Abstract
Cancer remains a significant cause of mortality, and the application of microarray technology has opened new avenues for cancer diagnosis and treatment. However, due to the challenges in sample acquisition, the genetic dimension of microarray data surpasses the sample dimension, resulting in high-dimensional small sample data. Effective feature selection is crucial for identifying biomarkers and facilitating further analysis. However, existing methods struggle to fully exploit the interdependencies among genes, such as regulatory networks and pathways, to guide the feature selection process and construct efficient classification models. In this paper, we propose a novel feature selection algorithm and classification model based on graph neural networks to address these challenges. Our proposed method employs a multidimensional graph to capture intricate gene interactions. We leverage link prediction techniques to enhance the graph structure relationships and employ a multidimensional node evaluator alongside a supernode discovery algorithm based on spectral clustering for initial node filtering. Subsequently, a hierarchical graph pooling technique based on downsampling is used to further refine node selection for feature extraction and model building. We evaluate the proposed method on nine publicly available microarray datasets, and the results demonstrate its superiority over both classical and advanced feature selection techniques in various evaluation metrics. This highlights the effectiveness and advancement of our proposed approach in addressing the complexities associated with microarray data analysis and cancer classification.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Cancer poses an escalating risk and mortality worldwide, posing a significant threat to human life and health. It has emerged as a leading cause of death in both developed and developing countries (Serrano et al. 2019). With the absence of a definitive treatment for cancer, the focus has shifted towards cancer prevention as a meaningful approach to mitigate the risk of this devastating disease (Liu et al. 2017).
The Cancer Genome Atlas (TCGA) database contains genetic microarray data from cancer patients necessary for cancer identification and analysis. Microarray data can detect tens of thousands of genes from patients at the same time, and the precise analysis of these genes can determine and discover biomarkers with diagnostic significance for characteristic diseases, which is of interest for early diagnosis of cancer, prognosis, and various other applications in such fields (Shukla 2020). However, the challenge of this study comes from two aspects. The first is the curse of the dimensionality of the data, which is used to integrate tens of thousands of expression levels simultaneously in a single experiment, so the feature dimension is much higher than the sample dimension (Agarwalla and Mukhopadhyay 2022). Another challenge comes from the sample size. The number of patients participating in the test cohort is usually much smaller than the feature dimension, which can easily lead to problems such as data overfitting and sample imbalance (Chen et al. 2013).
Feature selection is essential for analyzing large-scale high-dimensional data, such as microarray data and mass spectrometry analysis data, and building statistically significant models (Cheng et al. 2012). In the feature selection process, significant features can be analyzed from microarray data as biomarkers, and effective classification models that can be used to predict diseases can be built (Wang et al. 2021). Based on the evaluation metrics and how feature selection methods are combined with classification models, standard feature selection methods include Filter method, Wrapper method, and Embedded method (Ghosh et al. 2020).
Filter methods assess feature importance based on data properties such as feature redundancy and feature-label correlation (Pashaei and Pashaei 2022). While they efficiently eliminate redundant features, their precision can be further enhanced for fine-scale filtering. Notable filter methods include T-Test (Hua et al. 2009), Chi-squared test (Huang 2009), MIC (Li et al. 2020), and Fisher score (Zhang et al. 2021). In contrast, Wrapper methods integrate meta-heuristic optimization algorithms with classification models to seek optimal feature subsets, with techniques like GA (Oh et al. 2004), PSO (Xue et al. 2012), and ACO (Ghosh et al. 2020). However, they may suffer from high time complexity and risk of local optimality. Embedded methods, bridging machine learning models with feature selection, rank features based on their weights during model construction, achieving a balance between efficiency and accuracy (Rodriguez-Galiano et al. 2018). Common embedded techniques are DT (Zhou et al. 2021), RF (Zhou et al. 2016), Lasso (Muthukrishnan and Rohini 2016), and Ridge Regression (Xu et al. 2020).
Hybrid feature selection methods have been extensively studied and proven to be effective on microarray data to address the above issues (Momenzadeh et al. 2019; Lin et al. 2019; Wang et al. 2022; Wan et al. 2016; Ouadfel and Abd Elaziz 2022; Zhang et al. 2020; Annavarapu and Dara 2021; Peng et al. 2013). These reports will be described in detail in the related work section. The hybrid approach combines the computational efficiency advantage of the Filter method and the accuracy advantage of the Wrapper method, which is essential for biomarker selection and cancer diagnosis.
Many feature selection methods, especially wrapper-based ones applied to microarray data, assume data is independently and identically distributed (i.i.d). These methods often miss intrinsic feature dependencies, even when real-world gene dependencies, such as genetic pathways, exist. Recognizing these can augment biomarker identification and enhance the biological relevance of selected features. Such interactions are cataloged in databases like GeneMANIA and String, which record gene and protein interactions respectively (Warde-Farley et al. 2010).
Research indicates that embracing feature dependencies can bolster feature selection accuracy. Jl et al. (2020), Saranya and Pravin (2022), Bhuyan et al. (2021), and others have adopted various methodologies. Several regularization methods have also highlighted the advantages of utilizing feature dependencies (Yuan and Lin 2006; Zou and Hastie 2005; Efron et al. 2004; Fan and Li 2002; Zou and Zhang 2009; Liu et al. 2019; Zeng and Xie 2014). However, a gap persists in merging deep learning-focused dependencies and the unified application of ensemble techniques and graph neural networks.
Addressing this, our study introduces a graph neural network approach to microarray data feature selection. Although graph neural networks can capture intricate data dependencies, their direct application to microarray data feature selection remains pioneering. Earlier studies honed in on gene features using graph structures but often sidestepped higher-order graph data connectivity, emphasized by He et al. (2020). Our method leverages graph structures and neural networks, drawing from our previous work (Yu et al. 2021; Xie et al. 2022a, b).
Using gene data relationships from GeneMANIA, our approach employs multidimensional graph neural network techniques. After refining the relationship graph, we minimized isolated nodes, thereby improving spectral clustering efficiency. The feature selection mechanism used a node importance method, combined with spectral clustering to identify supernodes. This leads to the establishment of an effective graph-based classification model.
We conducted experiments on nine publicly available microarray datasets. The results show that the prediction models built by the features selected by the proposed method have high accuracy and outperform the classical feature selection methods and most of the compared state-of-the-art hybrid methods. In addition, our method also outperforms the graph neural network approach for feature selection on microarray data. The main innovations of the proposed approach are as follows.
-
1.
Innovative use of graph neural networks for feature selection on microarray data and constructing graph structures based on multiple prior knowledge bases, and using multidimensional graph neural network technology techniques to achieve node embedding representation and information aggregation.
-
2.
A multidimensional node importance assessment method is designed and combined with a supernode assessment method based on spectral clustering for coarse-scale filtering of nodes.
-
3.
A hierarchical pooling technique based on downsampling to learn an end-to-end hierarchical representation of data with fewer parameters to effectively learn the importance of nodes for fine-scale filtering and a classification model based on graph structure is designed.
-
4.
A complete comparison with six classical methods on nine microarray datasets and a comparison with 18 advanced algorithms on three datasets prove the effectiveness and advancement of the proposed method. In addition, we compare GNN-based feature selection methods to demonstrate the advancedness of the proposed method.
The rest of the paper is organized as follows: Sect. 2 presents the related work, Sect. 3 describes the proposed method and each module in detail, Sect. 4 lists the experimental results and comparison results, and Sect. 5 summarizes the work of this paper and lists future directions.
