diseased class is under-represented. The aim of this question is to identify how class imbalance can be reduced and in turn prevent over-fitting.
-
RQ4: What pre-processing techniques are used to deal with the neuroimaging biomarkers? Sometimes, some pre-processing needs to be performed to improve the accuracy obtained while training the model. This question aims to answer the appropriate pre-processing techniques for neuroimaging.
-
RQ5:What are the data management techniques used for training the models Depending on the extracted features, there are several approaches to input data management in order to feed to the model. This question aims to describe the different categories of data input management.
-
RQ6: Which TL models have been used for the detection of AD? The purpose of this question is to identify the different TL models used for the detection of AD. The architectures of the TL models will be described as an answer to this question.
-
RQ7: What performance metrics are being used in the experiments carried out on AD and what are the performances achieved? Different techniques lead to different results. There is a lack of standard in the research community due to the contrasting ways of how the experiments are carried out. As an answer to this question, the performance metrics used by some of the researchers will be described and analysed.
-
RQ8: Which software platforms are required for the pre-processing of the neuroimaging data and to implement the TL models? Researchers can utilise a variety of software programmes to undertake pre-processing of neuroimaging data and the development of TL models. This purpose of this question is to identify the software packages.
2.2 Search strategy
Through a search on search engines such as Google Scholar, Paperity and Science Direct, articles on the detection of AD are identified.
2.2.1 Search queries
The search queries are designed to capture articles that specifically address the detection of AD utilizing AI methodologies. Each query combines relevant terms related to AI techniques and Alzheimer’s disease. Some of the search queries are described as follows:
-
"Artificial Intelligence" AND "Alzheimer"-This query aims to retrieve articles that discuss the intersection of AI and Alzheimer’s disease without specifying any particular AI technique.
-
"Deep Learning" AND "Alzheimer"-This query focuses on articles that utilize deep learning techniques for AD detection. Deep learning is a subset of AI known for its ability to process complex data patterns, making it suitable for medical image analysis.
-
"Transfer Learning" AND "Alzheimer"-Transfer learning involves leveraging knowledge gained from one task to improve learning and performance on another related task. This query targets articles that employ transfer learning methods for AD detection.
-
"AlexNet" Or "VGG16" AND "Alzheimer"-AlexNet and VGG16 are specific deep learning architectures. These queries aim to find articles that utilize these architectures for AD detection, indicating a more specialized focus on neural network models.
-
"Medical images" AND "Data augmentation"-Medical images, such as MRI scans, are commonly used for AD diagnosis. Data augmentation techniques involve artificially increasing the size of a dataset by applying transformations to existing images. This query targets articles that discuss the application of data augmentation methods to medical image datasets for AD detection.
-
"MRI" AND "Pre-processing"-MRI is a widely used imaging modality for studying AD-related brain changes. Pre-processing refers to the steps taken to clean and enhance raw MRI data before analysis. This query seeks articles that focus on pre-processing techniques specifically applied to MRI data for AD detection.
In addition to directly searching for articles using the specified queries, the strategy includes leveraging the references cited in relevant articles. This approach helps to identify additional relevant literature that may not have been captured through initial searches.
By employing a combination of these search strategies, researchers can comprehensively explore the existing literature on AI-based approaches to AD detection, gaining insights into the latest developments, methodologies, and findings in the field.
2.3 Inclusion/exclusion criteria
The Inclusion/Exclusion Criteria section outlines the guidelines used to filter and select relevant papers from the initial pool of search results. Studies which were published from the year 2019 to 2024 were considered for this review.
-
Relevance to neuroimaging modalities: The primary focus of the search is on papers that discuss the detection of Alzheimer’s disease (AD) using neuroimaging modalities, particularly methods involving medical imaging data such as MRI scans. Papers that primarily focus on other diagnostic methods of AD, such as genetic markers or cognitive assessments, are considered irrelevant to the current study and are therefore excluded from further consideration.
-
Evaluation of model performance: To ensure that the selected papers provide meaningful insights into the effectiveness of AI models for AD detection, a criterion is established requiring that the performance of the model be evaluated using at least one metric. This criterion ensures that the selected papers not only propose AI-based approaches but also rigorously assess their performance using quantitative measures. Common evaluation metrics in this context included accuracy, Loss, F1 Score, Precision, ROC, AUC, Specificity, Confusion Matrix
By applying these inclusion/exclusion criteria, We aimed to focus on papers that are directly relevant to our objectives and provide valuable insights into the application of AI techniques for neuroimaging-based detection of Alzheimer’s disease. This systematic approach helps to ensure the quality and relevance of the literature included in the review process, facilitating a more comprehensive and informative analysis of the existing research landscape in this domain.
3 Discussion
Section 3 presents the findings of the research.
3.1 Neuroimaging modalities
According to the papers reviewed, several neuroimaging techniques are employed to identify AD, some of which are currently being researched. These neuroimaging approaches are discussed further below.
3.1.1 Structural magnetic resonance imaging (sMRI)
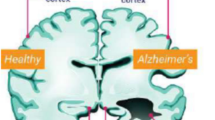
sMRI is a non-intrusive process that uses strong magnets to create magnetic waves to model two- or three-dimensional images of brain architecture without using radioactive substances (Ahmed et al. 2019). Hence, no harmful radiations are generated. Moreover, sMRI is used to research brain anatomy due to its ability to contrast soft tissue (Yamanakkanavar et al. 2020). According to Ahmed et al. (2019), sMRI allows researchers to assess functional and structural brain abnormalities. sMRI estimates of tissue damage or loss in generally sensitive brain regions, such as the hippocampus, have been proven to predict the progression from MCI to AD in multiple investigations (Frisoni et al. 2010). A benefit of sMRI is that the data can be acquired quickly.
3.1.2 Functional magnetic resonance imaging (fMRI)
fMRI is an MRI-based neuroimaging technique that utilises blood oxygenation levels to monitor brain activity (Forouzannezhad et al. 2019). fMRI is a non-intrusive and radiation-free imaging analytic approach for diagnosing, forecasting, and classifying disease stages in cross-sectional or longitudinal studies (Forouzannezhad et al. 2019). Furthermore, fMRI offers a high spatial and appropriate temporal resolution and may be performed concurrently with sMRI (Sperling 2011). Numerous research investigated the links between fMRI patterns and AD. There exist two types of fMRI, namely resting-state fMRI (rs-fMRI) and task-based fMRI (t-fMRI), described below.
Resting-state fMRI
The low-frequency oscillations of the fMRI signal are encapsulated in rs-fMRI (Fox and Greicius 2010). When these spontaneous variations are analysed, the authors (Fox and Greicius 2010) claim that functional connectivity (correlations between distant brain locations) is identified. The brain is monitored in the absence of overt task performance or stimulation. Subjects are typically asked to lie quietly in resting positions. The default mode network (DMN) is a one-of-a-kind brain network characterised by the rest state. The DMN is assumed to be active during waking daydreaming, introspection or thinking about the past or future, and any other time when there is no external stimulus (Forouzannezhad et al. 2019). rs-fMRI can detect minor functional abnormalities in brain networks (Agosta et al. 2012). Tuovinen et al. (2017) conducted a study with 23 AD patients and 25 healthy controls and found that the AD patients had lower functional connectivity in the posterior DMN.
Task-based fMRI
t-fMRI is an advanced methodology for assessing functional activity alternations and brain mapping in response to a given chore (Forouzannezhad et al. 2019). The study is carried out while the patient is performing a task that focuses on a domain such as vision, language, memory, motor, or sensory function (Kesavadas 2013). Li et al. (2015) examined the t-fMRI literature from January 1990 to January 2014 and discovered that MCI and AD patients have aberrant regional brain activation.
3.1.3 Diffusion tensor imaging (DTI)
Diffusion-weighted imaging is an MRI technique for seeing and quantifying the micro-motion of water molecules in living tissues, primarily Brownian motion or water diffusion (Yu et al. 2021). DTI is a technique for identifying AD by analysing water diffusion at the micro-structural level of the brain (Rathore et al. 2017). DTI detects the random movement of water molecules within tissues, allowing it to reflect structural features in the surrounding environment (Alves et al. 2015). The DTI metrics, such as fractional anisotropy (FA) and mean diffusivity (MD), describes changes in white matter structure (Yu et al. 2021). Sexton et al. (2011) examined 41 DTI investigations and concluded that FA was reduced and MD was increased in AD patients.
DTI believes diffusivity to be a Gaussian process and does not take diffusion hindrance into consideration (Arab et al. 2018). The environment of brain tissues, on the other hand, is characterised by non-Gaussian diffusion, according to the authors (Arab et al. 2018). Diffusion kurtosis imaging (DKI) was created as a DTI extension to examine the non-Gaussian dispersal of water diffusion. DKI can offer additional information on the severity of AD and can be utilised to detect the illness in its early stages (Yu et al. 2016).
3.1.4 Proton magnetic resonance spectroscopy (1H-MRS)
1H-MRS is a low-cost, non-intrusive neuroimaging technology which does not use radio-tracers (Murray et al. 2014). 1H-MRS monitors metabolic changes in the brain linked to AD (Talwar et al. 2021). H-MRS quantitatively analyses nuclei and molecules (Yu et al. 2021). 1H-MRS regionally assesses metabolites such as myo-inositol (ml), N-acetyl aspartate (NAA), choline (Cho), and creatine (Cr) (Gradd-Radford and Kantarci 2013). Klunk et al. (1992) discovered a lower amount of NAA in autopsy brain samples from AD patients than healthy controls.
3.1.5 Chemical exchange saturation transfer (CEST)
CEST is a newer MRI contrast enhancement technology which allows for the indirect identification of metabolites with exchangeable protons (Kogan et al. 2013). CEST identifies chemical information using a radio-frequency pulse that magnetically marks the exchangeable protons (Yu et al. 2021). Chemicals containing exchangeable protons are selectively saturated, and when this saturation is transmitted, it is detected indirectly with improved sensitivity via the water signal (van Zilj and Yadav 2011).
3.1.6 Positron emission tomography (PET)
PET can detect pathophysiological and metabolic changes in the brain (Khan 2016). PET employs positron emitter labelled radio-pharmaceuticals that interact with specific cells after administration (Kurth et al. 2013). Following the detection of tracer emissions, the patterns are reconstructed as tomographic pictures of protein levels or brain metabolic activity (Khan 2016). PET imaging can be used to study the interactivity of amyloid-beta and tau in vivo, as well as their impact on neurodegeneration (Khan 2016). PET scans provide clinicians with a visual interpretation of the results with a colour scale, as well as quantitative regional brain data that may be used to objectively assess diagnostic performance and treatment effectiveness (Nordberg et al. 2010). There are two types of PET imaging, namely amyloid-PET and 2-[18F]-fluoro-2-deoxy-d-glucose (18F-FDG) PET.
Amyloid-PET
Amyloid-beta imaging shows increased tracer binding in cortical areas with high amyloid plaque concentrations (Rowe and Villemagne 2013). PET amyloid imaging demonstrates that amyloid buildup in the brain begins before cognitive symptoms, and potentially even before modest memory loss (Nordberg et al. 2010). Amyloid accumulation occurs before tau pathology in the progression of AD, and hence amyloid-beta PET scanning could offer information about pre-symptomatic AD (Khan 2016). 11C-Pittsburgh Compound B (11C-PIB), and 18F-labelled amyloid-PET tracers (florbetapir, flutemetamol, and florbetaben), are used to determine amyloid burden (Rathore et al. 2017). The cortical and sub-cortical brain areas of 16 AD individuals were shown to have increased 11C-PIB retention compared to equivalent brain regions in healthy controls in the first PET investigation to employ this substance (Klunk et al. 2004).
2-[18F]-fluoro-2-deoxy-d-glucose (18F-FDG) PET
Neuronal degeneration causes reduced glucose metabolism in the afflicted brain areas (Kurth et al. 2013). The 18F-FDG can monitor brain glucose metabolism (Nordberg et al. 2010). AD patients exhibited lower 18F-FDG activity as compared to age-matched controls (Khan 2016). Zhang et al. (2012) conducted a meta-analysis in which they discovered that 18F-FDG PET shows a sensitivity of 78% and a specificity of 74% for predicting MCI to AD conversion within 1-3 years.
3.1.7 Single-photon emission computerised tomography (SPECT)
SPECT is a nuclear imaging modality which generates quantitative three-dimensional pictures using radio-labelled bio-molecules with short half-lives (Coimbra et al. 2006). Brain SPECT may investigate regional cerebral blood flow (CBF), which corresponds to resting glucose consumption and reflects neuronal activity (Ortiz-Terán et al. 2011). The authors (Ortiz-Terán et al. 2011) also stated that CBF accurately reflects neuronal activity in each brain area, allowing for an early diagnosis of functional issues before the onset of clinical illness. Jobst et al. (1998) discovered that SPECT had 89% sensitivity, 80% specificity, and 83% accuracy in 200 cases of dementia and 119 controls.
Figure 1 answers RQ1 by depicting the neuroimaging modalities used in the automated detection of AD.
Neuroimaging methods provide information about the structure, function, and metabolism of the brain and are essential for the detection and classification of Alzheimer’s disease (AD). In-depth pictures of cortical atrophy and hippocampus volume loss are provided by structural MRI, which helps in early identification and tracking the course of the disease. Functional MRI detects variations in brain activity to identify functional abnormalities and correlations of cognitive impairment. Diffusion Tensor Imaging evaluates the integrity of the white matter, exposing changes in connectivity that are critical in AD. While amyloid-beta plaque deposition and changes in metabolic activity are detected by PET and SPECT scans, biochemical alterations are identified by magnetic resonance spectroscopy. Amyloid-beta and tau protein aggregates are the focus of emerging PET imaging, which improves therapy monitoring and diagnostic precision. A thorough picture of AD pathophysiology is provided by integrating the results of different approaches, which supports diagnosis, prognosis, and the development of new therapies.
