Abstract
This study’s foremost objectives were to scrutinize how unexpected weather affects agricultural output and to assess how well AI-based machine learning and deep leaning algorithms work for spotting apple leaf diseases. The researchers carried out a bibliometric study to obtain understanding of the current research trends, citation patterns, ownership and partnership arrangements, publishing patterns, and other parameters related to early identification of apple illnesses. Comprehensive interdisciplinary scientific maps are limited because syndrome recognition is not restricted to any solitary arena of research, despite the fact that there have been many studies on the identification of apple diseases. By employing a scientometric technique and 109 publications from the Scopus database published between 2011 and 2022, this study attempted to assess the condition of the research area and combine knowledge frameworks. To find important journals, authors, nations, articles, and topics, the study used the automated processes of VOSviewer and Biblioshiny software. Patterns and trends were discovered using citation counts, social network analysis, and citation and co-citation studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The agriculture industry is of utmost significance since it significantly boosts the global economy and contributes to guaranteeing food security for the whole population (Stefanis 2014). Yet, there has been a sharp rise in plant disease infections as a result of the environment’s continuing deterioration. Hence, the entrance of diseases into plants causes the agricultural industry to sustain considerable losses each year (Bonkra et al. 2022b). The manual evaluation of trees to identify the illness is a time-consuming and arduous operation that needs a lot of resources, both human and financial. The demand for quick and accurate disease identification techniques is growing increasingly essential as plant diseases continue to pose problems for the agriculture sector. The utilisation of IoT devices and cloud-based collaboration (Bonkra and Dhiman 2021; Goyal et al. 2021; Kaur and Kaur 2022) is one possible approach. Farmers may access real-time data and analytics by utilising these technologies, allowing them to streamline their processes and react rapidly to changing conditions. Farmers are better able to avoid disease outbreaks and lessen their impacts when they can monitor and analyse their activities in real-time. This can boost the apple industry’s overall production and quality while lowering its financial losses.
By using such a system, farmers may minimise the detrimental effects of agriculture on the environment by using less resources, such as water and fertiliser, and applying them more precisely in accordance with the demands of the crop (Bashir et al. 2011). The traditional techniques of disease identification by visual examination and expert consultation, especially when dealing with large apple orchards, can be ineffective and impractical, however, with over 100 known apple diseases (Hasan et al. 2022), including Botryosphaeria obtusa (Abbasiet al. 2019), Venturia inaequalis (Abbasiet al. 2019), and Gymnosporangium juniperi-virginianae (Shaffer and Wrather 1996). In the agricultural sector, the application of machine learning and computer vision has grown in significance, especially for the detection and diagnosis of apple illnesses (Zhang et al. 2018; Bonkra et al. 2023a; Verma et al. 2019). The leaves serve as a dependable and essential source of information for disease identification as practically every apple disease affects them. Early disease detection can increase output, lower expenses, and improve the quality of the crop (Gawade 2021). Deep learning (Bonkra et al. 2022a) and machine learning methods have many uses in a assortment of fields, with processing of text (Prasanna and Tripathi 2022), detection of Deep Fake (Dhiman et al. 2023a), medical diagnostics (Chalabi et al. 2021; Heryawan et al. 2022), and others. Regulating authorities like the French government and the European Commission (Lefebvre et al. 2015) have been concocted inducement programmes focused at cutting back on use of chemical as a vital step to minimising environmental impact. Spraying plants with chemicals like fungicides, insecticides, and herbicides is a typical agricultural practise to manage weeds, pests, and diseases and assure great crop harvests (Hough et al. 1951). Yet, the use of pesticides in agriculture has sparked worries about the environment and public health, and the way that pesticides are now sprayed is not sustainable. Other strategies that are both more successful and environmentally benign are required to solve these issues. Reducing the need for chemical sprays requires the early identification and diagnosis of apple illnesses. The following section describes the several types of illnesses that harm Apple leaves.
1.1 Apple leaf disease’s categories
1.1.1 Leaf blotch from marsonina
An undesirable fungus known as apple Marsonina Blotch (AMB) causes early defoliation, shrinkage, and decreased starch development in fruit. The pathogen Diplocarpon mali, which causes AMB, was first discovered in Japan and has since spread fast over the entire world (Wöhner and Emeriewen 2019).
1.1.2 Apple crumbling leaves
Due to an imbalance in the soil’s moisture content, the amount of light, and the air’s temperature, the crop is affected physiologically (Kalwad et al. 2022).