2 Related work
In this section, we review the current advancements in various categories of feature selection methods, emphasizing their use in microarray data analysis. Based on insights from review articles (Remeseiro and Bolon-Canedo 2019; Alhenawi et al. 2022; Bolón-Canedo et al. 2014), these methods are categorized into Filter, Wrapper, Embedded, Hybrid, and Ensemble approaches. Owing to the exceptional efficacy of regularization methods with grouping effects for grouped feature evaluation, we treat it as a separate category. For conciseness, we group the Embedded and Hybrid methods under the Wrapper category due to their similarities (Xue et al. 2015), and this categorization is further elaborated in this section.
2.1 Filter method
Mazumder and Veilumuthu (2019) proposed an advanced filter for microarray cancer data classification. The method, aiming for improved accuracy in differentiating cancer types, showcased promise in handling the complexities of microarray datasets. Thabtah et al. (2020) introduced "Least Loss", a refined filter technique for feature selection. Central to a unique strategy, this method streamlines feature selection, evidencing its efficiency across diverse data analysis scenarios. Yuan et al. (2017) developed a feature selection method emphasizing correlation information maximization, especially for high-dimensional protein data. By harnessing correlation insights, the method enhanced the accuracy of protein data analysis, marking a pivotal contribution to bioinformatics. Lefakis and Fleuret (2016) unveiled a Gaussian modeling-based feature selection approach, targeting jointly informative features. Their methodology simplifies the otherwise complex task, underscoring the collective significance of features in machine learning. Li and Oh (2016) advocated for feature selection enhancement using pairwise pre-evaluation. Emphasizing feature collaboration, their method points towards optimized feature selection outcomes in bioinformatics.
2.2 Wrapper method
Wang et al. (2022) introduced an adaptive weighted differential evolutionary algorithm to select features from high-dimensional small sample data. This method, building on the differential evolution algorithm, uses an adaptive mechanism, leverages historical data, and applies a weighting model. Its effectiveness has been confirmed on 12 datasets, surpassing six comparable evolutionary algorithms.
Wan et al. (2016) combined a binary coded ant colony optimization algorithm with a genetic algorithm to refine feature selection. They introduced the visibility density model (VMBACO) and the pheromone density model (PMBACO), utilizing the genetic algorithm for initial data gathering. This method prioritizes features with low redundancy and high discriminative power and outperforms several other algorithms in effectiveness.
Ouadfel and Abd Elaziz (2022) developed a two-stage feature selection approach for high-dimensional small sample data. Initially, the ReliefF method preprocesses features, which is then followed by a balanced optimization algorithm. Pearson coefficients help in weeding out redundant features. The method’s success is evident on 26 varied datasets.
Zhang et al. (2020) unveiled a binary difference evolutionary algorithm with a self-learning strategy for multiple objective feature selection. It introduces a binary mutation operator and a one-bit purifying search operator. Furthermore, a non-dominated sorting operator cuts down the selection operator’s time consumption, with the method’s efficacy tested on 20 UCI datasets.
Annavarapu and Dara (2021) proposed a hybrid method for microarray data feature selection. Their approach combined a k-mean clustering algorithm with a signal-to-noise ratio ranking. The results on five datasets highlight its potential.
Peng et al. (2013) showcased a modified sequential forward floating selection algorithm for feature selection, emphasizing gene classification. This method pairs weighted Mahalanobis distance with a modified SFFS technique. Results from the Colon dataset validate its capabilities.
Aziz (2022) presented a hybrid machine learning framework maximizing classification accuracy. Merging the Cuckoo Search Algorithm and the Artificial Bee Colony, the method excels in exploration and exploitation phases, with Independent Component Analysis assisting in feature acquisition. Its prowess is evident across six gene expression datasets.
Shen and Zhang (2022) delivered a two-stage Grey Wolf Optimization algorithm for high-dimensional data. A multi-layer perceptron plays a central role in both the pre-screening and final selection stages. Its effectiveness is endorsed by results from ten gene expression datasets.
2.3 Embedded method
Momenzadeh et al. (2019) introduced a feature selection method for microarray data using the Hidden Markov Model, combining five metrics to enhance classification performance. This method’s efficacy was validated on three distinct datasets. Seijo-Pardo B et al. introduced thresholding methods in feature selection. In their first study (Seijo-Pardo et al. 2016), they utilized data complexity measures as thresholds. In another study (Seijo-Pardo et al. 2019), they aimed to develop an automatic threshold for ensemble-based feature selection, enhancing ensemble techniques’ accuracy. Bolón-Canedo et al. (2014) presented an ensemble method for data classification that blends multiple filters to achieve superior results, underscoring the ensemble approach’s potential in this domain. Abdulla and Khasawneh (2020) unveiled "G-Forest," designed for cost-sensitive feature selection in gene expression microarrays. Their innovative ensemble-based method balances cost considerations with accuracy, showcasing its applicability in microarray analysis. Ben Brahim and Limam (2018) proposed an ensemble feature selection technique for high-dimensional data. This approach consolidates the benefits of various feature selection methods, with validations emphasizing its effectiveness in high-dimensional datasets like genomics. Li F et al. introduced methods for molecular signature discovery. In their first work (Li et al. 2022a), ConSIG was presented to consistently identify molecular signatures across datasets. In another study (Li et al. 2022b), the POSREG method was crafted to discover proteomic signatures, optimizing both reproducibility and generalizability, addressing the rising demands in proteomics.
2.4 Regularization methods with grouping effects
Yuan and Lin (2006) discussed regression with grouped variables, introducing methods that enhance model accuracy and interpretability by considering group structures. Zou and Hastie (2005) introduced the "elastic net" to overcome challenges with Lasso and correlated predictors, combining L1 and L2 penalties. This approach, influential in statistical learning, manages multicollinearity in predictors. Efron et al. (2004) presented the LARS method for high-dimensional regression. LARS, efficient in computing Lasso regression paths, has become pivotal in statistical learning for large datasets. Fan and Li (2002) centered on variable selection in survival analysis, introducing nonparametric methods for the Cox’s proportional hazards and frailty models, aiming to enhance model flexibility and capture nuances. Zou and Zhang (2009) explored the adaptive elastic-net for high-dimensional data, adjusting penalties based on variable importance, optimizing variable selection in large datasets. Zeng and Xie (2014) proposed a group variable selection method using SCAD-L2, innovatively combining L2-norm adjustments to handle high-dimensional and group-structured data more effectively. Liu et al. (2019) showcased a novel regularization method for biomarker identification in cancer classification, merging computational and biological data to enhance precision and offering promising insights for bioinformatics in cancer diagnostics.
By summarizing the existing methods, it can be observed that the following improvement strategies are mainly applied to feature selection techniques for microarray data.
-
1.
Hybrid two-stage feature selection: This type of methods uses the Filter method as a pre-processing process to reduce the feature space at the coarse granularity and improve the efficiency of the algorithm. Subsequently, the wrapper method is used to further search for optimal features at a fine-grained scale. It is difficult for these research methods to effectively exploit the dependencies between features.
-
2.
Integration of multiple methods: In this type of methods, multiple feature evaluation methods are used to assess feature importance, and subsequently, a unified feature ranking list is generated based on the aggregation strategy.
-
3.
Combination of multiple evolutionary algorithms: In this strategy, an attempt is made to draw on the advantages of different evolutionary algorithms by two or more evolutionary algorithms combined into an evolutionary algorithm with better classification accuracy and operational efficiency for feature selection.
-
4.
Using feature dependencies: in this strategy, the algorithm is able to analyse the features based on the dependencies between the features so as to achieve feature selection based on the dependencies between different features.
Yet, many existing techniques fall short in effectively integrating reported and validated feature dependencies across various dimensions using deep learning. Moreover, to the best of our understanding, there hasn’t been any research that leverages graph neural networks for the amalgamation of such multi-dimensional features in tandem with ensemble evaluation strategies for feature selection. Therefore, to address these challenges, we present a feature selection algorithm grounded in graph neural networks. This model erects graph structures rooted in a priori knowledge, enabling thorough utilization of feature dependencies and capturing high-order feature connections, culminating in a more proficient and precise feature selection.