To sum up, every neuroimaging method provides distinct perspectives on the aetiology and development of Alzheimer’s disease, and when combined, they can yield a thorough comprehension of the illness’s course. In AD research and clinical practice, integrating results from many modalities can improve prognostic evaluation, treatment development, and diagnostic accuracy. Because a number of neuroimaging techniques can provide important insights into the structure, function, and pathology of the brain, they are frequently utilised in Alzheimer’s disease research and clinical treatment. From our analysis, structural magnetic resonance imaging (MRI) is one of the most widely used methods among them. It can be observed that, with the help of its precise photographs of brain architecture, one may see the structural alterations associated with Alzheimer’s disease, such as hippocampal volume reduction and cortical atrophy. Furthermore, a lot of PET (Positron Emission Tomography) imaging is used, especially Amyloid PET and FDG-PET (Fluorodeoxyglucose-PET). While Amyloid PET visualises the formation of amyloid-beta plaques, which helps in AD diagnosis and staging, FDG-PET assesses glucose metabolism, showing regions of lower metabolic activity associated with neurodegeneration in AD. CT is widely available and less expensive than MRI or PET. But overexposure to ionizing radiation is a concern, for repeated use. All imaging techniques are of different quality and resolution. It is important to pre-process the images to enhance their quality.
3.2 Existing datasets
The principal dataset leveraged by researchers for performing investigations on AD is Alzheimer’s Disease Neuroimaging Initiative (ADNI). ADNI, a North-American-based study, is a multi-centre, ongoing study tracking clinical, imaging, genetic, and biochemical indicators for AD in its early stages. The research expected to enrol 400 patients with early MCI, 200 people with early AD and 200 healthy controls (Weiner et al. 2012).
The Open Access Series of Imaging Studies (OASIS), a project aimed at making neuroimaging datasets freely accessible, is another used dataset. OASIS contains cross-sectional and longitudinal MRI data, whereby the former incorporates 416 patients aged from 18 to 98 while the latter includes data from 150 people aged from 60 to 90 (Marcus et al. 2010). The strength of OASIS dataset is that a detailed documentation about its processes is available.
Researchers also utilise databases such as Australian Imaging, Biomarkers and Lifestyle (AIBL) and Minimal Interval Resonance Imaging in Alzheimer’s Disease (MIRIAD) (Leandrou et al. 2018). The AIBL includes clinical and cognitive testing, MRI, PET, bio-specimens, and dietary/lifestyle assessments. The study recruited 1112 volunteers, with 211 having AD, 133 having MCI and 768 healthy controls (Ellis et al. 2009). AIBL contains sMRI and amyloid-PET tracers in conjunction, which makes it difficult to use on single modality studies. Moreover the size of the data is undersized compared to other datasets. On the other hand, the MIRIAD dataset includes 46 patients with mild-to-moderate AD and 23 healthy controls who had longitudinal T1 MRI scans. It consists of 708 scans conducted by the same radiographer with the same scanner at 2, 6, 14, 26, 38, and 52 weeks, 18 and 24 months from baseline, with gender, age, and MMSE scores recorded (Malone et al. 2013). Its drawback is that the number of scans is not enough for training models, so the images have to be augmented.
Kaggle, an open-source platform, has also been employed for providing AD images. The dataset contains 6400 brain images. The images are divided into four classes, namely Mild Demented, Moderate Demented, Non-Demented and Very Mild Demented. The data was collected manually from various websites, with every label verified. The dataset has already been partitioned into training and testing sets. The brain scans were acquired in the axial plane. The major drawbacks of Kaggle are:
-
1.
There exists class imbalance since Moderate Demented contains only 52 images for training,
-
2.
Only axial images are available,
-
3.
It is not possible to know whether the images were pre-processed since they were curated from different sources,
-
4.
There is a lack of information regarding the criteria for selecting the images.
Some researchers employed different datasets for their studies by training the models on one dataset and evaluating the robustness of their model by testing it on another dataset (Ding et al. 2019). Moreover, some researchers collected an independent dataset that they utilised.
Table 1 provides an overview on the different datasets available for the detection of AD and in turn answers RQ2. From the column chart in Fig. 2, it can be inferred that sMRI has the highest percentage in each database since it is easily accessible and cheap. Neuroimaging modalities such as CEST, 1H-MRS, DKI, and SPECT are unavailable in these databases.
To help in the creation and assessment of computer vision and machine learning models, a number of datasets have been employed. The Alzheimer’s Disease Neuroimaging Initiative (ADNI) is a well-known dataset that contains structural MRI, PET, and cerebrospinal fluid biomarker data from people who have AD, mild cognitive impairment (MCI), and healthy controls. ADNI has been widely applied to a number of tasks, such as cognitive decline prediction, stage-by-stage classification of AD, and segmentation of brain images. The Open Access Series of Imaging Studies (OASIS) is another frequently used dataset that includes MRI images from both AD patients and healthy controls. It is ideal for researching brain shrinkage and structural alterations linked to the advancement of AD.
Furthermore, freely accessible datasets like the Alzheimer’s Disease Image Dataset (ADNI-2) and the Medical Image Computing and Computer-Assisted Intervention (MICCAI) Grand Challenge on Alzheimer’s Disease offer important tools for comparing algorithms and encouraging teamwork in research projects. These datasets differ in terms of demographics, clinical annotations, imaging modalities, and sample sizes, which allows researchers to tackle a range of research topics and provide reliable computer vision solutions for prognosis and diagnosis of AD (Huang 2023). In our opinion and from the analysis made, to ensure the reliability and generalizability of findings in computer vision research on Alzheimer’s disease, standard evaluation metrics and data preprocessing techniques are crucial. These challenges include data heterogeneity, limited sample sizes, and variability in image acquisition protocols.
3.3 Data augmentation techniques
One of the issues in AD prediction studies is the lack of training datasets. Each categorical dataset is limited, and the success of computer-aided systems is dependent on a vast volume of high-quality information (Yang and Mohammed 2020). Data augmentation is a strategy that generates new data used to train the model and enhances performance when tested on an unknown dataset (Chlap et al. 2021). Data augmentation aids in the resolution of two issues: over-fitting and class imbalance. Over-fitting happens when a model cannot predict previously unseen cases due to its inability to generalise well to new testing data (Afzal et al. 2019). Class imbalance occurs when one or more of the classes to be classified are under-represented, driving the model to favour the over-represented class (Chlap et al. 2021). The safety of a data augmentation technique is defined by its possibility of retaining the label after transformation (Shorten and Khoshgoftaar 2019). Rotations and flips, for example, are typically safe on classification issues such as dog vs cat but dangerous on digit identification tasks such as 6 against 9 (Shorten and Khoshgoftaar 2019). Basic data augmentation techniques include geometric transformations, cropping, occlusion, intensity operations, noise injection, and filtering. However, a question arises as to which of these data augmentation methods is suitable for neuroimaging. The chosen techniques should not modify the neuroimage to such an extent that it loses the data that communicates the presence of AD. Methods considered to be suitable, depicted in Fig. 3, are described next.
3.3.1 Geometric transformations
A flip is created by reversing the pixels horizontally or vertically. Rotation augmentations are performed by rotating the image to the right or left on an axis ranging from \(1^\circ \) to \(359^\circ \) (Shorten and Khoshgoftaar 2019). The rotation degree parameter strongly influences the safety of rotation augmentations (Shorten and Khoshgoftaar 2019). Rotating \(90^\circ \) in the plane specified by axes could be considered for neuroimaging modalities. Ajagbe et al. (2021) used random flipping and zooming as data augmentation techniques in their study of multi-class classification of AD. Likewise, Hazarika et al. (2021) utilised rotation, mirroring and reflection for augmenting their datasets.
3.3.2 Intensity operations
Intensity operations change the pixel/voxel values in an image by adjusting the image’s brightness or contrast (Chlap et al. 2021). Gamma correction and histogram equalisation(HE) are standard approaches for adjusting image contrast (Chlap et al. 2021).
3.3.3 Noise injection
The goal of noise injection is to simulate noisy visuals (Chlap et al. 2021). The typical application of noise injection is when image intensities are changed by randomly sampling a Gaussian distribution (Chlap et al. 2021). Uniform noise shifts values by randomly selecting a uniform distribution (Chlap et al. 2021). Additionally, salt and pepper noise, which randomly sets pixels to black or white, can be employed (Chlap et al. 2021).
3.3.4 Filtering
Filtering, a popular image processing technique for sharpening and blurring images, functions by dragging an n*n matrix across an image (Shorten and Khoshgoftaar 2019). A Gaussian blur filter produces a blurrier image (Shorten and Khoshgoftaar 2019). On the other hand, a high contrast vertical or horizontal edge filter constructs a sharper image around the edges (Shorten and Khoshgoftaar 2019).
Figure 4 depicts some data augmentation techniques applied to an MRI scan in the sagittal plane.
Examples of some basic data augmentation techniques. Basic data augmentation techniques, namely flipping left-right, flipping up-down, rotating \(90^\circ \) on the (0,1) and (1,0) axes, gamma correction, histogram equalisation, Gaussian noise, uniform noise, salt and pepper noise, Gaussian blur and sharpen are illustrated
3.3.5 Generative adversarial networks (GAN)
GANs employ two competing neural networks, a discriminator and a generator. The generator attempts to generate realistic-looking phoney photos, while the discriminator attempts to determine if the data is genuine or phoney. The adversarial loss is critical to GAN’s success (Chlap et al. 2021). It forces the model to generate visualisations that are indistinguishable from real-world imagery (Chlap et al. 2021). A standard GAN architecture is shown in Fig. 5.
Bai et al. (2022) have developed a brain slice generative adversarial network for AD detection (BSGAN-ADD) to categorise the disease. The proposed technique merges GAN and CNN. BSGAN-ADD extracts the representative features of the brain based on an adversarial method. This is in contrast to the encoder technique since BSGAN-ADD takes feedback from the classifier, and thus, impacts the image reconstruction with more apparent category information. This follows by the use of deep CNN, which extracts the brain feature from the category-enhanced images. Consequently, the generator of the GAN offers more representative features for the classifier to have a more accurate diagnosis in terms of proper classification. Sajjad et al. (2022) have applied Deep Convolution Generative Adversarial Network (DCGAN) to produce synthetic PET images. In fact, DCGAN is an improved form of GAN, which consist of two stages namely: a learning phase, and a generation phase. The generator provides an input to the discriminator to generate a sample. Consequently, the generator is trained to deceive the discriminator, which determines whether a picture is fake or real. DCGAN has two features over GAN. One is the batchNorm, which normalises the extracted features. The other one is the LeakyRELU, which avoids vanishing gradients in the model. In addition, the max-pooling layer is being substituted by the convolutional strided layer. Batch Normalisation is utilised rather than the fully connected layer.
The effectiveness and generalizability of machine learning models for Alzheimer’s disease (AD) detection and diagnosis are greatly enhanced by data augmentation techniques. These techniques address problems arising from lack of data, unequal distribution of classes, and variability in medical imaging data by artificially increasing the training dataset. In our opinion, using geometric operations like translation, flipping, and rotation in conjunction with intensity modifications like contrast alteration and spatial deformations speeds up the augmentation process and improves the model’s resistance to anatomical changes and imaging artefacts. The dataset can be further enhanced by noise injection and domain-specific augmentation methods, such as patch extraction and 3D augmentation, with a variety of anatomical and contextual variables pertinent to AD pathophysiology.
An enhanced method of supplementing data that strengthens model robustness against adversarial examples is provided by adversarial perturbations. When combined, these tactics help to raise the precision and consistency of AD diagnosis models. The application of these augmentation strategies has been extensively documented in a number of papers, such as those by Shi et al. (2017) and Suk and Shen (2013), demonstrating how effective they are in improving the performance of AD detection systems. Though there is some preliminary work that has been conducted on data augmentation, it is sine-qua-non to further explore the different advanced data augmentation techniques. This can cater to the lack of annotated datasets available for medical images.
3.4 Pre-processing techniques
The importance of pre-processing in determining the effectiveness of any intelligent system cannot be overstated. Pre-processing cleans up the data by removing artefacts and noise to improve image quality. Figure 6 illustrates some of the pre-processing techniques for some of the neuroimaging modalities.
3.4.1 Intensity normalisation
Data is subjected to intensity normalisation to assure that every volume has the identical mean intensity (Ramzan et al. 2020). Because images from different scanners may have significant intensity differences, this method attempts to account for scanner-dependent differences (Sun et al. 2015). One of the benefits of intensity normalisation is the increase in processing speed. On the other hand, it may result in irreversible data loss if done incorrectly. Francis and Pandian (2021) performed intensity normalisation on their images. Likewise, Sharma et al. (2022) have standardised all the sMRI input images using normalisation. In fact, the intensity values of the ADNI images used were normalised by subtracting the average intensity values and eventually dividing the deviation in such a way that the images obtain a zero mean and unit variance.
3.4.2 Bias-field correction
A bias field is a smooth, low frequency, and unwanted signal which alters sMRI scans due to inhomogeneities in the machine’s magnetic fields (Juntu et al. 2005). Bias field blurs images, reducing the image’s high-frequency components such as contours and edges (Juntu et al. 2005). Furthermore, it modifies the intensity values of image pixels for the identical tissue having varying grey level distribution across the image (Juntu et al. 2005). On the contrary, it estimates the inhomogeneity field from one tissue and spreads it blindly throughout the image.