1.1.3 Leaf alternaria apple
The virus Physalospora obtusa causes the fungus, often known as “frogeye leaf spot,” or “Black Rot,” which can destroy different stages of crops (Madhu et al. 2020).
1.1.4 Microscopic mold
This apple disease often manifests itself while the plant is young and the buds are forming new leaves and stems. The telltale signs of this condition are clumps of white or greyish-chalky material on the undersides of the leaves, together with curled leaf edges (Salomão et al. 2014).
1.1.5 Scabby apple
Small sores on the undersides of leaves are the hallmark of the common and harmful fungal disease known as apple scab, which is brought on by the Venturia inaequalis fungus. The fruit may develop little black scabs as a result of it spreading there (Belete and Boyraz 2017).
1.1.6 Mosaic apple
A pathogenic positive-sense RNA disease called the apple mosaic virus causes intermittent patches or streaks of light yellow color on apple leaves in the season of spring, When the temperature is unstable, the virus spreads (Dursunoglu and Ertunc 2006).
The primary objectives of this work are as follows:
-
1.
This study offers helpful insights for creating the literature survey by helping to discover and analyse current trends in the research field.
-
2.
This research can contribute to mapping such knowledge gaps and assist in detecting gaps in the body of current literature.
-
3.
The purpose of this study is to monitor and assess changes in publishing trends throughout time. It offers perceptions into the changing environment for scholarly publishing.
-
4.
To pinpoint geographical areas or nations that have made significant contributions to the field.
-
5.
The major objective is to pinpoint trends via in-depth analysis of a variety of sources and to appropriately acknowledge authors who have made excellent and substantial contributions to the field of study.
-
6.
The major objective is to identify publication patterns by examining the writers’ affiliations with organisations or groups. This study looks for links between writers and their institutional affiliations in the context of scientific publications.
-
7.
The purpose of the study is to examine the citation frequency of publications by counting the number of times that they have been cited in academic works. This analysis examines the volume of citations that individual articles have in order to understand their influence and impact on the scholarly community.
The study is broken into many sections, with Part 2 including a thorough assessment of the literature on various segmentation and classification techniques for apple leaf diseases. The primary goals of Sect. 3 are to gather and analyse Scopus data, particularly using network and statistical analysis, which are the most used research approaches in this field. Section 4 presents the investigation’s findings, while Sect. 5 contains the paper’s conclusion and bibliography. Part 6 concludes with an overview of the study’s constraints.
2 Previous work
The development of automated systems that use computer vision, image processing, and machine learning (Kaur et al. 2022a) to detect and identify plant diseases has received a lot of attention recently. These systems main objective is to help agriculturalists and farm-workers to recognise plant illnesses-swiftly and accurately (Ouhami et al. 2021), which can help to slow the spread of disease and boost crop production. Several plant species have been proposed as potential candidates for automated disease detection systems, including tomato (Chowdhury et al. 2021), papaya (Islam et al. 2020), olive (Alruwaili et al. 2019), rice (Tian et al. 2021), cassava, and palm. While the particular methods used may differ, the majority of these systems examine photos of plants to find disease symptoms including discolouration, lesions, or other anomalies. In a recent study, a unique method for identifying apple leaf diseases was suggested using image processing and machine learning (Mohapatra et al. 2022) approaches. This process included dividing the disease zone into several sections, as well as photo enhancement, feature selection, and other critical procedures. To improve visibility and identify the locations of the illnesses in the photos, the authors combined the 3D-Box filter, de-correlation, 3D-Gaussian, and 3D-median filters (Cui et al. 2013). A reliable technique known as correlated pixel (SCP) was used to precisely detect and isolate the sick spots in the pictures. Incorporating the expectation-maximization (EM) method improved this strategy much further. After segmenting the disease zones, the researchers used local binary patterns (LBP) (Qiu et al. 2012) and colour histogram-based techniques to extract features from each region. The following step was applying a genetic algorithm (GA) (Qiu et al. 2012) to choose the qualities that were most relevant and unique for the categorization job. Many machine learning techniques (Kaur et al. 2022b) were used, including support vector machines (SVM), decision trees, segmentation (Kaushal et al. 2020, 2022), and random forests, to evaluate the effectiveness of the proposed strategy. The study found that using the histogram transformation approach improved the accuracy of illness detection, even when using test images that were not in the best quality. Also, the SVM method outperformed competing methods when the authors used