3 The proposed method
3.1 Overall framework of the proposed method
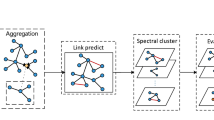
The overall framework of the proposed method is shown in Fig. 1. In our study, we designed a graph-based framework wherein every feature of the primary gene expression matrix acted as a node. Each sample was represented by an individual graph, with all graphs maintaining a consistent structure. Inter-node relationships were sourced from GeneMANIA’s multi-dimensional gene connections. Utilizing these connections and as delineated in Sect. 3.2, we employed a multi-dimensional graph construction method to derive multi-dimensional relationships, subsequently producing multiple relational graphs. These graphs depicted varied relational weights between nodes, as showcased in Fig. 1. A node’s preliminary data corresponds to the expression value of the related sample across diverse features. After removing superfluous connections to refine the graph, the multi-dimensional graph filtering method from Sect. 3.3 was invoked to harness multi-dimensional relationships for disseminating and collating node data. This led to the creation of a node feature matrix using multi-dimensional associations. Nodes linked in GeneMANIA received an edge, leading to our definitive representation graph.
Post execution of the above steps, we curated a graph-structured dataset for each sample, ensuring uniformity in structural details. Moreover, distinct samples had differing node feature vectors. To address the relationships GeneMANIA couldn’t discern, the link prediction method in Sect. 3.4 was adopted. Subsequently, the multi-dimensional node evaluation technique from Sect. 3.5 and the super-node identification algorithm from Sect. 3.6 were utilized for node appraisal, highlighting key nodes. In the end, the method from Sect. 3.7 focusing on Hierarchical graph pooling based on downsampling was employed to sieve through nodes, streamline feature selection, and devise a predictive model.
3.2 Multidimensional graph construction and graph purification
In a gene relationship network, there are multiple types of relationships between pairs of genes, including physical correlation, co-expression, co-localization and gene pathways, etc. We use multidimensional graphs to construct graph structure data to represent such complex relationships effectively. In a multidimensional graph, all dimensions share the same set of nodes, and each dimension can also have its unique structure. First, the microarray data from the GPL platform need to be transformed with probe and gene IDs, and then their correlation information is obtained from GeneMANIA using the gene IDs.
The mathematical representation of the above operations is as follows. Firstly, microarray data can be defined as two sets of samples and features, in which the sample set can be expressed as \(\textrm{S}=\left\{ \textrm{S}_{1},\,\textrm{S}_{2}, \ldots , \textrm{s}_{\textrm{m}}\right\}\), Where, each sample corresponds to a label \(y_{i} \in \{0,1\}(i=1,2, \ldots , m)\) (take the second category task as an example). The feature sets can be represented as \(\textrm{F}=\left\{ \textrm{f}_{1}, \textrm{f}_{2}, \ldots , \textrm{f}_{\textrm{n}}\right\}\), The feature dependency obtained from GeneMANIA can be expressed as matrix R, which contains the dependency \(r_{\left( f_{i} f_{j}\right) }^{k}(k=1,2, \ldots k)\) between any \(f_{i}\) and \(f_{j}\). Where k represents the different dependencies included.
Subsequently, we can define a set of graphs \(\textrm{G}=\left\{ \textrm{g}_1, \,\textrm{g}_2, \ldots , \textrm{g}_{\textrm{k}}\right\}\), where \(\textrm{g}=(\textrm{V}, \textrm{E})\), where \(\textrm{V}=\left( \textrm{v}_{1}, \textrm{v}_{2}, \ldots , \textrm{v}_{\textrm{n}}\right)\) corresponding to the feature set F, set E contains the connection weights between any two nodes \(\left( \textrm{e}_{\textrm{v}_{\textrm{i}}}, \textrm{e}_{\textrm{v}_{\textrm{j}}}\right) =r_{\left( f_{i}, f_{j}\right) }\).
Since the complex relations obtained by GeneMANIA are uncertain, we introduce the sum graph purification operation. Specifically, for the set of graphs \(\textrm{G}=\left\{ \textrm{g}_1, \,\textrm{g}_2, \ldots , \textrm{g}_{\textrm{k}}\right\}\) defined above, in each graph g, we compute the adjacency matrix \(A_{gi}\) corresponding to each graph and decompose it using singular value decomposition (SVD), keeping only top-k singular values to reconstruct the adjacency matrix (experimentally 90% of the overall number is kept), and the reconstructed adjacency matrix can be approximated as the purified graph structure for the following operations.
3.3 Multi-dimensional graph filter
When designing multidimensional graph filters, intra-dimensional and inter-dimensional interactions must be considered. Specifically, intra-dimensional interactions are reflected in the connections between nodes in the same dimension. In contrast, inter-dimensional interactions are reflected in the "copies" of the same node in different dimensions. Therefore, we use a graph filter that captures both intra-dimensional and inter-dimensional information to filter the graph structure data in different dimensions.
First we give the process of node information propagation and aggregation in a one-dimensional graph structure. We define the L-layer hidden state vector of any node \(v_i\) as \(h_{v_{i}}^{L}\), define \(N_{\left( v_{i}\right) }\) to represent the first-order neighborhood of node \(v_i\). The graph neural network can be implemented by introducing spatial filters to propagate and aggregate node information to obtain hidden state vectors that can have the ability to characterize the global situation. The aggregation process is represented as shown in Eq. 1, by which we get the hidden state vector of the node’s L layer.
where \(A G G R E G A T E_{L}\) denotes the aggregation process of nodes from \(L-1\) layer hidden state to L layer hidden state, and h represents the function’s output, which is obtained by aggregating all the first-order neighborhood node information of node \(v_i\).
where \(A G G R E G A T E_{L}\) denotes the aggregation process of nodes from \(L-1\) layer hidden state to L layer hidden state, and h represents the function’s output, which is obtained by aggregating all the first-order neighborhood node information of node \(v_i\). Subsequently, stitch the vectors of nodes using the Eq. 2, and normalize the node vectors using the Eq. 3 to obtain the Embedding information of each node.
Next, we extend the above procedure to the multidimensional graph, where two types of neighbours are defined: intra-dimensional neighbours and cross-dimensional neighbours. For a given node \(v_i\) in dimension d, an intra-dimensional neighbour is a node directly connected to node \(v_i\) in dimension d. Moreover, cross-dimensional neighbours consist of copies of node \(v_i\) in other dimensions, and we define the intra-dimensional neighbours of a node on dimension d to be denoted as \(N_d\left( v_i\right)\). As shown in Fig. 2, node \(v_2\)’s intra-dimensional neighbours in the orange dimension are nodes \({v_1,v_3,v_4,v_5}\). In addition, all dimensions share the same node \(v_2\), which can be considered copies of the same node in different dimensions. These nodes in different dimensions are implicitly connected. So they are the cross-dimensional neighbours of node \(v_2\). The cross-dimensional neighbours of node \(v_2\) in Fig. 2 are its copies of node \(v_2\) in the blue and green dimensions.
Using these two neighbors, we can give the graph filtering operation for the multidimensional graph structure, which is shown in Eqs. 4 to 8.
In Eq. 4 and Eq. 5, the representation of the intra-dimensional neighbours of the nodes \(v_i\) from the previous layer (layer \(l-1\)) is mapped to dimension d by \(\Theta _d^{(l-1)}\), where \(\sigma (*)\) is a nonlinear activation function. Similarly, the representations of node \(v_i\) from the previous layer are mapped to different dimensions, where D is the total number of dimensions in the multidimensional graph. Inter-dimensional aggregation is performed in Eq. 6, which generates intra-dimensional representations for node \(v_i\) in layer l. Cross-dimensional information aggregation is executed in Eq. 7, where \(\beta _{g, d}^{(l-1)}\) is the attention score, which indicates the influence of dimension g on dimension d. It is calculated as shown in Eq. 9.
where \(W^{(l-1)}\) is the parameter matrix to be learned. Finally, the intra-dimensional and cross-dimensional representations of the nodes are combined in Eq. 8 to generate the representation \(\varvec{F}_i^{(l)}\) of the node \(v_i\) updated after layer l, where parameter \(\eta\) is the hyperparameter used to balance these two components.