3.4.3 Anterior commissure (AC) - posterior commissure (PC) alignment
The AC-PC line is used as a reference plane for axial imaging to compare various participants (Murphy 2018). In an AC-PC aligned brain, the AC and PC are in the same axial plane. An advantage of aligning the AC-PC line is that the image quality is consistent from patient to patient.
3.4.4 Contrast enhancement
The variation between the maximum and minimum pixel intensities is referred to as contrast enhancement (Arafa et al. 2022). It enhances picture quality and raises the contrast of image boundaries, allowing us to distinguish between organs (Arafa et al. 2022). Techniques employed to improve the contrast of a medical image are discussed next.
Histogram equalisation
Histogram Equalisation (HE) increases contrast by generating a uniform histogram and may be applied to the entire image or just a portion of it Dawood and Abood (2018). Based on the image histogram, this approach seeks to scatter the grey levels in a picture and reallocates the brightness value of pixels (Dawood and Abood 2018). This approach works best on a low contrast image.
Adaptive histogram equalisation (AHE)
AHE is a strategy that uses localised HE to consider a local window for every individual pixel and calculates the new intensity value based on the local histogram defined in the local window (Dawood and Abood 2018). Even though various rapid approaches for updating the local histograms exist, adaptive features can produce superior results (Dawood and Abood 2018). This technique improves the image’s contrast by altering the values in the intensity picture. AHE tries to address the constraints of global HE by delivering all of the necessary data in one picture (Dawood and Abood 2018).
Contrast limited adaptive histogram equalisation (CLAHE)
CLAHE is an adaptive hybrid of HE and AHE in which the histogram is levelled in blocks with a defined clip limit (Dawood and Abood 2018). CLAHE does not work on the entire picture but rather on the image’s discrete portions known as tiles (Dawood and Abood 2018). In fact, CLAHE employs partial histogram displacement expansion to prevent additional contrast discrepancies after the enhancement process. The pixels in an image are enhanced uniformly when using CLAHE and eventually the histogram range can be adjusted (Chen et al. 2015).
3.4.5 Mean filter
The mean filter is a straightforward linear filter that smooths pictures (Rajeshwari and Sharmila 2013). The mean filter minimises noise and the amount of intensity fluctuation from pixel to pixel (Rajeshwari and Sharmila 2013). The mean filter operates by iterating over the image pixel by pixel, substituting every value with the average value of the pixels around it, including itself. Two potential concerns with applying the mean filter are:
-
1.
A pixel having an unrepresentative value can seriously vary the average of all the pixels in its vicinity.
-
2.
When the filter neighbourhood crosses an edge, the filter interpolates new pixels values on the edge, causing it to blur. This might be an issue if the output requires crisp edges.
3.4.6 Gaussian filter
The Gaussian smoothing operator is a two-dimensional convolution operator which removes noise from pictures (Arafa et al. 2022). In this aspect, it is similar to the mean filter, but it uses a different kernel that resembles the shape of a Gaussian hump. The Gaussian kernel is separable, allowing for quick computing. On the contrary, when applying Gaussian filters, image brightness may be lost, and it is ineffective at reducing salt and pepper noise.
3.4.7 Median filter
The median filter is a method for reducing noise without causing edge blur. The median filter detects noise in pixels by comparing every pixel in the picture to its neighbours (Arafa et al. 2022). It operates via pixel-by-pixel traversal of the image, substituting every value with the median value of adjacent pixels (Arafa et al. 2022). The median value is computed by ordering the values of the adjacent pixels and then substituting the pixel with the matching middle pixel value (Arafa et al. 2022). The median filter works well in removing salt and pepper noise.
3.4.8 Wiener filter
The Wiener filter is useful for reducing the amount of noise in a picture by comparing it to an estimate of the desired noiseless signal (Rajeshwari and Sharmila 2013). Inverse filtering allows a picture to be recovered if it is obscured by a known low pass filter (Rajeshwari and Sharmila 2013). However, the inverse filtering is extremely sensitive to additive noise (Rajeshwari and Sharmila 2013). Wiener filtering strikes the optimum compromise between inverse filtering and noise smoothing, by eliminating the additive noise while also inverting the blurring (Rajeshwari and Sharmila 2013).
3.4.9 Non-local methods
Non-local methods approach image noise removal from a different angle. Non-local methods use redundancy information in images rather than denoising based on local information, such as linear filtering and median filtering (Wang et al. 2021). The picture is partitioned into multiple sections. The methods will explore other blocks with comparable structures and calculate a weighted average estimation as the denoised pixel value to eliminate noise in a specific block (Wang et al. 2021).
3.4.10 Skull stripping
A brain scan includes not just the brain but also skin, skull, eyes, meninges, cranial nerves, and facial muscles (Tax et al. 2022). Skull stripping is a scan method that removes non-brain tissues such as skull tissues and neck (Ramzan et al. 2020). CSF is utilised to perform skull stripping (Arafa et al. 2022). Hazarika et al. (2021) pre-processed their images using the skull stripping technique.
3.4.11 Motion correction
Subjects’ head motion can have multiple effects on the quality of the data obtained. To name a few (Chen and Glover 2016):
-
1.
Motion combines signals from neighbouring voxels, resulting in substantial signal fluctuation at the boundaries of different cortical regions.
-
2.
Motion interacts with field inhomogeneity and slice excitation, introducing more complicated noisy fluctuations.
Hence, motion correction is a technique for eliminating and correcting the influence of people’s head movements during data acquisition procedures (Ramzan et al. 2020). Motion correction is valid on all current field strengths as well as clinical scanners.
3.4.12 Spatial smoothing
Spatial smoothing employs a full-width at half maximum (FWHM) Gaussian filter with a width of 5-8 mm to minimise noise while keeping the underlying signal intact (Ramzan et al. 2020). The magnitude of the underlying signal determines the extent of spatial smoothing (Ramzan et al. 2020). Its primary benefit is that it increases the signal-to-noise ratio (Chen and Glover 2016). However, when the activation peaks are less than twice the FWHM apart, they are considered a single activation rather than two discrete ones.
3.4.13 High pass filtering
High-pass filtering withdraws low-frequency noise signals induced by psychological anomalies such as breathing, heartbeat, or scanner drift (Ramzan et al. 2020). A benefit of high-pass filtering is that it sharpens the image. Nevertheless, unwanted phase shifts in certain frequencies emerge.
3.4.14 Spatial normalisation
Spatial normalisation entails mapping a subject’s neuroimage to a reference brain region, allowing the comparison of subjects with different anatomy. It is commonly accomplished in two stages:
-
1.
co-registration of functional and structural pictures, and
-
2.
registration of the anatomical picture to a high-resolution structural template.
The former prevents inconsistent geometric distortions caused by distinct imaging contrasts, whilst the latter seems more resilient owing to increased structural image resolution and quality - the use of which is dependent on the specific scanning environment and imaging protocols (Chen and Glover 2016).
3.4.15 Wavelet denoising
Wavelet transform decomposes a signal into multiple frequency components at various resolution scales, showing the spatial and frequency properties of a picture concurrently (Rajeshwari and Sharmila 2013). Wavelet-based denoising is computationally efficient and needs minimal manual involvement because there is no parameter adjustment (Wang et al. 2021). Further, wavelet approaches can preserve edges (Wang et al. 2021).
3.4.16 Tissue segmentation
Tissue segmentation is the process of dividing a neuroimaging modality into segments that correspond to different tissues (grey matter (GM), white matter (WM), and CSF), probability maps, and metabolic intensities of the brain region. An issue with tissue segmentation is that post-processing is required for third-dimensional images.
Because preprocessing approaches are designed to address issues with data quality and feature extraction from medical imaging data, they are essential to the construction of machine learning models for the diagnosis and classification of Alzheimer’s disease (AD). To ensure uniformity in image intensities across many participants and scanners and reduce variability that could skew classification findings, intensity normalisation is crucial. Nevertheless, biases in the data may be introduced by factors like image capture parameters and scanner features, which might affect how successful normalisation procedures are.
We have seen that spatial normalization helps align images to a common template space, facilitating inter-subject comparisons; however, it may lead to information loss or distortion if not performed accurately. Noise reduction techniques, such as Gaussian smoothing or filtering, are crucial for suppressing image artifacts and enhancing the clarity of brain structures, yet excessive filtering can blur important details and impact diagnostic accuracy. Skull stripping is essential for removing non-brain tissues, but inaccurate skull stripping can result in the loss of relevant anatomical information or introduce errors into subsequent analyses. Advanced feature extraction methods offer valuable insights into AD-related structural changes; however, the choice of features and extraction algorithms can significantly impact model performance and generalisability.
3.5 Input data management
Feature extraction approaches evaluate photos to extract the most discernible characteristics that reflect the many classes of objects (Leandrou et al. 2018). Based on neuroimaging data, feature extraction approaches strive to build a quantifiable collection of accurate data such as the texture, form, and volume of distinct brain areas. The input data management approach may be classified into four groups based on the extracted features: slice-based, voxel-based, ROI-based, and patch-based. The categories of input data management are detailed as follows:
3.5.1 Voxel-based
Statistics on voxel distributions on brain tissues such as GM, WM, and CSF are frequently used to create voxel-based characteristics (Zheng et al. 2016). This approach often necessitates spatial normalisation, in which individual brain pictures are normalised to a standard three-dimensional space. This method can gather three-dimensional data from a brain scan. Nonetheless, it disregards the local information provided by the neuroimaging modalities and considers each voxel individually. Ding et al. (2019) used the Otsu threshold method to select the voxels.
3.5.2 Slice-based
Two-dimensional slices are extracted from three-dimensional brain scans in this technique (Agarwal et al. 2021). This approach is implemented to reduce the number of super parameters. On the other hand, complete brain analysis is challenging since two-dimensional picture slices cannot hold all of the information from brain scans (Agarwal et al. 2021). A majority of the studies used slice-based data management during their investigation.
3.5.3 Patch-based
A patch is a three-dimensional cube. The patch-based approach can capture disease-related patterns in the brain by extracting attributes from tiny image patches (Agarwal et al. 2021). The patch-based approach is sensitive to small changes. Additionally, it does not require ROI identification. Nevertheless, it is challenging to determine the most informative image patches.
3.5.4 ROI-based
ROI studies concentrate on the regions deemed as informative. The ROI method focuses on particular areas of the brain known to be impaired with AD, such as the hippocampus. In ROI-based approaches, ROI segmentation is often performed prior to feature extraction (Zheng et al. 2016). The ROI-based procedure can be interpreted easily. Moreover, few features can mirror the entire brain. However, there exists limited knowledge about the brain regions implicated in AD.
From the analysis of the different pre-processing techniques, it can be observed that each of them has their own merits and drawbacks. It also depends on the type and quality of images. However, pre-processing techniques collectively enhance image quality, consistency, and relevance. We have observed that most research work apply pre-processing techniques prior to the application of any ML or DL techniques.
3.6 Architectures of transfer learning models
Artificial Neural Networks are sported loosely on a more undersized scale of a network of neurons in the mammalian cortex (Sharma et al. 2020). Neural networks are composed of numerous layers, each consisting of several interconnected nodes with activation functions (Sharma et al. 2020). Information is delivered into the network via the input layer, which communicates with other layers and processes the incoming data using a weighted connection mechanism (Sharma et al. 2020). The output layer then obtains the processed data.
3.6.1 Brief overview of CNN
CNN is a type of feed-forward neural network that utilises convolution structures to retrieve features from data (Li et al. 2020). Yann Le Cunn, who developed a seven-level convolutional network named LeNet-5 that included back-propagation and adaptive weights for various parameters, was a major contributor to the advancement of CNN (Ajit et al. 2020). All of the primary architecture in use today are variants of LeNet-5 (Ajit et al. 2020). CNNs have several advantages over generic artificial neural networks (Li et al. 2020):
-
1.
Local connections: Each neuron is no longer linked to all neurons in the preceding layer, but only to a subset of them, which aids in parameter reduction and accelerates convergence.
-
2.
Weight sharing: A collection of links might share the same weights, thereby minimising parameters.
-
3.
Dimensionality reduction by down-sampling.
Figure 7 illustrates a standard CNN architecture. Some key concepts related to CNN are detailed next.
Convolution layers
Convolution is a critical phase in the feature extraction process (Li et al. 2020). It essentially convolves or multiplies the pixel matrix generated for the provided input to build an activation map for the given image (Ajit et al. 2020). The fundamental advantage of activation maps is that they store all of the differentiating qualities of a given image while minimising the quantity of data that has to be processed (Ajit et al. 2020). The data is convolved using a matrix, known as a feature detector, which is essentially a set of values with which the machine is compatible (Ajit et al. 2020). Various image versions are created by varying the value of the feature detector (Ajit et al. 2020). When appointing a convolution kernel of a given dimension, information will be lost in the edges (Li et al. 2020). As a result, padding is created to increase the input with a zero value to keep the size of the input and output volume constant (Ajit et al. 2020). Stride is also used to alter the density of convolving (Li et al. 2020). Stride sets how much the distance stride will shift after each convolution (Ajit et al. 2020). It is required to configure stride so that the output volume is an integer (Ajit et al. 2020). Smaller strides produce better results (Ajit et al. 2020), and greater strides create lesser density (Li et al. 2020). A convolution operation with and without padding is depicted in Fig. 8.
Activation functions
Activation functions are used to transform an input signal into an output signal, which is subsequently passed on to the next layer in the stack (Sharma et al. 2020). A neural network lacking an activation function functions as a linear regression model with limited performance and power (Sharma et al. 2020). It is preferable if a neural network not only learns and computes a linear function but also performs complicated tasks (Sharma et al. 2020). Moreover, the type of activation function utilised determines the prediction accuracy of a neural network (Sharma et al. 2020). Some types of activation functions are as follows (Sharma et al. 2020):
-
1.