the edited photographs to look for illnesses (Rodriguez-Galiano et al. 2015). Numerous studies back a third strategy that uses a hybrid contrast stretching technique that combines adaptive contrast stretching and histogram equalisation to improve the brightness and contrast of pictures. When it comes to enhancing the aesthetic appeal of images, this technique has demonstrated encouraging outcomes. This makes it simpler to distinguish between unhealthy and healthy leaf sections. The scientists used a amended version of the Mask R-CNN model (Wang et al. 2020) to identify the ill areas. For segmenting and identifying objects in pictures, this model represents the most advanced deep learning technique (Dhiman et al. 2023b). Some suggested systems involve a number of important steps, such as photo capture, feature extraction, pre-processing, and classification (Cohen 2021; Trivedi et al. 2021). The photographs were taken using high-resolution cameras to photograph leaves that were afflicted with various ailments. During the picture preparation step, morphological processing and Gaussian filtering were applied to enhance image quality and reduce noise. The authors then used the extraction of feature’s methods Local Binary Patterns (LBP) (Dubey and Jalal 2012) and Gray Level Co-occurrence Matrix to identify significant features from the preprocessed pictures (GLCM) (Omrani et al. 2014). A deep learning-based strategy for classifying apple leaf diseases was proposed by Sanal et al. (2019) in 2019. In order to categorise apple leaf illnesses according to their symptoms, the authors utilised a convolutional neural network (CNN) (Kaushal et al. 2022; Khiabani et al. 2022). The usefulness of deep learning approaches for this problem was demonstrated by the proposed method’s accuracy of 98%. In order to classify apple leaf disease, Jiang et al. (2019) also employed a CNN. A transfer learning-based strategy was suggested by the authors, in which a CNN that had already been trained to classify apple leaf disease was further refined. The accuracy rate for the suggested approach was 96.9%, proving the value of transfer learning in this context. The diagnosis of apple leaf diseases has made substantial use of machine learning techniques. In order to categorise the disorders, a research was done to evaluate how well several machine learning algorithms, such as support vector machine (SVM) (Evgeniou and Pontil 2001), decision tree, k-nearest neighbours (KNN), and random forest, performed. For the categorization of apple leaf disease, Liu et al. (2017) also employed SVM and random forest methods. The authors used a hybrid feature extraction technique that incorporated discrete wavelet transform (DWT) and principal component analysis (PCA) to extract distinguishing characteristics from photos of apple leaves. This approach attempted to efficiently capture differentiating qualities. A CNN-based method for identifying apple leaf disease was suggested by Yadav et al. (2020). In order to extract and classify features, the authors used a deep residual network (ResNet) architecture. Deep learning approaches (Bhardwaj et al. 2022) were successful in this challenge, as evidenced by the proposed method’s accuracy of 94.2%. The researchers also used a Convolutional Neural Networks (CNN)-based approach to identify and categorise apple leaf illnesses (Di and Li 2022). The authors suggested a deep learning-based approach that improved a pre-trained CNN’s ability to recognise apple leaf disease. The segmentation techniques used on different datasets are summarised in Table 1.
3 Research techniques and data gathering
The gathering and selection of data for the study will continue. The search technique was chosen using the PRISMA approach, which is shown in Fig. 1, in order to create a thorough paper. PRISMA, or Preferred Reporting Items for Systematic Reviews and Meta-Analyses, is an acronym for systematic reviews and meta-analyses and offers a formal framework for performing them. It is an example of an evidence-based reporting guideline that offers authors a structured method for publishing the outcomes of a meta-analysis or systematic review. Since its creation in 2009, the PRISMA statement has been revised to include a checklist and flow diagram to enable transparent reporting of a systematic review or meta-analysis. PRISMA followed four basic stages for data collection, data analysis and data visualization. The serach string used for searching the optimum collection of sources. Then by using Identification, Screening, Eligibility and Included keywords collection is refined as per requirements from the total 414 documents and after proceeding of various stages final 109 artciles are utilized for this study.
3.1 Data analysis tools
3.1.1 Bibliometrix
The Bibliometrix program was designed using the R language and a logical bibliometric methodology. This programme can do a variety of bibliometric studies, such as co-citation analysis, coupling analysis, scientific cooperation analysis, and co-word analysis, by creating data matrices (Aria and Cuccurullo 2017). The Bibliometrix software can import bibliographic information from several databases, such as Scopus, PubMed, Digital Science Dimensions, Web of Science, and Cochrane (Lages et al. 2018). People without coding skills can use bibliometrics for analysis with the help of the Biblioshiny web app.