3.4 Link prediction
Genes not involved in the prior knowledge will form isolated nodes during the establishment of the graph structure. However, isolated nodes are likely to be critical non-redundant features, so link prediction of the established graph structure establishes connections between isolated nodes and other nodes to achieve further information sharing and to lay a good foundation for discovering supernodes.
In the link prediction process, positive and negative samples must be constructed first. Taking node \(v_i\) as an example, we destroy any head-to-tail link connected to node \(v_i\) in graph G, randomly sample some new edges \(e_n\) with \(v_i\) as the center node, and mark them as positive samples if \(e_n \in E\); otherwise, mark them as negative samples. We then record the similarity between the node with \(v_i\) as the center node and all new edges connected to it, which is calculated as shown in Eq. 10.
where \(z_{v_i}\) denotes the value of the feature vector z, w denotes the dimensionality of the feature vector. We set the set of positive samples as Pos and the set of negative samples as Neg, and establish the loss function as shown in Eq. 11.
where \(L_{link}\) denotes the loss value of the loss function, \(E\left( v_j, v_r\right) \in \textrm{Pos}\) denotes the edge of any set of positive samples of data, \(E\left( v_j, v_r\right) \in \textrm{Neg}\) denotes the edge of any set of positive samples of data, and \(\sigma\) is the nonlinear activation function. The model is trained using the stochastic gradient descent algorithm, and the loss value \(L_{link}\) in training is retained, and training is stopped when the difference between the loss values of two training sessions is less than B. Meanwhile, we calculate the Mean reciprocal rank (MRR) of each predicted graph in training, calculated as shown in Eq. 12, and select the optimal graph as the final result according to the MRR.
MRR represents the average reciprocal rank, and rank represents the rank number of the scores from highest to lowest when the \(\varepsilon -th\) edge in the positive sample set scores the corresponding \(\tau -th\) edge in the negative sample set.
3.5 Multi-dimensional node evaluator
In order to evaluate the performance of nodes corresponding to features from multiple dimensions for a more comprehensive feature selection, we also added a multidimensional evaluation score for each node, which a multidimensional feature evaluator obtained we designed to perform a more accurate node evaluation based on the multidimensional evaluation results for the feature selection task. The multidimensional feature evaluator is determined jointly using feature selection evaluation methods with the ability to evaluate features. We used a feature ranking fusion method to combine the results of each evaluator in the following procedure.
Feature ranking fusion can fuse the evaluation results of features from multiple evaluators and generate a unified ranking list. This method is effective in producing fair and objective unified rankings. Although the advantages of feature ranking fusion have been reported in studies, these methods are not fully applicable to fusing the evaluation results of features from different evaluators because the noise of the original data cannot be handled effectively. In the proposed method, we employ a new robust rank aggregation (RRA) method, which detects features with higher than expected rank under the null hypothesis that they are not correlated between feature preference lists as well as within feature preference lists, and assigns a significance score to each feature, which is ranked from smallest to largest to obtain the final ranking of the features. The probabilistic model at the bottom of the algorithm is robust to outliers, noise and errors (Kolde et al. 2012).
The flow of the feature ranking fusion algorithm is as follows, for a given dataset \(D_{M*N}\), where M denotes the number of features and N denotes the number of samples. For any evaluation model \(E V_k(*)\), the nodes go through the evaluation model and generate the corresponding feature ranking vector \(R^{(i)}\) as shown in Eq. 13.
where \(r_1<r_2<\cdots <r_M\), \(r_i\)corresponds to a feature in the original data. In this paper, we use L1 regularization, L2 regularization, linear regression, stability selection, random forest, correlation coefficient and the maximal information coefficien, respectively, as feature evaluation methods. The set \(E V S=\left\{ E V_1, E V_2, \cdots E V_7\right\}\) corresponds to each evaluation method. Each individual generates a corresponding feature ranking vector after the feature evaluation set, and we input these vectors together into the feature ranking model to finally obtain a final score list after feature fusion ranking.
3.6 Supernode discovery algorithms
In order to perform initial feature selection from the original graph structure and screen features with rich information and low redundancy, we use a supernode discovery algorithm for initial feature screening. The set composed of all supernodes constitutes a feature subset after feature coarse-grained screening. We use a spectral clustering-based approach to generate the supernodes and form the graph structure and node features of the coarsened graph.
The goal of subgraph cutting is to generate subgraphs with "the lowest possible sum of edge weights between subgraphs and the highest possible sum of edge weights within subgraphs" so that nodes within the generated subgraphs have more edge weights (more redundancy between features) and nodes between subgraphs have fewer edge weights (less redundancy between features), thus generating a subgraph based on a given number k of features.
First, given the set of all nodes \(V=\left\{ v_1, v_2, \cdots , v_i\right\}\) to be clustered and the number of clusters K. Next, the fully connected method is used to construct the similarity matrix W, which is a similarity matrix composed of \(W_{ij}\) and is calculated as shown in Eq. 14.
It is assumed that any two samples have similarities, but the more distant samples have lower similarities. \(\sigma\) is used to control the width of the neighbourhood of the sample points. Then the degree matrix D is calculated, where the similarity D is the \(n*n\) diagonal matrix composed of \(d_i\), and the \(d_i\) is calculated as shown in Eq. 15.
Subsequently, the Laplacian matrix \(L=D-W\) is calculated and the symmetrized Laplacian matrix representation is used to obtain the normalized Laplacian matrix \(L_{sym}\) as shown in Eq. 16.
Our target is to minimize the loss function as shown in Eq. 17. Next, the eigenvalues of \(L_{sym}\) are calculated and sorted from smallest to largest, and the first k eigenvalues are taken according to the number of clusters k and their eigenvectors \(u_1, u_2, \cdots , u_k\) are calculated. and the k eigenvectors are made into a matrix \(U=u_1, u_2, \ldots , u_k, U \in R^{n * k}\). In this paper, k is set according to the elbow rule.
Subsequently, let \(y_i \in R^k\) be the i-th row feature vector of U (\(i=1,2, \cdots , n\)),and cluster the new sample points \(Y=y_1, y_2, \ldots , y_n\) into clusters \(C = C_1, C_2, \ldots , C_k\) using the K-Means clustering algorithm.
After spectral clustering we divide its nodes into K groups: \(V=\left[ V_1, V_2, \cdots V_k\right]\), \(V_t\) represents the set of nodes in the t-th group, so we can obtain k subgraphs as shown in Eq. 18.
where each \(\mathcal {E}_t\) contains a connection between nodes \(V_t\). If the adjacency matrix of graph \(G_t\) is represented by A, the adjacency matrix is decomposed into a \(k^2\) matrix as shown in Eq. 19 after performing the partition.
where each diagonal block \(A_{tt}\) is the \(V_t * V_t\) adjacency matrix that contains all the connections between graphs \(G_t\), \(\bar{A}\) is the adjacency matrix of graph \(G*\), and \(A_{mn}\) represents the connections between the \(V_m\) and \(V_n\) components. \(\Delta\) is the matrix consisting of all non-diagonal blocks of matrix A.