Linear: The linear activation function correlates directly to the input. It can be defined as:
$$\begin{aligned} f(x) = ax \end{aligned}$$where a is a constant value chosen by the user. The gradient is a constant number independent of the input value x, suggesting that the weights and biases will be modified during the back-propagation phase, albeit with the same updating factor. No such advantage exists to employing a linear function because the neural network would not improve the error because the gradient value would be the same for every iteration. Additionally, the network will be unable to discern complicated data patterns. As a result, linear functions are suited for basic tasks that need interpretability.
-
2.
Sigmoid: The sigmoid activation function is the most commonly employed activation function because it is a non-linear function. The sigmoid function changes the values from zero to one. It is defined as:
$$\begin{aligned} f(x) = \frac{1}{(1 + e^{-x})} \end{aligned}$$The sigmoid function is not symmetric about zero, meaning that the signs of every neuron output values will be identical. Scaling the sigmoid function can help with this problem.
-
3.
ReLU: ReLU is an abbreviation for rectified linear unit, and it is a non-linear activation function commonly employed in neural networks. The advantage of utilising the ReLU function is that not all neurons are stimulated simultaneously, meaning that a neuron will be deactivated only if the linear transformation output is zero. It can be expressed as:
$$\begin{aligned} f(x) = max(0,x) \end{aligned}$$which means that f(x) = x if x\(>=0\) and f(x) = 0 when x\(<0\). ReLU is more efficient than other functions because only a subset of neurons is active at a time. Regardless, the gradient value is zero in some circumstances, implying that the weights and biases are not adjusted during the back-propagation stage of neural network training.
-
4.
Leaky ReLU: Leaky ReLU is an improved variant of the ReLU function where the ReLU function’s value is defined as an extremely tiny linear component of x rather than zero for negative values of x. It can be defined as:
$$\begin{aligned} f(x)= & {} 0.01x, x<0 \\ f(x)= & {} x, x>=0 \end{aligned}$$ -
5.
SoftMax:The softmax function is a concatenation of several sigmoid functions. Since a sigmoid function returns values from zero to one, these may be considered probabilities of data points from a specific class. The softmax function is used for multi-class classification. It can be expressed as:
$$\begin{aligned} \sigma (\textbf{z})_i = \frac{e^{z_i}}{\sum _{j=1}^{K} e^{z_j}} \end{aligned}$$
Pooling layers
Pooling is a key step in further lowering the dimensions of the activation map, retaining only the important elements while reducing spatial in-variance (Ajit et al. 2020). As a result, the model’s learn-able features are reduced, contributing to the resolution of the over-fitting issue (Ajit et al. 2020). Pooling enables CNN to absorb all of the distinct dimensions of an image, allowing it to effectively recognise the provided object even if its shape is distorted or at a different angle (Ajit et al. 2020). Although there are various types of pooling, CNN utilises two types of pooling, namely average and max pooling due to their computational efficiency (Akhtar and Ragavendran 2019). Max pooling takes the greatest value from each sub matrix of the activation map and creates a new matrix from it Ajit et al. (2020). Although max pooling has produced outstanding empirical results, it does not ensure generalisation on test data (Akhtar and Ragavendran 2019). In comparison, the average pooling method takes all of the elements’ low and high magnitudes and computes the mean value of all elements in the pooling zone Akhtar and Ragavendran (2019). Nonetheless, it may fail if there are numerous zeros in the pooling region (Akhtar and Ragavendran 2019). A max and average pooling operation is illustrated on Fig. 9.
Fully connected layer
The fully connected layer is the last layer that is fed into the neural network (Ajit et al. 2020). In general, matrices are flattened before being passed on to neurons (Ajit et al. 2020). It is difficult to follow the data after this point due to the inclusion of various hidden layers with varied weights for each neuron’s output (Ajit et al. 2020).
Dropout
Dropout is a regularisation approach that zeroes out the activity levels of randomly picked neurons during training (Shorten and Khoshgoftaar 2019). This limitation drives the network to assimilate more robust properties instead of depending on a small subset of neurons in the network’s predictive capability (Shorten and Khoshgoftaar 2019).
Batch normalisation
Batch normalisation is another regularisation method that normalises the set of activation in a layer (Shorten and Khoshgoftaar 2019). Normalisation is accomplished by removing the batch mean from every activation and dividing the result by the batch standard deviation (Shorten and Khoshgoftaar 2019). This normalisation approach, coupled with standardisation, is a customary approach in pixel value processing (Shorten and Khoshgoftaar 2019).
3.6.2 Brief overview of TL
TL operates by first training a network on a large dataset, such as ImageNet, and then utilising the weights as the starting weights in a novel classification task (Shorten and Khoshgoftaar 2019). In most cases, only the weights in the convolutional layers are duplicated rather than the complete network, including fully connected layers (Shorten and Khoshgoftaar 2019). Doing so is highly useful since many image datasets contain low-level spatial properties which can be taught more effectively with enormous data (Shorten and Khoshgoftaar 2019).
TL and pre-training are conceptually very similar. However, the network architecture is defined in pre-training and then trained on a large dataset such as ImageNet (Shorten and Khoshgoftaar 2019). Pre-training is different from TL since, in TL, the network design, such as VGG-16 or ResNet, including the weights, must be transferred (Shorten and Khoshgoftaar 2019). Pre-training allows establishing the weights with large datasets while maintaining flexibility in the network architecture design (Shorten and Khoshgoftaar 2019). Figure 10 illustrates a timeline of some TL models.
3.6.3 LeNet-5
In 1998, LeNet-5 was presented, an efficient CNN trained with the back-propagation technique for handwritten character recognition. LeNet-5 was a relatively simple network trained on 32* 32 grey-scale images. LeNet-5 incorporates seven layers, namely two convolution layers, two pooling layers, two fully connected layers and an output layer. Although LeNet-5 is effective in recognising handwritten characters, it falls short of the classic support vector machine and boosting techniques (Li et al. 2020). Figure 11 depicts a standard architecture of LeNet-5.
Aderghal et al. (2020) have used the pre-trained LeNet architecture and applied it on the brain images from the ADNI dataset. On the other side, Sarraf and Tofighi (2016) have achieved an accuracy of 96.86% using CNN DL LeNet architecture.
3.6.4 AlexNet
AlexNet expands on LeNet’s concepts by applying the fundamental concepts of CNN to a deep and broad network (Li et al. 2020). AlexNet includes five convolution layers, two pooling layers, two fully connected layers and an output layer. The following are AlexNet’s primary innovations (Li et al. 2020):
-
1.
AlexNet employs ReLU as the activation function, which alleviates the issue of gradient vanishing when the network is deep. Despite the fact that ReLU was suggested prior to AlexNet, it was not extended until AlexNet.
-
2.
AlexNet employs dropout to avoid over-fitting.
-
3.
AlexNet creates a group convolution structure that can be trained on two different GPUs since one GPU has a processing resource restriction. Then, as the final output, two feature maps created by the two GPUs are blended.
-
4.
In training, AlexNet employs two techniques of data augmentation, one of which is Principal Component Analysis (PCA), used to alter the RGB values of the training set. AlexNet demonstrates how data augmentation can significantly reduce over-fitting and enhance generalisation capacity.
Figure 12 depicts a standard architecture of AlexNet.
Kumar et al. (2022) have used AlexNet for transfer learning for the early detection of AD using sMRI brain images.The architecture was based on the AlexNet 2012 ImageNet neuron network. The architecture comprised of 5 convolutional layers and 3 more classical neural layers. An accuracy of 98.61% was achieved. Likewise, Fu’adah et al. (2021) have used the pre-trained AlexNet model to categorise images into very mild demented, mild demented and moderate demented and have reached an accuracy of 95%.
3.6.5 VGG-16
VGG-16 incorporates 13 convolution layers, five pooling layers, three fully connected layers and one output layer. The authors (Simonyan and Zisserman 2014) raised the network’s depth by adding more convolutional layers, which is possible due to relatively small 3*3 filters, resulting in a more accurate architecture. This streamlined design facilitates ease of implementation and understanding, particularly in comparison to more complex architectures such as Inception or ResNet. The model’s considerable depth, encompassing 16 layers, surpasses that of earlier models like AlexNet, thereby enabling the extraction of more complex features and yielding enhanced performance in large-scale image recognition tasks. Furthermore, VGG16 is extensively utilized for transfer learning. Its pre-trained weights, derived from large datasets such as ImageNet, provide a solid foundation for a variety of computer vision tasks, significantly reducing training time and improving performance on new datasets. Fig. 13 illustrates a standard architecture of VGG-16.
3.6.6 VGG-19
VGG-19 can be considered an extension of VGG-16, whereby blocks three, four and five contain an additional convolution layer. In this way, VGG-19 includes 16 convolutional layers, five pooling layers, three fully connected layers and one output layer. Some authors have used the VGG-19 pre-trained model to classify brain images. Mehmood et al. (2021) have adopted the pre-trained VGG-19 for the early detection of AD. Initially data augmentation was applied on the sMRI and PET images taken from the ADNI dataset. The last two fully connected layers of the architecture were modified and the hyper-parameters were fine-tuned. In another work conducted by Naz et al. (2022), the VGG-19 architecture was applied on the pre-processed brain images and outperformed the other pre-trained models like AlexNet, GoogLeNet, ResNet, DenseNet, InceptionV3 and InceptionResNet.
3.6.7 GoogLeNet
GoogLeNet, also known as InceptionV1, comprises 22 layers using numerous inception models. The network has 50 convolution layers in total, with each module having 1*1, 3*3 and 5*5 convolutional layers, including one 3*3 max-pooling layer (Hazarika et al. 2021). GoogLeNet aimed for efficiency and practicality by using 12 times fewer parameters than AlexNet and a processing cost that is less than two times that of AlexNet while being significantly more accurate (Szegedy et al. 2014). The 1*1 convolutional layers are mostly employed as dimension reduction modules to overcome computational bottlenecks that would otherwise restrain the size of the network (Szegedy et al. 2014). In turn, the depth and width of the network can be increased without sustaining considerable performance penalties (Szegedy et al. 2014). Figure 14 shows a sample inception module block. Shanmugam et al. (2022) have used the GoogLeNet architecture for the training of their model. This architecture was trained on the ImageNet dataset, which consist of large amount of data. Consequently, it tends to have a generalisation ability and was able to achieve a good performance on the ADNI sMRI data.
3.6.8 InceptionV2
In InceptionV2, the output of each layer is normalised to a normal distribution, which increases the model’s robustness and allows the model to be trained at a relatively high learning rate (Li et al. 2020). InceptionV2 also demonstrates that a single 5*5 convolutional layer can be substituted by two 3*3 ones (Li et al. 2020). In the same breath, one n*n convolutional layer can be substituted with one 1*n and one n*1 convolutional layer (Li et al. 2020). However, the original research states that factorisation is ineffective in the early stages (Li et al. 2020). Figure 15 shows a sample InceptionV2 module block.
3.6.9 InceptionV3
InceptionV3 is an improved version of GoogLeNet and InceptionV2. The goal of InceptionV3 was to lower the computational cost of deep networks while maintaining generalisation (Khan et al. 2020). InceptionV3 factorises 5*5 and 3*3 convolution kernels into two one-dimensional ones (Li et al. 2020). This procedure speeds up the training process while increasing network depth and non-linearity (Li et al. 2020). Furthermore, the network’s input size was adjusted from 224*224 to 299*299 (Li et al. 2020).
3.6.10 ResNet
As the number of layers increases in neural networks, there are more likely to have issues such as vanishing gradient (Li et al. 2020). By introducing the notion of residual learning in CNNs and establishing an efficient framework for deep network training, ResNet revolutionised the CNN architectural race (Khan et al. 2020). The notion of a residual block is proposed to overcome the vanishing gradient (Hazarika et al. 2021). The fundamental idea is to create a shortcut bridge that allows the bypassing of one or more layers (Hazarika et al. 2021). The structure of a three-layer residual block is depicted in Fig. 16.
3.6.11 InceptionResNet
InceptionResNet combined the capabilities of residual learning and the inception block (Khan et al. 2020). As a result, the residual connection took the role of filter concatenation (Khan et al. 2020). The inception modules are improved to fine-tune the layers and detect more relevant aspects (Hazarika et al. 2021). Batch normalisation is not used in this model to reduce network size and make it more efficient for a single GPU (Hazarika et al. 2021).
3.6.12 SqueezeNet
SqueezeNet delivers the accuracy of AlexNet with five times fewer parameters (Ashraf et al. 2021). SqueezeNet’s core design consists of a module known as a fire module. A fire module is made up of a squeeze convolution layer with 1*1 filters, onward to extend layer with both 1*1 and 3*3 convolution filters (Ashraf et al. 2021). The SqueezeNet design begins with standalone convolution, then moves to eight fire modules, and finally to the final convolution layer (Ashraf et al. 2021). Figure 17 depicts the SqueezeNet architecture along with a fire module.
3.6.13 Xception
Xception is an extreme variant of Inception. Xception is a linear stack of depth-wise separable convolution layers with residual connections, making it easy to define and alter (Chollet 2016). A depth-wise separable convolution applies each convolution kernel to only one channel of input, and then a 1*1 convolution is used to aggregate the output of depth-wise convolution (Li et al. 2020). The Xception architecture contains 36 convolutional layers forming the feature extraction base of the network (Chollet 2016). The model is separated into three flows; an entry flow, a middle flow with eight repetitions of the same block and an exit flow. Figure 18 illustrates the architecture of Xception.
3.6.14 MobileNetV1
The depth-wise separable convolutions suggested in Xception are used in MobileNetV1. MobileNetV1 includes the width multiplier, which reduces the number of channels in each layer (Li et al. 2020). In the same breath, it introduces the resolution multiplier, which diminishes the resolution of the input image (Li et al. 2020).
3.6.15 MobileNetV2
MobileNetV2 improves on MobileNetV1 by introducing two new features, namely inverted residual blocks and linear bottleneck modules (Li et al. 2020). An inverted residual block’s input is convoluted by 1*1 convolution filters for channel expansion, then by 3*3 depth-wise separable convolution, and finally by 1*1 convolution kernels to condense the number of channels back (Li et al. 2020). Since depth-wise separable convolution cannot modify the number of channels, the number of input channels restricts feature extraction (Li et al. 2020). Inverted residual blocks are used to rectify this problem. The information is easily destroyed when the ReLU activation function is used after depth-wise convolution. As a result, the ReLU activation following the second 1*1 convolution of inverted residual blocks is deleted, and a linear transformation is utilised (Li et al. 2020). Therefore, this architecture is known as the linear bottleneck module.
3.6.16 DenseNet
Like ResNet, DenseNet was proposed to rectify the vanishing gradient issue. The issue with ResNet was that it explicitly retains information through additive identity transformations, which resulted in multiple layers contributing very little or no information (Khan et al. 2020). DenseNet uses cross-layer connectivity in a modified form to address this issue. DenseNet utilised a feed-forward connection to connect each preceding layer to the next layer (Khan et al. 2020). Hence, feature maps from all prior layers were used as inputs into all successive layers (Khan et al. 2020). DenseNet concatenated the previous layer’s characteristics rather than adding them Khan et al. (2020). Hence, the network can explicitly distinguish between information introduced or preserved (Khan et al. 2020). Figure 19 depicts the architecture of DenseNet. Islam and Zhang (2017) have explored DenseNet on sMRI images obtained from the OASIS dataset. The authors have considered patches from three physical planes of imaging namely axial or horizontal plane, coronal or frontal plane, and sagittal or median plane for each sMRI image. These patches were then fed into the CNN. An accuracy of 93.18% were achieved.
3.6.17 NASNet
NASNet designed a new search space that allows transferability. This transferability was achieved in such a way that the architecture’s complexity is independent of the network depth and the size of input images (Zoph et al. 2017). All convolutional networks in the search space are made up of convolutional cells with the same structure but different weights (Zoph et al. 2017). Thus, the quest for the best convolutional architecture is minimised to the search for the best cell structure. Looking for the optimal cell structure has two major advantages: it is significantly quicker than looking for the best network architecture, and the cell itself is more likely to generalise to other issues (Zoph et al. 2017). To develop scalable architectures for images of any size, the researchers (Zoph et al. 2017) used two types of convolutional cells, namely the Normal Cell and the Reduction Cell. The Normal Cell returns a feature map of the exact dimension, whereas the Reduction Cell yields a feature map where its height and width are lessened by a factor of two Zoph et al. (2017). Additionally, NASNet utilises the reinforcement learning search method to optimise architecture arrangements. Finally, NASNet improves generalisation by using a new regularisation technique called ScheduledDropPath, which drops out each path in the cell with a probability that increases linearly during training (Zoph et al. 2017).
3.6.18 EfficientNet
It is customary in prior work to scale only one of the three dimensions: depth, width and image size (Tan and Le 2020). Resolution scaling is performed to obtain more complex features and fine-grained patterns. Depth scaling is done to handle high-resolution images, while width scaling is performed to increase the number of channels. Although it is possible to freely scale two or three dimensions, uncontrolled scaling involves time-consuming manual adjustment and typically leads in sub-optimal accuracy and efficiency (Tan and Le 2020). EfficientNet scales all the dimensions of depth, width and resolution using a compound coefficient (Tan and Le 2020).
TL techniques significantly enhance AD diagnostic models by adapting pre-trained architectures like EfficientNet, VGG16, ResNet, AlexNet, and InceptionV3 to medical imaging tasks (Gupta et al. 2024). EfficientNet offers superior performance and efficiency, making it ideal for detailed analyses with limited resources. VGG16 captures intricate features but demands high computational power, while ResNet’s residual connections allow for deep and effective feature learning, improving diagnosis accuracy. AlexNet, although foundational, is less suited for nuanced tasks compared to modern models. InceptionV3’s multi-scale feature extraction balances complexity and efficiency, making it effective for identifying diverse AD manifestations. Each model’s unique strengths cater to different aspects of AD diagnosis, enhancing early detection and treatment strategies. Shanmugam et al. (2022) and Raza et al. (2023) have explicitly discussed on the different TL techniques and have reported their outstanding performance.
3.7 Potentials and limitations of transfer learning in AD
Transfer learning has emerged as a crucial methodology within Alzheimer’s disease research, particularly in the analysis of medical imaging data (Nanni et al. 2020; Kora et al. 2022; Pradhan et al. 2024). TL leverages the knowledge embedded within pre-trained models, initially developed for tasks such as image classification, and adapts it to the specific objective of identifying AD-related anomalies in medical images, such as MRI scans or PET scans (Kim et al. 2022; Shanmugam et al. 2022). By utilizing feature representations derived from extensive datasets like ImageNet, TL models effectively identify intricate patterns and deviations in brain images indicative of AD progression. This facet of feature representation acquisition through TL is especially advantageous for detecting subtle variations in brain morphology or function, even in scenarios where labeled medical imaging data relevant to AD diagnosis is limited or costly to obtain (Yi et al. 2023).
Moreover, TL significantly reduces the dependence on labeled data for AD detection in medical imaging by facilitating knowledge transfer from similar tasks. This feature is particularly important in medical settings where acquiring annotated anomaly data for AD diagnosis is challenging due to the rarity of anomalies and the need for specialized annotations. Additionally, TL accelerates the convergence during model training by initializing the AD model with pre-trained model weights, thereby expediting the adaptation process and reducing the computational resources required for model optimization (Salehi et al. 2023). This expedited convergence trajectory promotes the timely development of accurate and reliable diagnostic tools for AD, which is crucial for early intervention and personalized treatment strategies, especially in medical imaging where timely diagnosis is vital for patient care.
However, the effectiveness of TL in AD detection from medical imaging data depends on addressing several domain-specific challenges, including task alignment, catastrophic forgetting, model interpretability, and data bias (Radhika Shetty and Antony 2023; Dixit 2023). Ensuring alignment between the source and target tasks is essential for optimal performance, while preventing catastrophic forgetting requires the implementation of sophisticated fine-tuning strategies to effectively preserve and transfer relevant knowledge. Additionally, addressing interpretability concerns is important for understanding model decisions and gaining insights into disease etiology, while mitigating data bias inherited from pre-trained models is necessary for developing fair and unbiased diagnostic tools for AD in medical imaging. In conclusion, TL presents a promising avenue for advancing AD detection and diagnosis in medical imaging, offering opportunities to develop more accurate, efficient, and interpretable tools for early detection and disease management.
3.8 Performance metrics
For comparing classification performance, balanced accuracy and sensitivity/specificity or precision/recall should be supplied (Rathore et al. 2017). Some performance metrics are detailed next.
3.8.1 Accuracy
Accuracy is the proportion of correct forecasts to total predictions. It can be defined as:
where TP, TN, FP, and FN represent the True Positive, True Negative, False Positive, and False Negative values. A true positive outcome is one in which the model predicts the positive class correctly. A true negative, on the other hand, is a result in which the model predicts the negative class properly. A false positive is when the model predicts the positive class incorrectly. A false negative is an outcome in which the model incorrectly predicts the negative class.
3.8.2 Loss
Loss is a value that indicates how inaccurate the model’s forecast was on a single example. For binary classification, it can be expressed as:
where y is the actual value and p is the predicted value. For multi-class classification, loss can be defined as:
where M is the number of classes, l is the loss value, and p is the predicted value.
3.8.3 F1 Score
F1 score is the arithmetic mean of precision and recall, having a range of [0, 1]. The model’s performance improves as the F1 Score increases. It can be expressed as:
3.8.4 Recall
Recall, also known as True Positive Rate(TPR) or sensitivity, is the accurate positive result amount divided by the total number of relevant sample amounts. It can be defined as:
Precision
Precision is the amount of positive results that match to the quantity of positive results predicted by the classifier. It can be expressed as:
3.8.5 The receiver operating curve (ROC) and Area under the curve (AUC)
It selects a reasonable cut-off Threshold for the model by showing TPR versus False Positive Rate (FPR) for various Threshold values in the range of [0, 1]. FPR, also known as specificity, can be expressed as:
AUC is the area under the ROC curve. For example, if AUC = 0.7, it suggests that the model has an 70% chance of distinguishing between the positive and negative classifications.
3.8.6 Confusion Matrix
A confusion matrix is a summary of classification problem prediction outcomes. The number of correct and incorrect predictions is totalled and split by class.
From Table 2, it can be insinuated that accuracy is the most commonly employed performance metric while specificity is not used as much. Most researchers have employed more than two performance metrics to evaluate their model.
3.9 Critical appraisal of literature review
Detection of Alzheimer’s disease (AD) in MRI images using deep learning
Pradhan et al. (2021) utilised VGG-19 and DenseNet169 architectures for multi-class classification. The sMRI images were obtained from Kaggle. The dataset was augmented using the Image Data Generator function of Keras. However, the authors did not explicitly discuss the augmentations performed. The DenseNet169 model acquired an accuracy of about 80% and an AUC of about 82% in the test data. On the other hand, the VGG-19 model demonstrated accuracy and AUC of 82.6% and 86.7% on the test dataset. The authors hold the view that VGG-19 performs better than DenseNet169. Nonetheless, the models were trained for a different number of epochs, with VGG-19 having the higher one.
Classification of Alzheimer’s disease in MobileNet
Lu et al. (2019) compared the performance of VGG-16 and MobileNet for binary sMRI images categorisation. They used the OASIS dataset. The following steps were done for the data pre-processing:
-
1.
The three-dimensional scan was cut into 176 two-dimensional images.
-
2.
The authors chose the 32 most informative photos based on the image entropy.
-
3.
Finally, the chosen jpg images were resized.
Dropout was employed to avoid over-fitting. However, they did not specify the dropout rate. VGG-16 converges to 92% on the validation set, while MobileNet converges to 94% when run for 200 epochs.
Deep learning based binary classification for Alzheimer’s disease detection using brain MRI images
Hussain et al. (2020) compared a 12-layer CNN model and pre-trained CNN models, namely InceptionV3, Xception, MobileNetV2 and VGG-19. They used 112 T1-weighted sMRI scans without any data augmentation from OASIS dataset for binary classification. The images were resized and denoised in the pre-processing step. Neither to what size were the images resized nor the type of denoising performed were mentioned in the paper. Moreover, the authors did not explain what padding and stride values were used for the proposed CNN model. Accuracy, precision, recall, f1 score and area under the ROC curve were computed as performance metrics, with the proposed CNN model having the highest accuracy of 97.75%.
Early detection of Alzheimer’s Disease using ensemble of pre-trained models
Francis and Pandian (2021) first compared the performance of some pre-trained models, namely ResNet50, VGG-16, DenseNet121, NasNetMobile, EfficientNetB0, Xception and MobileNet. The authors used the middle slice from three-dimensional sMRI scans from the ADNI dataset. The two-dimensional images were normalised. A total of five transformations were applied to the dataset as a data augmentation technique. Conversely, the authors did not specify the type of transformation techniques employed. Xception and MobileNet performed the best on the dataset, with an accuracy of 89.23% and 89.89%, respectively. The models were concatenated to obtain an ensemble model, which gave an accuracy of 91.3%. The authors mentioned the hyper-parameter values in the paper. The research would have been more relevant if binary cross-entropy instead of categorical cross-entropy was used since the classification is a binary one.
Alzheimer’s disease classification using transfer learning
Acharya et al. (2021) sought to classify sMRI images from Kaggle of AD patients into multiple classes using CNN, VGG-16, ResNet50 and modified AlexNet. The images were resized to 127*127. The performance metrics employed were accuracy, precision, recall and f1 score. The modified AlexNet model performed best, according to the authors. However, this claim is debatable since the number of epochs was not the same for each model. Some shortfalls of the research are as follows:
-
1.
They failed to describe the CNN layers and the modified AlexNet explicitly.
-
2.
No data augmentation was mentioned. Hence, there is a lack in understanding of how the authors dealt with class imbalance and over-fitting.
-
3.
No description of the software tools employed.
Classification of Alzheimer’s Disease Using Traditional Classifiers with Pre-Trained CNN
Almadhoun and Abu-Naser (2021) utilised sMRI images for multi-class classification. Nonetheless, they did not mention from where they obtained their dataset. The authors resized the photos and trained them on Xception architecture for 20 epochs. The accuracy achieved was 97.74%. The authors failed to specify the techniques employed to reduce over-fitting and the software tools used.
Classification of Alzheimer’s disease MRI images with CNN based hybrid method
Yildirim and Cinar (2020) engaged sMRI images from Kaggle for the multi-class classification. They cited the software tools employed. However, data augmentation and pre-processing techniques were unspecified. The main weakness of the study is the failure to address how class imbalance and over-fitting should be catered for, since Moderate Demented had 52 images while Non-Demented had 2560 images. Nevertheless, in the description and structure provided for the layers of a hybrid model, it was noticed that it incorporated dropout (rate missing) and batch normalisation. They compared the performance of AlexNet, ResNet50, DenseNet201, VGG-16 and a hybrid model based on ResNet50. Accuracy, precision, recall and f1 score were stated for the hybrid model.On the other hand, confusion matrix and accuracy were given for the remaining models. The hybrid model achieved an accuracy of 90%.