This gives us k Clusters, the connectivity matrix \(\hat{A}\) within a Cluster, and the connectivity matrix \(\Delta\) between Clusters. It can be assumed that the features characterized by the nodes within each Cluster have high redundancy, while the features characterized by the nodes between each Cluster have low redundancy. Subsequently, we evaluate supernodes in each Cluster as a subset of features in coarse filtering mode and capture the adjacency matrix between supernodes from the original graph structure to form a graph structure that is used as input for the next step of downsampling hierarchical graph pooling for further feature selection. The supernode evaluation score is calculated as shown in Eq. 20.
where \(v_{edge}\) denotes the degree of the node, \(\gamma\) is the hyperparameter used to balance the relationship between the two components, and \(R^i\) is the node’s corresponding multidimensional evaluator ranking.
3.7 Hierarchical graph pooling based on downsampling
After obtaining the graph structure composed of supernodes, we use a hierarchical graph pooling method based on downsampling for further feature selection. Specifically, we first obtain the importance score of each node by evaluating the node importance and then rank the nodes to retain the top-k nodes. Finally, based on the retained nodes, a classification model based on the graph structure is built to evaluate the feature selection performance and achieve the final classification task. In our graph pooling model, we use cross entropy as loss function, Adam as optimiser, ReLU as activation function and learning rate is set to 0.001, in addition Dropout technique is used in the network to prevent overfitting.
In the process of evaluating the important nodes, we use a downsampling strategy to perform a hierarchical pooling operation on the whole graph, and the importance of the nodes is learned from the features \(\varvec{F}_i^{(i p)}\) of the input nodes, as shown in Eq. 21.
where \(\varvec{F}_i^{(i p)} \in \mathbb {R}^{N_{i p} \times d_{i p}}\) denotes the input feature of the i-th node and \(\varvec{p} \in \mathbb {R}^{d_{i p}}\) denotes the feature vector to be learned. After obtaining the importance score y, all nodes can be sorted and the top n most important nodes are selected, and the selected node index idx is shown in Eq. 22.
Subsequently, to avoid introducing information about nodes that have not been selected in the new graph, we extract the adjacency matrix after node selection from the adjacency matrix of the original graph as the structural information of the new graph operated as shown in Eq. 23.
Finally, we re-perform the filtering operation on the obtained graph and build the classification model. The overall flow of the hierarchical graph pooling process based on downsampling is shown in Fig. 3.
4 Data set and experimental results
4.1 Data sets and pre-processing
The 9 publicly available datasets used in this experiment are from the GEO (Gene Expression Omnibus) database, namely ALL2, ALL3, ALL4, DLBCL, Leukemia, Prostate, CNS and Myeloma. All datasets can be obtained from NCBI (https://www.ncbi.nlm.nih.gov/geo/) and the details of the datasets are shown in Table 1.
We first use prior knowledge to build gene network. GeneMANIA provides a large amount of functional association data that can help us find other genes related to a set of input genes. These association data include interactions, pathways, co-expression, co-localization, and protein domain similarity (Warde-Farley et al. 2010).
We use the method described in Sect. 3 to weighted average these relations, combining the microarray data expression values to the graph structure data. All results are averaged with five-fold cross-validation as the final evaluation result. Specifically, the dataset was partitioned into five subsets. In each iteration, four subsets were utilized for training, while the remaining subset was used for testing. This process was repeated five times to ensure each subset had an opportunity to serve as the test set. This method provides a more comprehensive evaluation of our model’s performance.
4.2 Analysis of the results of the proposed method feature selection
In this subsection, we perform a detailed analysis of the feature ranking results of the proposed method on night publicly available microarray datasets, as shown in Fig. 4. We select the number of features from 1 to 50 according to the feature ranking results, input them into different classification models, and evaluate the performance of the feature set using the average classification accuracy of the five-fold cross-validation as the final evaluation index. These classification models include Support Vector Machine (SVM), Decision Tree (DT), K-Nearest Neighbor (KNN), Navie Bayes (NB), Logistic Regression (LR), and our Graph Neural Network based method (GNN), Figure 4a–i correspond to the datasets ALL2, ALL3, ALL4, DLBCL, Leukemia, Prostate, CNS, Myeloma and Lymphoma respectively.
Based on the analysis of the results of Fig. 4, we can conclude the following. The proposed method uses a graph neural network-based classifier with a definite advantage among all classifiers, especially when the number of features is relatively small (up to 10) and can obtain a higher classification accuracy. This is because the graph neural network-based classifier can effectively exploit the dependencies between features. In addition, we found that in the feature selection task for microarray data, the classification accuracy does not improve significantly as the number of features increases. There is even a decreasing trend in the classification accuracy on some datasets due to the introduction of too many redundant features, which justifies our proposed method for feature selection, i.e., removing redundant features by multiple dependencies between features.
Moreover, from the practical application point of view, the sample size needs to be expanded for clinical validation and application after performing the feature selection task to select valid biomarkers. Too many features will incur a considerable cost and cause a waste of resources. We believe selecting less than 20 features as biomarkers is usually reasonable.
4.3 Comparison with classical feature selection methods
To demonstrate the effectiveness of the proposed methods, in this subsection, we compare the proposed methods with a variety of classical feature selection methods, including L1 regularization (Lasso) based methods, random forest (RF), linear regression (LR), L2 regularization (Ridge) based methods, correlation coefficient (Corr) methods, and decision trees (DT). Additionally, we compared four feature selection methods based on group information, namely Group Lasso (GL), Sparse Group Lasso (SGL), Group Elastic Net (GEN), and Least Angle Regression (LARS). Among these methods, we employed the KMeans algorithm to define the grouping strategy and utilized the elbow method to determine the optimal number of clusters. Considering that the data include unbalanced datasets, we compared four evaluation metrics separately, including the average Acc, Pre, Rec and F1 for five-fold cross-validatio. To ensure the fairness of the results, we used SVM as the classification model and set the number of features to 10. The comparison model uses the default parameters of scikit-learn 0.24.2, and the regularization-based model achieves a constraint on the number of features by controlling the penalty term coefficients.
Table 2 shows the results of the proposed method compared with the classical methods mentioned above in terms of classification accuracy, from the results it can be found that the proposed method outperforms the classical feature selection methods on all datasets, where the proposed method improves the average classification accuracy over all classical feature selection methods by 21.12% on the ALL2 dataset, 9.31% on the ALL3 dataset, 4.72% on the ALL4 dataset, on the DLBCL dataset by 5.58%, on the Leukaemia dataset by 4.83%, on the Prostate dataset by 9.04%, on the CNS dataset by 12.3%, on the Myeloma dataset by 8.88%, and on the Lymphoma dataset by 9.3%. Overall, the features selected by the proposed method improved on average 9.46% in classification accuracy compared to the classical method, which proves the effectiveness of the features selected by the proposed method.
Table 3 shows the results of the proposed method compared with the classical method mentioned above on the F1 Score. From the results it can be found that the proposed method outperforms the classical feature selection method on all datasets, where the proposed method improves the average classification accuracy over the classical method by 13.5% on the ALL2 dataset, 22.49% on the ALL3 dataset, 7.89% on the ALL4 dataset, on the DLBCL dataset by 6.91%, on the Leukemia dataset by 7.17%, on the Prostate dataset by 8.51%, on the CNS dataset by 19.68%, and on the Myeloma dataset by 5.49% and on the Lymphoma dataset by 10.82%. Overall, the features selected by the proposed method improved on average by 11.38% in classification accuracy compared to the classical method, which proves that the proposed method is equally effective on the imbalanced dataset.
Table 4 shows the results of the proposed method compared with the classical method mentioned above on the Pre Score. From the results it can be found that the proposed method outperforms the classical feature selection method on all datasets, where the proposed method improves the average Pre over the classical method by 24.80% on the ALL2 dataset, 20.00% on the ALL3 dataset, 7.06% on the ALL4 dataset, on the DLBCL dataset by 7.34%, on the Leukemia dataset by 3.84%, on the Prostate dataset by 9.20%, on the CNS dataset by 13.83%, and on the Myeloma dataset by 10.48% and on the Lymphoma dataset by 9.53%. Overall, the features selected by the proposed method improved on average by 11.79% in Pre compared to the classical method, which proves that the proposed method is equally effective on the imbalanced dataset.