CNN model performance analysis on MRI images of an OASIS dataset for distinction between healthy and Alzheimer’s patients
Khagi and Kwon (2019) used the cross-sectional data of sMRI images from the OASIS dataset for binary classification. They selected the last 30 slices from axial scans. No pre-processing was conducted. The authors modified the models, namely AlexNet, GoogLeNet, ResNet50 and a proposed CNN, in four ways. The proposed CNN model yielded the highest accuracy of 98.5%. Some strengths of the research are as follows:
-
1.
The authors gave justifications of their followed steps.
-
2.
The software tools employed were cited.
-
3.
The researchers explained the proposed CNN layers.
-
4.
The hyper-parameter values were given.
-
5.
They trained the models for the same number of epochs.
Automated classification of Alzheimer’s Disease Based on MRI Image Processing using Convolutional Neural Network (CNN) with AlexNet Architecture
Fu’adah et al. (2021) used sMRI images for multi-class classification. They failed to mention the dataset. Moreover, the authors overlooked data augmentation and pre-processing. AlexNet with different learning rates was the model used. Precision, recall and f1 score were the performance metrics provided.The best learning rate was 0.0001, yielding an accuracy of 95%.
Multi-classification of Alzheimer’s Disease on magnetic resonance images (MRI) using deep convolutional neural network (DCNN) approaches
Ajagbe et al. (2021) employed sMRI images from Kaggle for multi-class classification, performing random flipping and zooming as data augmentation techniques. No pre-processing was done. A proposed CNN, VGG-16 and VGG-19 were the architectures used. The models were trained on CPU for a small number of epochs which led to a high computational time. Accuracy, AUC, f1 score, precision and recall with their graphical representations were provided. The results obtained were described but not explained. VGG-19 obtained the highest accuracy of 77.7%.
AlexNet approach for early stage Alzheimer’s Disease detection from MRI brain images
Kumar et al. (2021) compared the performance of AlexNet, VGG-16, GoogLeNet and ResNet18 for the multi-class classification using sMRI images from OASIS dataset. No data augmentation or pre-processing were discussed. Accuracy and loss with their graphs were included in the paper. AlexNet produced the highest accuracy of 97.53%.
An experimental analysis of different Deep Learning based Models for Alzheimer’s Disease classification using brain magnetic resonance images
Hazarika et al. (2021) opted for T1-weighted MP-RAGE sMRI images from ADNI for multi-class classification. Rotation, mirror and reflection are the transformation techniques used to augment the data. Skull stripping was then performed as a pre-processing step. Several models were used for the training phase, including AlexNet, VGG-16, VGG-19 and GoogLeNet. The hyper-parameters used to train the model are highlighted in the paper. A table having accuracy, precision, recall and f1 score as headers are built up along with graphs to evaluate the model performance. Moreover, the paper has provided a detailed description of the various software tools.
A deep learning model to predict a diagnosis of Alzheimer’s disease by using 18F-FDG PET of the brain
Ding et al. (2019) employed 18F-FDG PET scans from ADNI and a small independent test data. However, the independent test data was not collected from a clinical trial. The authors used Otsu threshold method to select the voxels. Random height and width shift and zooming were applied as data augmentation techniques. InceptionV3 was used as the model with a dropout rate of 0.6 included. The hyper-parameter values were also noted.The authors aimed for a multi-class classification and mentioned the sensitivity, specificity, precision and f1 score in the paper. Moreover, they discussed about their findings and limitations.
Table 3 provides a summary of the investigations performed by researchers. As mentioned before, only (Ding et al. 2019) utilised voxel-based while the others used sliced based data management. That is why the input data management category is not present in the table.
3.10 Software platforms
To assist researchers, a variety of software tools are available. They are discussed next.
3.10.1 Pre-processing software
FSL
This is a complete set of tools for analysing neuroimaging data from sMRI, fMRI, and DTI. The vast majority of programmes may be executed through command line or graphical user interface.
SPM12
This is meant for the processing of neuroimaging data sequences, which might include a succession of images from multiple sources. The most recent edition is designed exclusively for the processing of fMRI and PET scans.
Nipype
The Nipy community created this open-source Python project to give a common interface to the currently available brain imaging technologies while also allowing communication across numerous packages inside a single flow. One of the most significant benefits is that it allows one’s research to be replicated and allows one to share one’s processing methods with the scientific community.
MATLAB
The image processing toolkit allows you to automate standard image processing procedures. The researchers can segment image data, do batch processing on big datasets, register photos, and create histograms.
NifTi\(\_\) 2014 toolkit
This is an application for analysing and processing neuroimaging pictures that can be imported by the MRIcron software and MATLAB2015b.
3.10.2 Software packages
TensorFlow
TensorFlow is an open source ML platform in its entirety. It comprises a comprehensive, flexible ecosystem of tools, libraries, and community resources that enable academics to push the boundaries of ML while also allowing developers to easily build and deploy ML-powered products.
Keras
Keras is a DL framework built on TensorFlow2. Keras adheres to best practises for lowering cognitive load by offering consistent and simple APIs, minimising the amount of user steps required for typical use cases, and giving clear and actionable error signals.
Theano
Theano is a Python library and compiler that allows you to manipulate and analyse mathematical expressions, particularly matrix-valued expressions. Theano computations are described in a NumPy-like syntax and are designed to perform efficiently on either CPU or GPU architectures.
Caffe
Caffe is a DL framework that prioritises expressiveness, performance, and modularity. Caffe is used in university research, startup prototypes, and large-scale industrial vision, speech, and multimedia applications.
Pytorch
PyTorch is an open source DL framework designed for research but with the reliability and support necessary for production deployment. PyTorch is a Python package that offers high-level features such as tensor computation (akin to NumPy) with substantial GPU acceleration.
4 Research perspective
Alzheimer’s disease remains a global health challenge impacting millions of people worldwide. Early detection improves the outcomes, yet the accuracies can be hampered by manual interpretation. Misdiagnosis of dementia can be attributed to several scientific factors. Firstly, the intricate nature of dementia subtypes presents challenges in accurately delineating subtle deviations in brain structure and function from imaging data, thereby contributing to potential misinterpretations. Secondly, inherent limitations in the sensitivity and specificity of existing imaging modalities, such as MRI or PET scans, may yield false-positive or false-negative results, complicating diagnostic precision. With the introduction of the latest technologies in image processing and AI, there are more avenues for developing automated applications that can aid in AD diagnosis. Nevertheless, the development of such applications also necessitates the appropriate requirements in terms of datasets and algorithms. Besides, the development of AI models is very complex, and necessary adjustments have to be made to several parameters to achieve a good performance. Access to large, annotated datasets is vital for training AI models to make critical decisions, diagnose AD, or predict market trends. From the analysis conducted, it is confirmed that there is a lack of annotated datasets for the medical field. To contribute to the development of AI models for the detection of AD, researchers, and medical practitioners have to work together to build large annotated datasets.
Developing AI models from scratch often requires substantial computational resources, including powerful GPUs or TPUs, as well as significant time for training iterations, making it inaccessible for individuals or organizations with limited resources. The type of imaging used significantly affects the performance and applicability of Alzheimer’s disease diagnostic systems. MRI and PET provide detailed anatomical and molecular information that is highly valuable but come with higher costs and complexity. CT offers a more accessible and quicker option but with lower resolution. Functional imaging like fMRI and SPECT adds important data on brain activity but also presents challenges in terms of cost, complexity, and resolution. AI systems must leverage the strengths of each imaging modality while addressing their limitations through advanced data integration, pre-processing, and analysis techniques. A multimodal approach, combining different types of imaging data, is likely the most promising path forward for developing accurate and reliable Alzheimer’s disease diagnostic tools. For AD diagnosis, the most appropriate pre-processing techniques include image normalisation, registration, segmentation, skull stripping, smoothing and denoising, intensity correction, and data augmentation. Normalisation and intensity correction standardize image intensities across different scans, while registration aligns images to a common coordinate system, crucial for accurate longitudinal and multimodal analysis. Segmentation and skull stripping isolate relevant brain regions, reducing irrelevant data and computational load. Smoothing and denoising improve the signal-to-noise ratio, aiding in detecting subtle changes. Data augmentation increases training dataset size, enhancing model robustness and generalization. These pre-processing steps ensure the reliability and accuracy of machine learning models in diagnosing AD. TL has emerged as a helping hand, whereby, trained models can be adopted instead of developing models from scratch. TL has gained much popularity lately since it allows models to leverage knowledge learned from one task or domain and apply it to another, often requiring less data for fine-tuning. From our opinion and experience, we have seen that TL is more beneficial rather than developing our own AI models. There have been some improvements in the work carried out lately using TL in terms of datasets and optimisation of techniques. Different researchers have adopted different TL techniques and have analysed their perfomance. In addition to this, various data augmentation techniques have been used. In the first instance, traditional data augmentations like rotation, shear and other translations have been adopted. Later on, other advanced techniques like GAN are being deployed. Nevertheless, there is a need to investigate further on other types of GAN like Least Squares GAN (LSGAN), Wasserstein GAN (WGAN) among others. From the analysis conducted, it can observe that data augmentation improves the performance of automated systems. Different authors have developed different TL techniques. The latter differ in how they reuse knowledge from the source task for the target task. Transfer learning techniques offer powerful tools for leveraging pre-existing knowledge in neural network layers, thereby enhancing performance and efficiency in new tasks with limited data. Feature extraction enables efficient reuse of generalized representations from initial layers, reducing computational demands by focusing new training on added task-specific layers. Fine-tuning balances between retaining learned features and adapting to new tasks, providing flexibility based on data similarity. Domain adaptation fine-tunes layers to align feature distributions across different but related domains, while multi-task learning optimizes shared and task-specific layers to benefit from commonalities among tasks. Model-based transfer initialises new models with pre-trained weights, expediting convergence, whereas parameter transfer selectively freezes or fine-tunes layers to maintain beneficial features. Task-specific transfer learning tailors layer modifications to meet unique task requirements, enhancing model adaptability. Collectively, these techniques underscore the strategic manipulation of neural network layers to effectively transfer and optimise knowledge across diverse applications, significantly advancing the field of deep learning. In our opinion, TL has significantly helped in the development of automated application of AD and has greatly improved the performance.
Nevertheless, the adoption of automated AD applications by medical practitioners has been slow due to a combination of technical, clinical, ethical, and logistical challenges. Deep learning models are considered as black boxes. Clinicians are hesitant to trust decisions made by these AI models. In addition, integrating automated detection systems into existing healthcare IT infrastructure is complex and requires significant time and resources. To overcome this problem, it is high time to explore explainable AI (XAI). The refers to a set of processes and methods that allow human users to understand and trust the results and outputs created by machine learning algorithms. XAI aims to make these processes more transparent, interpretable, and comprehensible. Techniques such as attention models and vision transformers have to be explored to have a clear demarcation on the factors contributing to the decision output by AI models. There is a need to use visualisation techniques such as Local Interpretable Model-agnostic Explanations (LIME), SHapley Additive exPlanations (SHAP) and Gradient-weighted Class Activation Mapping (Grad-CAM) to obtain unique insights into the decision-making processes.
5 Conclusion
The availability of assisted AD detection and classification application will definitely be beneficial to doctors and the society at large. However, its development require careful attention, while catering for the high challenges involved in the deployment of such applications. In terms of images, this study reveals that sMRI is the preferred neuroimaging modality of researchers. As for data augmentation, it is not being widely explored though it is known to prevent over-fitting and class imbalance. To pre-process brain images, resizing is the most common techniques. While the other pre-processing techniques are very beneficial in terms of improving the quality of the images, researchers have preferred not to pre-process the data before applying ML or DL techniques. Various architectures of the TL models have been discussed and appraised. The most popular technique is the VGG-16 and has been adopted in several research work. It can be observed that VGG-16 has been the TL that has been widely used in the development of automated AD applications. This is because VGG16 is a simple architecture compared to other complex architectures like Inception or ResNet. In addition, VGG-16 has deeper layers allowing it to capture more complex features and achieve better performance on large-scale image recognition tasks while also reducing the training time and improving performance on new datasets. However, from the analysis conducted, it can be deduced that VGG-16 does not always result into better performance compared to other TL techniques. This paper has provided an analysis on the other TL models which can potentially be used in the development of automated AD detection and classification. All models are evaluated using performance metrics and it was reported that performance accuracy is the most widely used though the others also have their own importance. Nevertheless, current research on Alzheimer’s disease diagnosis using AI is hindered by a lack of large, annotated datasets and the opaque nature of deep learning models, which diminishes trust among clinicians. Future studies should focus on building comprehensive datasets and developing explainable AI techniques to enhance model transparency and interpretability. Additionally, integrating AI systems into clinical practice requires creating user-friendly, interoperable tools that seamlessly blend into existing healthcare workflows.
Data availability
No datasets were generated or analysed during the current study.
References
Acharya H, Mehta R, Singh DK (2021) Alzheimer disease classification using transfer learning. In: 2021 5th international conference on computing methodologies and communication (ICCMC). IEEE, Erode https://doi.org/10.1109/ICCMC51019.2021.9418294
Aderghal K, Afdel K, Benois-Pineau J et al (2020) Improving alzheimer’s stage categorization with convolutional neural network using transfer learning and different magnetic resonance imaging modalities. Heliyon 6:e05652. https://doi.org/10.1016/j.heliyon.2020.e05652
Afzal S, Maqsood M, Nazir F et al (2019) A data augmentation-based framework to handle class imbalance problem for alzheimer’s stage detection. IEEE Access 7:115528–115539. https://doi.org/10.1109/ACCESS.2019.2932786
Agarwal D, Marques G, de la Torre-Díez I et al (2021) Transfer learning for alzheimer’s disease through neuroimaging biomarkers: a systematic review. Sensors 21:7259. https://doi.org/10.3390/s21217259
Agosta F, Pievani M, Geroldi C et al (2012) Resting state fmri in alzheimer’s disease: beyond the default mode network. Neurobiol Aging 33:1564–1578. https://doi.org/10.1016/j.neurobiolaging.2011.06.007
Ahmed R, Zhang Y, Feng Z et al (2019) Neuroimaging and machine learning for dementia diagnosis: recent advancements and future prospects. IEEE Rev Biomed Eng 12:19–33. https://doi.org/10.1109/RBME.2018.2886237
Ajagbe SA, Amuda KA, Oladipupo MA et al (2021) Multi-classification of Azheimer disease on magnetic resonance images using deep convolutional neural network approaches. Int J Adv Comput Res. https://doi.org/10.19101/IJACR.2021.1152001
Ajit A, Acharya K, Samanta A (2020) A review of convolutional neural networks. In: 2020 International conference on emerging trends in information technology and engineering (ic-ETITE). IEEE, Vellore https://doi.org/10.1109/ic-ETITE47903.2020.049
Akhtar N, Ragavendran U (2019) Interpretation of intelligence in cnn-pooling processes: a methodological survey. Neural Comput Appl. https://doi.org/10.1007/s00521-019-04296-5
Akkus Z, Galimzianova A, Hoogi A et al (2017) Deep learning for brain mri segmentation: state of the art and future directions. J Digital Imaging 30:449–459. https://doi.org/10.1007/s10278-017-9983-4
Almadhoun HR, Abu-Naser SS (2021) Classification of Alzheimer’s disease using traditional classifiers with pre-trained cnn. Int J Acad Health Med Res (IJAHMR) 5:17–21
Alves GS, Knöchel VO, Knöchel C et al (2015) Integrating retrogenesis theory to Azheimer’s disease pathology: insight from dti-tbss investigation of the white matter microstructural integrity. BioMed Res Int 2015:1–11. https://doi.org/10.1155/2015/291658
Arab A, Wojna-Pelczar A, Khairnar A et al (2018) Principles of diffusion kurtosis imaging and its role in early diagnosis of neurodegenerative disorders. Brain Res Bull 139:91–98. https://doi.org/10.1016/j.brainresbull.2018.01.015
Arafa DA, Moustafa HED, Ali-Eldin AMT et al (2022) Early detection of Alzheimer’s disease based on the state-of-the-art deep learning approach: a comprehensive survey. Multimed Tools Appl. https://doi.org/10.1007/s11042-022-11925-0
Ashraf A, Naz S, Shirazi SH et al (2021) Deep transfer learning for alzheimer neurological disorder detection. Multimed Tools Appl 80:30117–30142. https://doi.org/10.1007/s11042-020-10331-8
Association A (2018) (2018) 2018 alzheimer’s disease facts and figures:includes a special report on the financial and personal benefits of early diagnosis. Alzheimers Dement 14:367–429
Bai T, Du M, Zhang L et al (2022) A novel Alzheimer’s disease detection approach using gan-based brain slice image enhancement. Neurocomputing 492:353–369. https://doi.org/10.1016/j.neucom.2022.04.012
Belleville S, Fouquet C, Duchesne S et al (2014) Detecting early preclinical alzheimer’s disease via cognition, neuropsychiatry, and neuroimaging: Qualitative review and recommendations for testing. J Alzheimer’s Dis 42:S375-s382. https://doi.org/10.3233/JAD-141470
Blennow K, Zetterberg H (2018) Biomarkers for Alzheimer’s disease: current status and prospects for the future. J Intern Med 284:643–663. https://doi.org/10.1111/joim.12816
Buerger K, Ewers M, Pirttilä T et al (2006) Csf phosphorylated tau protein correlates with neocortical neurofibrillary pathology in Alzheimer’s disease. Brain 129:3035–3041. https://doi.org/10.1093/brain/awl269
Chen CM, Chen CC, Wu MC et al (2015) Automatic contrast enhancement of brain mr images using hierarchical correlation histogram analysis. J Med Biol Eng 35:724–734. https://doi.org/10.1007/s40846-015-0096-6
Chen JE, Glover GH (2016) Functional magnetic resonance imaging methods. Neuropsychol Rev 25:289–313. https://doi.org/10.1007/s11065-015-9294-9
Chlap P, Min H, Vandenberg N et al (2021) A review of medical image data augmentation techniques for deep learning applications. J Med Imaging Radiat Oncol 65:545–563. https://doi.org/10.1111/1754-9485.13261
Chollet F (2016) Xception: deep learning with depthwise separable convolutions. CoRR abs/1610.02357. arXiv:1610.02357
Morris CJ, (1993) The clinical dementia rating (cdr): current version and scoring rules. Neurology 43:2412–2412. https://doi.org/10.1212/WNL.43.11.2412-a
Coimbra A, Williams DS, Hostetler ED (2006) The role of mri and pet/spect in Alzheimers disease. Curr Top Med Chem 6:629–647. https://doi.org/10.2174/156802606776743075
Craig-Schapiro R, Fagan AM, Holtzman DM (2009) Biomarkers of alzheimer’s disease. Neurobiol Dis 35:128–140. https://doi.org/10.1016/j.nbd.2008.10.003
Dawood FA, Abood ZM (2018) The importance of contrast enhancement in medical images analysis and diagnosis. Int J Eng Res Technol 7:12
Ding Y, Sohn JH, Kawczynski MG et al (2019) A deep learning model to predict a diagnosis of Alzheimer disease by using 18f-fdg pet of the brain. Radiology 290:456–464. https://doi.org/10.1148/radiol.2018180958
Dixit P (2023) Transfer learning in image recognition: leveraging pre-trained models for improved performance. Edu J Int Aff Res ISSN: 2583-9993 2(4):31–37
Ellis KA, Bush AI, Darby D et al (2009) The Australian imaging, biomarkers and lifestyle (aibl) study of aging: methodology and baseline characteristics of 1112 individuals recruited for a longitudinal study of Alzheimer’s disease. Int Psychogeriatr 21:672–687. https://doi.org/10.1017/S1041610209009405
Falahati F, Westman E, Simons A (2014) Multivariate data analysis and machine learning in Alzheimer’s disease with a focus on structural magnetic resonance imaging. J Alzheimer’s Dis 41:685–708. https://doi.org/10.3233/JAD-131928
Folstein MF, Folstein SE, McHugh PR (1975) Mini-mental state. J Psychiatr Res 12:189–198. https://doi.org/10.1016/0022-3956(75)90026-6
Forouzannezhad P, Abbaspour A, Fang C et al (2019) A survey on applications and analysis methods of functional magnetic resonance imaging for Alzheimer’s disease. J Neurosci Methods 317:121–140. https://doi.org/10.1016/j.jneumeth.2018.12.012
Fox MD, Greicius M (2010) Clinical applications of resting state functional connectivity. Front Syst Neurosci. https://doi.org/10.3389/fnsys.2010.00019
Francis A, Pandian IA (2021) Early detection of Alzheimer’s disease using ensemble of pre-trained models. In: 2021 International conference on artificial intelligence and smart systems (ICAIS). IEEE, Coimbatore https://doi.org/10.1109/ICAIS50930.2021.9395988
Frisoni GB, Fox NC, CRJ, et al (2010) The clinical use of structural MRI in Alzheimer disease. Nat Rev Neurol 6:67–77. https://doi.org/10.1038/nrneurol.2009.215
Fu’adah YN, Wijayanto I, Pratiwi NKC et al (2021) Automated classification of Alzheimer’s disease based on MRI image processing using convolutional neural network (cnn) with alexnet architecture. Journal of physics: conference series. IOP Publishing, Bristol
Gauthier S, Rosa-Neto P, Morais JA et al (2021) World Alzheimer report 2021: journey through the diagnosis of dementia. Report. Alzheimer’s Disease International, London
Gradd-Radford J, Kantarci K (2013) Magnetic resonance spectroscopy in Alzheimer’s disease. Neuropsychiatr Dis Treat 9:687–696. https://doi.org/10.2147/NDT.S35440
Gupta M, Kumar R, Abraham A (2024) Adversarial network-based classification for Alzheimer’s disease using multimodal brain images: a critical analysis. IEEE Access. https://doi.org/10.1109/ACCESS.2024.3381956
Harbi Z, Hicks Y, Setchi R (2017) Clock drawing test interpretation system. Proced Comput Sci 112:1641–1650. https://doi.org/10.1016/j.procs.2017.08.259
Hazarika RA, Kandar D, Maji AK (2021) An experimental analysis of different deep learning based models for Alzheimer’s disease classification using brain magnetic resonance images. J King Saud Univ Comput Inf Sci. https://doi.org/10.1016/j.jksuci.2021.09.003
H. Feldman H, Jacova C, Robillard A, et al (2008) Diagnosis and treatment of dementia: 2 diagnosis. Can Med Assoc J 178:825–836. https://doi.org/10.1503/cmaj.070798
Huang W (2023) Multimodal contrastive learning and tabular attention for automated alzheimer’s disease prediction. In: Proceedings of the IEEE/CVF international conference on computer vision, pp 2473–2482
Hussain E, Hasan M, Hassan SZ, et al (2020) Deep learning based binary classification for Alzheimer’s disease detection using brain MRI images. In: 2020 15th IEEE conference on industrial electronics and applications (ICIEA). IEEE, Kristiansand, Norway, pp 1115—1120, https://doi.org/10.1109/ICIEA48937.2020.9248213
Illakiya T, Karthik R (2024) A deep feature fusion network with global context and cross-dimensional dependencies for classification of mild cognitive impairment from brain MRI. Image Vis Comput 144:104967. https://doi.org/10.1016/j.imavis.2024.104967
Illakiya T, Ramamurthy K, Siddharth M et al (2023) Ahanet: adaptive hybrid attention network for Alzheimer’s disease classification using brain magnetic resonance imaging. Bioengineering 10(6):714
Islam J, Zhang Y (2017) An ensemble of deep convolutional neural networks for alzheimer’s disease detection and classification. CoRR abs/1712.01675.arXiv:1712.01675
Jo T, Nho K, Saykin AJ (2019) Deep learning in Alzheimer’s disease: diagnostic classification and prognostic prediction using neuroimaging data. Front Aging Neurosci 11:220. https://doi.org/10.3389/fnagi.2019.00220
Jobst KA, Barnestson LP, Shepstone BJ (1998) Accurate prediction of histologically confirmed Alzheimer’s disease and the differential diagnosis of dementia: the use of nincds-adrda and dsm-111-r criteria, spect, x-ray ct, and apo e4 in medial temporal lobe dementias. Int Psychogeriatr Assoc 10:271–302
Juntu J, Sijbers J, Dyck DV et al (eds) (2005) Bias field correction for MRI images. Springer, Berlin
Kesavadas C (2013) Resting state functional magnetic resonance imaging: an emerging clinical tool. Neurol India 61:103. https://doi.org/10.4103/0028-3886.111107
Khagi B, Kwon GR (2019) CNN model performance analysis on MRI images of an oasis dataset for distinction between healthy and Alzheimer’s patients. IEIE Trans Smart Process Comput. https://doi.org/10.5573/IEIESPC.2019.8.4.272
Khan A, Sohail A, Zahoora U et al (2020) A survey of the recent architectures of deep convolutional neural networks. Artif Intell Rev 53:5455–5516. https://doi.org/10.1007/s10462-020-09825-6
Khan R, Akbar S, Mehmood A et al (2023) A transfer learning approach for multiclass classification of Alzheimer’s disease using MRI images. Front Neurosci 16:1050777
Khan TK (ed) (2016) Biomarkers in Alzheimer’s disease. Academic Press, London
Kim HE, Cosa-Linan A, Santhanam N et al (2022) Transfer learning for medical image classification: a literature review. BMC Med Imaging 22(1):69
Klunk W, Panchalingam K, Moossy J et al (1992) N-acetyl-l-aspartate and other amino acid metabolites in Alzheimer’s disease brain: a preliminary proton nuclear magnetic resonance study. Neurology 42:1578–1585. https://doi.org/10.1212/wnl.42.8.1578
Klunk WE, Engler H, Nordberg A et al (2004) Imaging brain amyloid in Alzheimer’s disease with Pittsburgh compound-b: Imaging amyloid in ad with pib. Ann Neurol 55:306–319. https://doi.org/10.1002/ana.20009
Knopman D, DeKosky S, Cummings J et al (2001) Practice parameter: diagnosis of dementia (an evidence-based review). Neurology 56:2412–2412. https://doi.org/10.1212/WNL.56.9.1143
Kogan F, Hariharan H, Reddy R (2013) Chemical exchange saturation transfer (cest) imaging: description of technique and potential clinical applications. Curr Radiol Rep. https://doi.org/10.1007/s40134-013-0010-3
Kora P, Ooi CP, Faust O et al (2022) Transfer learning techniques for medical image analysis: a review. Biocybern Biomed Eng 42(1):79–107