Table 5 shows the results of the proposed method compared with the classical method mentioned above on the Rec Score. From the results it can be found that the proposed method outperforms the classical feature selection method on all datasets, where the proposed method improves the average Rec over the classical method by 16.37% on the ALL2 dataset, 24.60% on the ALL3 dataset, 13.34% on the ALL4 dataset, on the DLBCL dataset by 9.76%, on the Leukemia dataset by 7.07%, on the Prostate dataset by 8.99%, on the CNS dataset by 16.55%, and on the Myeloma dataset by 7.90% and on the Lymphoma dataset by 10.38%. Overall, the features selected by the proposed method improved on average by 12.77% in Rec compared to the classical method, which proves that the proposed method is equally effective on the imbalanced dataset.
4.4 Comparison with graph neural network-based methods
We compared feature selection methods based on graph neural network techniques to demonstrate the advancedness of the proposed methods. These methods include the GNNSC method based on graph neural networks and spectral clustering (Yu et al. 2021), the GNNGR method based on graph neural networks and feature correlation coefficients (Xie et al. 2022b), and the SAGPOOL method via classical hierarchical graph pooling (Lee et al. 2019). The first two of these comparison methods can be directly applied to our dataset. In contrast, for the SAGPOOL method, we use the pooling part of the SAGPOOL method as a feature selector and use the nodes after pooling as the feature selection result. We adjusted the parameters of the above methods so that all feature selection methods guaranteed the same number of features and compared them on four publicly available datasets. The results are shown in Fig. 5.
The results in Fig. 5 show that the proposed method pair has some advantages over the compared methods on different datasets. Among them, the proposed method improves the average classification accuracy by 3.14% compared to the GNNSC method, 2.58% compared to the GNNGR method, and 7.81% compared to the SAGPOOL method. The proposed method is proved to be advanced among graph neural network-based methods and suitable for the feature selection task of processing microarray data.
4.5 Comparison with advanced feature selection methods
To demonstrate the advancedness of the proposed method, in this subsection, we compare the proposed method with some advanced feature selection methods used for microarray data. The detailed results are shown in Table 6, where MFDPSO means novel multi-fitness discrete PSO, BLABC means blended Laplacian ABC algorithm, TSMO means two-phase sequential minimal optimization, ICA means Independent component analysis, RST means rough set theory, NEB means neighborhood entropy-based, IQMI means iterative qualitative mutual information, MMBDE means min-Redundancy and Max-Relevance and Binary Differential Evolution, EF means Ensemble filter, DEAV means Differential Evolution and African vultures. The two most effective methods are shown in bold in the table.
It is clear from the results that the proposed method can obtain higher classification accuracy using a smaller number of features. It is important to note that the proposed method outperforms the hybrid feature selection method and outperforms the feature dependency-based method. For example, in the DLBCL dataset, the Hidden Markov Decision-based feature selection method achieves a classification accuracy of 0.945 on five features, which is better than all the compared hybrid methods. However, the proposed method improves the classification accuracy by 2.9% while reducing the number of features by 67%. The proposed method improved the classification accuracy by 3.6% over the method for the same number of features on the Prostate dataset. Sun et al.’s information entropy-based method on the Leukemia dataset also outperformed the hybrid feature selection method. However, the proposed method improved the classification accuracy by 1.08% compared to this method despite adding one feature. When the same number of features is used, the proposed method still has a 5.9% improvement in classification accuracy, proving the proposed method’s advancedness. Collectively, the proposed method can outperform feature selection methods based on information entropy, Markov decision-making, and meta-heuristic algorithms, proving the advancedness of the proposed method.
4.6 Feature analysis selected by the proposed method
In this subsection, to demonstrate the effectiveness of the features selected by the proposed method, we analyze the first three features selected by the proposed method on the Lymphoma dataset, which corresponds to the probe IDs GENE3314X, GENE3332X and GENE3264X. First we analyze the heat map of these three features, and the results are shown in Fig. 6. It can be seen that there is a significant difference in the distribution of the features selected by the proposed method on the positive and negative samples, which indicates that the three features selected by the proposed method can effectively distinguish between positive and negative samples. We also plotted the distribution of these three features in the three-dimensional spatial structure, and the results are shown in Fig. 7, where black for positive samples and in red for negative samples. It can be seen that the distribution features of the three features are equally effective in distinguishing between positive and negative samples.
In addition, we also analyzed the Pearson correlation coefficients of these three features, and the results are shown in Fig. 8. It can be seen that none of the selected features has a strong correlation, so we consider that the features selected by the proposed method have a low redundancy.
Finally, we analyzed the statistical significance of the three probes, including the multiplicity of differences and p-values shown in Table 7. As can be seen, where * indicates \(p < 0.05\), ** indicates \(p < 0.01\), and *** indicates \(p < 0.001\). from the p-value, it can be seen that all features are significantly significant. From the difference multiples, we can see that GENE3264X and GENE3314X are significantly different in positive and negative samples, and the difference multiples are close to the absolute value of 2. Moreover, GENE3332X has a very significant difference, with a difference multiple reaching an absolute value of 14.
5 Conclusion and future work
This article proposes a method for biomarker selection and classification tasks in microarray data, using multi-dimensional graph neural networks to establish and capture gene multi-dimensional relationships. The method combines multi-dimensional node evaluation, spectral clustering, and super-node discovery algorithms to achieve biomarker selection and classification model construction. Experimental results demonstrate that the proposed method, employing graph neural network technology, effectively utilizes feature dependencies, improves feature selection, enhances classification model performance, and reduces the number of features. Furthermore, analysis of the selected features indicates that the proposed method identifies features with significant statistical differences and holds potential as viable biomarkers.
In future applications and research, for both genomics and proteomics data, we can utilize existing tools such as GeneMANIA (Warde-Farley et al. 2010) or STRING (Damian et al. 2015) to obtain the dependencies between features. By integrating this information with the expression profiles of omics data, we can effectively analyze potential biomarkers and establish diagnostic models. Clinical cohort data can also be analyzed to assist clinical researchers in identifying potential biomarkers. Moving forward, for the future research direction of the model, we can effectively combine graph neural network-based feature selection methods with multi-objective evolutionary algorithms. In recent years, multi-objective evolutionary algorithms have been proven to be highly capable in feature selection tasks (Han et al. 2021). Furthermore, some studies have already been reported concerning network biomarkers (Tang et al. 2022; Zhang et al. 2022), and combining these studies with graph neural network-based feature selection algorithms can be a promising avenue for future research.
Data availability
All datasets used in this paper can be obtained from NCBI (https://www.ncbi.nlm.nih.gov/geo/)
Code availability
More information about the code can be obtained by contacting the corresponding author.