Kumar L, Hariharasitaraman S, Narayanasamy K et al (2022) Alexnet approach for early stage Alzheimer’s disease detection from MRI brain images. Mater Today 51:58–65. https://doi.org/10.1016/j.matpr.2021.04.415
Kumar LS, Hariharasitaraman S, Narayanasamy K et al (2021) Alexnet approach for early stage Alzheimer’s disease detection from MRI brain images. Mater Today Proceed. https://doi.org/10.1016/j.matpr.2021.04.415
Kurth J, Sakretz M, Teipel S et al (2013) Molecular imaging of dementia. Geriatr Ment Health care 1:56–62. https://doi.org/10.1016/j.gmhc.2013.04.006
Lazli L, Boukadoum M, Mohamed OA (2020) A survey on computer-aided diagnosis of brain disorders through MRI based on machine learning and data mining methodologies with an emphasis on Alzheimer disease diagnosis and the contribution of the multimodal fusion. Appl Sci 10:1894. https://doi.org/10.3390/app10051894
Leandrou S, Petroudi S, Kyriacou PA et al (2018) Quantitative MRI brain studies in mild cognitive impairment and Alzheimer’s disease: a methodological review. IEEE Rev Biomed Eng 11:97–111. https://doi.org/10.1109/RBME.2018.2796598
Li HJ, Hou XH, Liu HH et al (2015) Toward systems neuroscience in mild cognitive impairment and alzheimer’s disease: A meta-analysis of 75 fmri studies: Neural networks in mci and ad. Human Brain Mapp 36:1217–1232. https://doi.org/10.1002/hbm.22689
Li Z, Yang W, Peng S, et al (2020) A survey of convolutional neural networks: analysis, applications, and prospects. https://doi.org/10.48550/ARXIV.2004.02806, https://arxiv.org/abs/2004.02806
Lu X, Wu H, Zeng Y (2019) Classification of Alzheimer’s disease in MobileNet. J Phys Conf Ser 1345(4):042012. https://doi.org/10.1088/1742-6596/1345/4/042012
Malone IB, Cash D, Ridgway GR et al (2013) Miriad-public release of a multiple time point Alzheimer’s MR imaging dataset. NeuroImage 70:33–36. https://doi.org/10.1016/j.neuroimage.2012.12.044
Marcus DS, Fotenos AF, Csernansky JG et al (2010) Open access series of imaging studies: longitudinal MRI data in nondemented and demented older adults. J Cogn Neurosci 22:2677–2684. https://doi.org/10.1162/jocn.2009.21407
Mehmood A, Yang S, Feng Z et al (2021) A transfer learning approach for early diagnosis of Alzheimer’s disease on MRI images. Neuroscience 460:43–52. https://doi.org/10.1016/j.neuroscience.2021.01.002
Murphy A (2018) Anterior commissure - posterior commissure line. https://radiopaedia.org/articles/anterior-commissure-posterior-commissure-line-1
Murray ME, Przybelski SA, Lesnick TG et al (2014) Early Alzheimer’s disease neuropathology detected by proton MR spectroscopy. J Neurosci 34:16247–16255. https://doi.org/10.1523/JNEUROSCI.2027-14.2014
Nanni L, Interlenghi M, Brahnam S et al (2020) Comparison of transfer learning and conventional machine learning applied to structural brain MRI for the early diagnosis and prognosis of Alzheimer’s disease. Front neurol 11:576194
Naz S, Ashraf A, Zaib A (2022) Transfer learning using freeze features for Alzheimer neurological disorder detection using adni dataset. Multimed Syst 28:85–94. https://doi.org/10.1007/s00530-021-00797-3
Nordberg A, Rinne JO, Kadir A et al (2010) The use of pet in Alzheimer disease. Nat Rev Neurol 6:78–87. https://doi.org/10.1038/nrneurol.2009.217
Oh K, Chung YC, Kim KW et al (2019) Classification and visualization of Alzheimer’s disease using volumetric convolutional neural network and transfer learning. Sci Rep 9:18150. https://doi.org/10.1038/s41598-019-54548-6
Olsson B, Lautner R, Andreasson U et al (2016) Csf and blood biomarkers for the diagnosis of Alzheimer’s disease: a systematic review and meta-analysis. Lancet Neurol 15:673–684. https://doi.org/10.1016/S1474-4422(16)00070-3
Ortiz-Terán L, Santos JM, de las Nieves Cabrera Martín M, et al (2011) Currently available neuroimaging approaches in Alzheimer disease (ad) early diagnosis. The clinical spectrum of Alzheimer’s disease -the charge toward comprehensive diagnostic and therapeutic strategies. InTech, London
Pradhan A, Gige J, Eliazer M (2021) Detection of Alzheimer’s disease (ad) in MRI images using deep learning. Int J Eng Res Technol (IJERT) 10:580
Pradhan N, Sagar S, Singh AS (2024) Machine learning and deep learning algorithms for Alzheimer disease detection and its implication in society 5.0. In: John V (ed) Digital transformation: industry 4.0 to Society 5.0. Springer, Berlin, pp 285–305
Prasath T, Sumathi V (2024) Pipelined deep learning architecture for the detection of Alzheimer’s disease. Biomed Signal Process Control 87:105442
Radhika Shetty D, Antony P (2023) Transfer learning techniques in medical image classification. International conference on information and communication technology for competitive strategies. Springer, Berlin, pp 239–248
Rajeshwari S, Sharmila TS (2013) Efficient quality analysis of mri image using preprocessing techniques. In: Proceedings of 2013 IEEE conference on information and communication technologies (ICT 2013), Tamil Nadu pp 391–396
Ramzan F, Khan MUG, Rehmat A et al (2020) A deep learning approach for automated diagnosis and multi-class classification of Alzheimer’s disease stages using resting-state fmri and residual neural networks. J Med Syst 44:37. https://doi.org/10.1007/s10916-019-1475-2
Rathore S, Habes M, Iftikhar MA et al (2017) A review on neuroimaging-based classification studies and associated feature extraction methods for Alzheimer’s disease and its prodromal stages. NeuroImage 155:530–548. https://doi.org/10.1016/j.neuroimage.2017.03.057
Raza N, Naseer A, Tamoor M et al (2023) Alzheimer disease classification through transfer learning approach. Diagnostics 13(4):801
Rodney DV (ed) (1994) Clinician’s guide to neuropsychological assessment, 1st edn. Psychology Press, London
Rowe CC, Villemagne VL (2013) Amyloid imaging with pet in early Alzheimer disease diagnosis. Med Clin N Am 97:377–398. https://doi.org/10.1016/j.mcna.2012.12.017
Sajjad M, Ramzan F, Khan MUG et al (2022) Deep convolutional generative adversarial network for Alzheimer’s disease classification using positron emission tomography (pet) and synthetic data augmentation. Microscopy Research and Technique 84:3023–3034. https://doi.org/10.1002/jemt.23861
Salehi AW, Khan S, Gupta G et al (2023) A study of CNN and transfer learning in medical imaging: advantages, challenges, future scope. Sustainability 15(7):5930
Sarraf S, Tofighi G (2016) Deep learning-based pipeline to recognize Alzheimer’s disease using FMRI data. Future technologies conference(FTC) 2016. IEEE, San Francisco, pp 816–820
Sexton CE, Kalu UG, Filippini N et al (2011) A meta-analysis of diffusion tensor imaging in mild cognitive impairment and Alzheimer’s disease. Neurobiol Aging 32:2322.e5-2322.e18. https://doi.org/10.1016/j.neurobiolaging.2010.05.019
Shanmugam JV, Duraisamy B, Simon BC et al (2022) Alzheimer’s disease classification using pre-trained deep networks. Biomed Signal Process Control 71:1.0321710321710322e+55
Sharma N, Singh AN (2016) Exploring biomarkers for Alzheimer’s disease. J Clin Diagn Res. https://doi.org/10.7860/JCDR/2016/18828.8166
Sharma R, Goel T, Tanveer M et al (2022) Fdn-adnet: fuzzy ls-twsvm based deep learning network for prognosis of the Alzheimer’s disease using the sagittal plane of MRI scans. Appl Soft Comput 115:108099. https://doi.org/10.1016/j.asoc.2021.108099
Sharma S, Sharma S, Athaiya A (2020) Activation functions in neural networks. Int J Eng Appl Sci Technol 4:310–316
Shi J, Zheng X, Li Y et al (2017) Multimodal neuroimaging feature learning with multimodal stacked deep polynomial networks for diagnosis of Alzheimer’s disease. IEEE J Biomed Health inf 22(1):173–183
Shorten C, Khoshgoftaar TM (2019) A survey on image data augmentation for deep learning. J Big Data 6:1–48. https://doi.org/10.1186/s40537-019-0197-0
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. https://doi.org/10.48550/ARXIV.1409.1556, https://arxiv.org/abs/1409.1556
Sperling R (2011) The potential of functional MRI as a biomarker in early Alzheimer’s disease. Neurobiol Aging 32:S37–S43. https://doi.org/10.1016/j.neurobiolaging.2011.09.009
Stomrud E, Hansson O, Blennow K et al (2007) Cerebrospinal fluid biomarkers predict decline in subjective cognitive function over 3 years in healthy elderly. Dement Geriatr Cognit Disord 24:118–124. https://doi.org/10.1159/000105017
Suk HI, Shen D (2013) Deep learning-based feature representation for ad/mci classification. In: Medical image computing and computer-assisted intervention–MICCAI 2013: 16th international conference, Nagoya, Japan, September 22-26, 2013, Proceedings, Part II 16, Springer, pp 583–590
Sun X, Shi L, Luo Y et al (2015) Histogram-based normalization technique on human brain magnetic resonance images from different acquisitions. BioMed Eng OnLine. https://doi.org/10.1186/s12938-015-0064-y
Szegedy C, Liu W, Jia Y, et al (2014) Going deeper with convolutions. https://doi.org/10.48550/ARXIV.1409.4842, https://arxiv.org/abs/1409.4842
Talwar P, Kushwaha S, Chaturvedi M et al (2021) Systematic review of different neuroimaging correlates in mild cognitive impairment and Alzheimer’s disease. Clin Neuroradiol 31:953–967. https://doi.org/10.1007/s00062-021-01057-7
Tan M, Le QV (2020) Efficientnet: Rethinking model scaling for convolutional neural networks. arXiv:1905.11946
Tax CM, Bastiani M, Veraart J et al (2022) What’s new and what’s next in diffusion MRI preprocessing. NeuroImage 249:118830. https://doi.org/10.1016/j.neuroimage.2021.118830
Tuovinen T, Rytty R, Moilanen V et al (2017) The effect of gray matter ICA and coefficient of variation mapping of bold data on the detection of functional connectivity changes in Alzheimer’s disease and bvftd. Front Human Neurosci. https://doi.org/10.3389/fnhum.2016.00680
van Zilj PCM, Yadav NN (2011) Chemical exchange saturation transfer (cest): What is in a name and what isn’t?: Cest: what is in a name and what isn’t? Magn Reson Med 65:927–948. https://doi.org/10.1002/mrm.22761
Wang S, Celebi ME, Zhang YD et al (2021) Advances in data preprocessing for biomedical data fusion: An overview of the methods, challenges, and prospects. Inf Fusion 76:376–421. https://doi.org/10.1016/j.inffus.2021.07.001
Weiner MW, Veitch DP, Aisen PS et al (2012) The Alzheimer’s disease neuroimaging initiative: a review of papers published since its inception. Alzheimer’s Dement 8:S1–S68. https://doi.org/10.1016/j.jalz.2011.09.172
Yamanakkanavar N, Choi JY, Lee B (2020) MRI segmentation and classification of human brain using deep learning for diagnosis of Alzheimer’s disease: A survey. Sensors 20:3243. https://doi.org/10.3390/s20113243
Yang K, Mohammed EA (2020) A review of artificial intelligence technologies for early prediction of Alzheimer’s disease. https://arxiv.org/abs/2101.01781v1
Yi F, Zhang Y, Yuan J et al (2023) Identifying underlying patterns in Alzheimer’s disease trajectory: a deep learning approach and mendelian randomization analysis. Eclinicalmedicine 1:64
Yildirim M, Cinar A (2020) Classification of Alzheimer’s disease MRI images with CNN based hybrid method. Ingénierie des Systèmes d’Information 25:413–418. https://doi.org/10.18280/isi.250402
Yu B, Shan Y, Ding J (2021) A literature review of MRI techniques used to detect amyloid-beta plaques in Alzheimer’s disease patients. Ann Palliat Med 10:10062–10074. https://doi.org/10.21037/apm-21-825
Yu L, Sun M, Chen Y et al (2016) Non-gaussian diffusion alterations on diffusion kurtosis imaging in patients with early Alzheimer’s disease. Neurosci Lett 616:11–18. https://doi.org/10.1016/j.neulet.2016.01.021
Zhang S, Han D, Tan X et al (2012) Diagnostic accuracy of 18f-fdg and 11c-pib-pet for prediction of short-term conversion to Alzheimer’s disease in subjects with mild cognitive impairment: pet for prediction. Int J Clin Pract 66:185–198. https://doi.org/10.1111/j.1742-1241.2011.02845.x
Zheng C, Xia Y, Pan Y et al (2016) Automated identification of dementia using medical imaging: a survey from a pattern classification perspective. Brain Inf 3:17–27. https://doi.org/10.1007/s40708-015-0027-x
Zoph B, Vasudevan V, Shlens J, et al (2017) Learning transferable architectures for scalable image recognition. CoRR abs/1707.07012. arXiv:1707.07012
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Heenaye-Mamode Khan, M., Reesaul, P., Auzine, M.M. et al. Detection of Alzheimer’s disease using pre-trained deep learning models through transfer learning: a review. Artif Intell Rev 57, 275 (2024). https://doi.org/10.1007/s10462-024-10914-z
Accepted:
Published:
DOI: https://doi.org/10.1007/s10462-024-10914-z