References
Abdulla M, Khasawneh MT (2020) G-forest: an ensemble method for cost-sensitive feature selection in gene expression microarrays. Artif Intell Med 108:101941
Agarwalla P, Mukhopadhyay S (2017) Bi-stage hierarchical selection of pathway genes for cancer progression using a swarm based computational approach. Appl Soft Comput 62:230–250
Agarwalla P, Mukhopadhyay S (2022) Genemops: Supervised feature selection from high dimensional biomedical dataset. Appl Soft Comput 123:108963
Alhenawi E, Al-Sayyed R, Hudaib A, Mirjalili S (2022) Feature selection methods on gene expression microarray data for cancer classification: a systematic review. Comput Biol Med 140:105051
Annavarapu CS (2021) Clustering-based hybrid feature selection approach for high dimensional microarray data. Chemom Intell Lab Syst 213:104305. https://doi.org/10.1016/j.chemolab.2021.104305
Annavarapu CSR, Dara S et al (2021) Clustering-based hybrid feature selection approach for high dimensional microarray data. Chemom Intell Lab Syst 213:104305
Apolloni J, Leguizamón G, Alba E (2016) Two hybrid wrapper-filter feature selection algorithms applied to high-dimensional microarray experiments. Appl Soft Comput 38:922–932
Aziz RM (2022) Nature-inspired metaheuristics model for gene selection and classification of biomedical microarray data. Med Biol Eng Comput 60(6):1627–1646
Ben Brahim A, Limam M (2018) Ensemble feature selection for high dimensional data: a new method and a comparative study. Adv Data Anal Classif 12:937–952
Bhuyan HK, Chakraborty C, Pani SK, Ravi V (2021) Feature and subfeature selection for classification using correlation coefficient and fuzzy model. IEEE Trans Eng Manag
Bolón-Canedo V, Sánchez-Maroño N, Alonso-Betanzos A (2012) An ensemble of filters and classifiers for microarray data classification. Pattern Recognit 45(1):531–539
Bolón-Canedo V, Sánchez-Marono N, Alonso-Betanzos A, Benítez JM, Herrera F (2014) A review of microarray datasets and applied feature selection methods. Inform Sci 282:111–135
Bolón-Canedo V, Sánchez-Marono N, Alonso-Betanzos A (2014) Data classification using an ensemble of filters. Neurocomputing 135:13–20
Chen M, Zang M, Wang X, Xiao G (2013) A powerful Bayesian meta-analysis method to integrate multiple gene set enrichment studies. Bioinformatics 29(7):862–869
Cheng T, Wang Y, Bryant SH (2012) Fselector: a ruby gem for feature selection. Bioinformatics 28(21):2851–2852
Damian S, Andrea F, Stefan W, Kristoffer F, Davide H, Jaime HC, Milan S, Alexander R, Alberto S, Tsafou KP (2015) String v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43(1):D447–D452
Efron B, Hastie T, Johnstone I, Tibshirani R (2004) Least angle regression. Ann Stat 32(2):407–499
Fan J, Li R (2002) Variable selection for cox’s proportional hazards model and frailty model. Ann Stat 30(1):74–99
Ghosh M, Guha R, Sarkar R, Abraham A (2020) A wrapper-filter feature selection technique based on ant colony optimization. Neural Comput Appl 32(12):7839–7857
Han F, Chen W-T, Ling Q-H, Han H (2021) Multi-objective particle swarm optimization with adaptive strategies for feature selection. Swarm Evolut Comput 62:100847
He X, Deng K, Wang X, Li Y, Zhang Y, Wang M (2020) Lightgcn: Simplifying and powering graph convolution network for recommendation. In: Proceedings of the 43rd International ACM SIGIR Conference on Research and Development in Information Retrieval, pp 639–648
Hua J, Tembe WD, Dougherty ER (2009) Performance of feature-selection methods in the classification of high-dimension data. Pattern Recognit 42(3):409–424
Huang L-T (2009) An integrated method for cancer classification and rule extraction from microarray data. J Biomed Sci 16(1):1–10
Jian T, Zhou S (2016) A new approach for feature selection from microarray data based on mutual information. IEEE/ACM Trans Comput Biol Bioinform 13(6):1–1
Jinthanasatian P, Auephanwiriyakul S, Theera-Umpon N (2018) Microarray data classification using neuro-fuzzy classifier with firefly algorithm. In: 2017 IEEE Symposium series on computational intelligence (SSCI)
Jl A, Iyc B, Chj C (2020) An efficient multivariate feature ranking method for gene selection in high-dimensional microarray data. Expert Syst Appl 166:113971
Khani E, Mahmoodian H (2020) Phase diagram and ridge logistic regression in stable gene selection. Biocybern Biomed Eng 40(3):78
Kolde R, Laur S, Adler P, Vilo J (2012) Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics 4:573
Lee J, Lee I, Kang J (2019) Self-attention graph pooling. In: International conference on machine learning, pp 3734–3743. PMLR
Lefakis L, Fleuret F (2016) Jointly informative feature selection made tractable by Gaussian modeling. J Mach Learn Res 17(1):6314–6352
Li S, Oh S (2016) Improving feature selection performance using pairwise pre-evaluation. BMC Bioinform 17:1–13
Li Y, Dai Z, Cao D, Luo F, Chen Y, Yuan Z (2020) Chi-mic-share: a new feature selection algorithm for quantitative structure-activity relationship models. RSC Adv 10(34):19852–19860
Li F, Yin J, Lu M, Yang Q, Zeng Z, Zhang B, Li Z, Qiu Y, Dai H, Chen Y et al (2022) Consig: consistent discovery of molecular signature from omic data. Brief Bioinform 23(4):253
Li F, Zhou Y, Zhang Y, Yin J, Qiu Y, Gao J, Zhu F (2022) Posreg: proteomic signature discovered by simultaneously optimizing its reproducibility and generalizability. Brief Bioinform 23(2):040
Li W, Chi Y, Yu K, Xie W (2023) A two-stage hybrid biomarker selection method based on ensemble filter and binary differential evolution incorporating binary african vultures optimization. BMC Bioinform 24(1):1–27
Lin S, Xz A, Yq C, Jx A, Sz A (2019) Feature selection using neighborhood entropy-based uncertainty measures for gene expression data classification. Inform Sci 502:18–41
Liu S-M, Ou S-Y, Huang H-H (2017) Green tea polyphenols induce cell death in breast cancer mcf-7 cells through induction of cell cycle arrest and mitochondrial-mediated apoptosis. J Zhejiang Univ Sci B 18(2):89–98
Liu X-Y, Wang S, Zhang H, Zhang H, Yang Z-Y, Liang Y (2019) Novel regularization method for biomarker selection and cancer classification. IEEE/ACM Trans Comput Biol Bioinform 17(4):1329–1340
Lu H, Chen J, Yan K, Jin Q, Xue Y, Gao Z (2017) A hybrid feature selection algorithm for gene expression data classification. Neurocomputing 256:56–62
Mazumder DH, Veilumuthu R (2019) An enhanced feature selection filter for classification of microarray cancer data. ETRI J 41(3):358–370
Medjahed SA, Saadi TA, Benyettou A, Ouali M (2016) Kernel-based learning and feature selection analysis for cancer diagnosis. Appl Soft Comput 51:39–48
Momenzadeh M, Sehhati M, Rabbani H (2019) A novel feature selection method for microarray data classification based on hidden Markov model. J Biomed Inform 95:103213
Musheer RA, Verma C, Srivastava N (2019) Novel machine learning approach for classification of high-dimensional microarray data. Soft Comput 23(24):13409–13421
Muthukrishnan R, Rohini R (2016) Lasso: A feature selection technique in predictive modeling for machine learning. In: 2016 IEEE international conference on advances in computer applications (ICACA), pp 18–20. IEEE
Nagpal A, Singh V (2019) Feature selection from high dimensional data based on iterative qualitative mutual information. J Intell Fuzzy Syst 36(6):5845–5856
Oh I-S, Lee J-S, Moon B-R (2004) Hybrid genetic algorithms for feature selection. IEEE Trans Pattern Anal Mach Intell 26(11):1424–1437
Ouadfel S, Abd Elaziz M (2022) Efficient high-dimension feature selection based on enhanced equilibrium optimizer. Expert Syst Appl 187:115882
Pashaei E, Pashaei E (2022) An efficient binary chimp optimization algorithm for feature selection in biomedical data classification. Neural Comput Appl 34(8):6427–6451
Peng H, Fu Y, Liu J, Fang X, Jiang C (2013) Optimal gene subset selection using the modified sffs algorithm for tumor classification. Neural Comput Appl 23(6):1531–1538
Remeseiro B, Bolon-Canedo V (2019) A review of feature selection methods in medical applications. Comput Biol Med 112:103375
Rodriguez-Galiano VF, Luque-Espinar JA, Chica-Olmo M, Mendes MP (2018) Feature selection approaches for predictive modelling of groundwater nitrate pollution: an evaluation of filters, embedded and wrapper methods. Sci Total Environ 624:661–672
Salem H, Attiya G, El-Fishawy N (2016) Classification of human cancer diseases by gene expression profiles. Appl Soft Comput 50:124–134
Saranya G, Pravin A (2022) A novel feature selection approach with integrated feature sensitivity and feature correlation for improved prediction of heart disease. J Ambient Intell Hum Comput 89:1–15
Seijo-Pardo B, Bolón-Canedo V, Alonso-Betanzos A (2019) On developing an automatic threshold applied to feature selection ensembles. Inform Fusion 45:227–245
Seijo-Pardo B, Bolón-Canedo V, Alonso-Betanzos A (2016) Using data complexity measures for thresholding in feature selection rankers. In: Advances in artificial intelligence: 17th conference of the spanish association for artificial intelligence, CAEPIA 2016, Salamanca, Spain, September 14–16, 2016. Proceedings 17, pp 121–131. Springer
Serrano D, Bonanni B, Brown K (2019) Therapeutic cancer prevention: achievements and ongoing challenges-a focus on breast and colorectal cancer. Mol Oncol 13(3):579–590
Shen C, Shen K (2022) Two-stage improved grey wolf optimization algorithm for feature selection on high-dimensional classification. Complex Intell Syst 45:1–21
Shukla AK (2020) Multi-population adaptive genetic algorithm for selection of microarray biomarkers. Neural Comput Appl 32(15):11897–11918
Sun L, Zhang XY, Qian YH, Xu JC, Zhang SG (2019) Joint neighborhood entropy-based gene selection method with fisher score for tumor classification. Appl Intell 1245(1259):49
Tang S, Yuan K, Chen L (2022) Molecular biomarkers, network biomarkers, and dynamic network biomarkers for diagnosis and prediction of rare diseases. Fundam Res
Thabtah F, Kamalov F, Hammoud S, Shahamiri SR (2020) Least loss: aD simplified filter method for feature selection. Inform Sci 534:1–15
Tumuluru P, Ravi B (2017) Goa-based dbn: grasshopper optimization algorithm-based deep belief neural networks for cancer classification. Int J Appl Eng Res 12:14218–14231
Wan Y, Wang M, Ye Z, Lai X (2016) A feature selection method based on modified binary coded ant colony optimization algorithm. Appl Soft Comput 49:248–258
Wang A, An N, Chen G, Li L, Alterovitz G (2015) Accelerating wrapper-based feature selection with k-nearest-neighbor. Knowled-Based Syst 83:81–91
Wang A, An N, Yang J, Chen G, Li L, Alterovitz G (2017) Wrapper-based gene selection with Markov blanket. Comput Biol Med 81:11–23
Wang T, Shao W, Huang Z, Tang H, Zhang J, Ding Z, Huang K (2021) Mogonet integrates multi-omics data using graph convolutional networks allowing patient classification and biomarker identification. Nat Commun 12(1):1–13
Wang X, Wang Y, Wong K-C, Li X (2022) A self-adaptive weighted differential evolution approach for large-scale feature selection. Knowl-Based Syst 235:107633
Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT et al (2010) The Genemania prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38(2):214–220
Wu SJ, Pham VH, Nguyen TN (2017) Two-phase optimization for support vectors and parameter selection of support vector machines: two-class classification. Appl Soft Comput 59:129–142
Xie W, Fang Y, Yu K, Min X, Li W (2022) Mfrag: multi-fitness rankaggreg genetic algorithm for biomarker selection from microarray data. Chemom Intell Lab Syst 226:104573
Xie W, Li W, Zhang S, Wang L, Yang J, Zhao D (2022) A novel biomarker selection method combining graph neural network and gene relationships applied to microarray data. BMC Bioinform 23(1):1–18
Xie W, Chi Y, Wang L, Yu K, Li W (2021) Mmbde: a two-stage hybrid feature selection method from microarray data. In: 2021 ieee international conference on bioinformatics and biomedicine (BIBM), pp. 2346–2351. IEEE
Xu W, Liu X, Leng F, Li W (2020) Blood-based multi-tissue gene expression inference with Bayesian ridge regression. Bioinformatics 36(12):3788–3794
Xue B, Zhang M, Browne WN (2012) Particle swarm optimization for feature selection in classification: a multi-objective approach. IEEE Trans Cybern 43(6):1656–1671
Xue B, Zhang M, Browne WN, Yao X (2015) A survey on evolutionary computation approaches to feature selection. IEEE Trans Evolut Comput 20(4):606–626
Yu K, Xie W, Wang L, Zhang S, Li W (2021) Determination of biomarkers from microarray data using graph neural network and spectral clustering. Sci Rep 11(1):1–11
Yuan M, Lin Y (2006) Model selection and estimation in regression with grouped variables. J R Stat Soc Ser B 68(1):49–67
Yuan M, Yang Z, Huang G, Ji G (2017) Feature selection by maximizing correlation information for integrated high-dimensional protein data. Pattern Recognit Lett 92:17–24
Zeng L, Xie J (2014) Group variable selection via scad-l 2. Statistics 48(1):49–66
Zhang Y, Gong D-W, Gao X-Z, Tian T, Sun X-Y (2020) Binary differential evolution with self-learning for multi-objective feature selection. Inform Sci 507:67–85
Zhang J, Xu D, Hao K, Zhang Y, Chen W, Liu J, Gao R, Wu C, De Marinis Y (2021) Fs-gbdt: identification multicancer-risk module via a feature selection algorithm by integrating fisher score and gbdt. Brief Bioinform 22(3):189
Zhang Y, Chang X, Xia J, Huang Y, Sun S, Chen L, Liu X (2022) Identifying network biomarkers of cancer by sample-specific differential network. BMC Bioinform 23(1):230
Zhou Q, Zhou H, Li T (2016) Cost-sensitive feature selection using random forest: selecting low-cost subsets of informative features. Knowl-Based Syst 95:1–11
Zhou H, Zhang J, Zhou Y, Guo X, Ma Y (2021) A feature selection algorithm of decision tree based on feature weight. Expert Syst Appl 164:113842
Zou H, Hastie T (2005) Regularization and variable selection via the elastic net. J R Stat Soc Ser B 67(2):301–320
Zou H, Zhang HH (2009) On the adaptive elastic-net with a diverging number of parameters. Ann Stat 37(4):1733
Acknowledgements
The authors would like to thank Dazhe Zhao of Nursoft Medical System for helpful discussions on topics related to this work.
Funding
This research was funded by National Key Research and Development Program of China (2021YFC2701003). Natural Science Foundation of Liaoning Province under grant 2022JH2/101300075. National Frontiers Science Center for Industrial Intelligence and Systems Optimization, Shenyang 110819, the 111 Project (B16009).
Author information
Authors and Affiliations
Contributions
WDX proposed experimental ideas, evaluated experimental data, and drafted manuscripts. SJZ designs experimental procedures collects data, and assists in manuscript writing. KY and LJW proposed the overall structure of the article and supplemented the experimental chart. WL revises the manuscript and evaluates the data. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Not applied.
Ethical approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xie, W., Zhang, S., Wang, L. et al. Feature selection of microarray data using multidimensional graph neural network and supernode hierarchical clustering. Artif Intell Rev 57, 63 (2024). https://doi.org/10.1007/s10462-023-10700-3
Accepted:
Published:
DOI: https://doi.org/10.1007/s10462-023-10700-3