3.1.2 VOSviewer
Bibliometric networks may be seen and analysed using VOSviewer (Van Eck and Waltman 2010, 2011), a potent tool. Based on co-citation or bibliographic coupling ties between publications, the VOSviewer programme, created by academics at Leiden University, enables users to generate maps of scientific literature. VOSviewer (Bonkra et al. 2023b) is a well-liked tool for studying and visualising complicated networks of academic literature among scholars, librarians, and information workers because of its user-friendly interface and sophisticated features. Scientific articles, patents, and grants are just a few examples of the sorts of data that may be analysed using VOSviewer (Pradhan 2017). By choosing various colour schemes and representations, as well as by filtering and grouping their data, users may personalise their maps.
3.1.3 SCI2
An all-inclusive software programme entitled the Sci2 Tool was created aimed at use in systematic research in the fields of informatics and data science. The Cyberinfrastructure for Network Science Center at Indiana University created this open-source programme, which offers a variety of analytical and visualisation capabilities for handling and studying huge and complicated information. Researchers in a variety of fields, including social science, biology, computer science, and library science, now frequently use the Sci2 Tool (Börner 2011) because of its modular architecture and user-friendly interface. Its versatility and adaptability make it a useful tool that can be tailored to meet various research objectives and procedures.
3.1.4 Copal red
The academic group EC3 at the University of Granada in Spain created the potent software tool known as Copal Red (Bonkra et al. 2023a). This commercial program’s main job is to do co-word analysis on scientific literature using keywords to draw out significant conclusions and provide new information. Users may quickly convert unprocessed data into useful insights thanks to the program’s learning mechanism, which can gather and analyse information from a variety of sources. Comma-Separated Values (CSV) files, which are frequently generated by benchmark supervisor applications, may be read and processed by Copal Red, which is one of its main advantages. Users may simply import data from a number of sources with this capability, which also streamlines the data entering process.
3.1.5 CiteSpace
CiteSpace (Synnestvedt et al. 2005) is a part of software for examining and displaying trends and patterns in scientific publications. CiteSpace, an application created by Chaomei Chen at Drexel University, uses bibliometric methods to find and examine citation networks, co-citation links, and co-occurrence patterns in scientific publications. CiteSpace is well-liked among scholars, librarians, and information specialists as a tool for studying and visualising intricate scientific networks because of its user-friendly design and cutting-edge capabilities. The VOSviewer and Biblioshiny software tools were used by the researchers to speed up processing and improve comprehension of the study data. These tools were helpful in achieving the best results and streamlining the data analysis procedure.
3.2 Preliminary data gathering
A database is a set of organised data that is electronically stored and retrieved. Databases may store and handle text, numbers, images, and other types of information in addition to multimedia content. To handle and analyse massive volumes of data, they are frequently utilised in enterprises, governments, and organisations. An comprehensive database called Scopus contains references and abstracts from a variety of scholarly works. It includes social sciences, humanities, as well as technological and medical sciences, and it encompasses a wide range of academic disciplines. It has about 75 million records, including more than 25 million patents, and is owned and maintained by Elsevier, one of the biggest academic publishers. In order to help researchers track, analyse, and visualise research trends, Scopus provides a number of tools. These techniques, which allow users to recognise significant research trends and developing fields of study, include author profiling, co-citation networks, and citation analysis (Hardcastle 2011). The methodological part of the study is divided into two steps: gathering the necessary information and selecting a search strategy. A bibliographic evaluation is employed in this work and is based on the Scopus database (https://www.scopus.com; accessed April 10, 2023). (“Apple” OR “Apple tree” OR “Malus” AND “Leaf” OR “Leaves” AND “Disease” OR “Infection” OR “Pathogen” AND “Detection” OR “Diagnosis” OR “Classification” OR “Segmentation” AND (“AI” OR “ML” OR “DL” OR “Neural network”). The dataset consists of papers that were located using the database’s search terms in the search title. 414 documents are discovered when all publications are taken into account, and 109 matches are discovered when only articles are examined. The study concentrated on reviewing 109 publications that were written between 2011 and 2022. The researchers used the “Export Records to CSV File” option to gather these publications, choosing only the texts that included “Logbook Content: Full Record and Cited References.” Through this procedure, the researchers were able to download the 109 articles in Comma-Separated File (.CSV) format for use in the second portion of the study’s analysis. It’s important to note that the system can only produce 109 documents at once, which results in the creation of a single “.csv” file.
4 Bibliometric evaluation and outcomes
Bibliometrics is a technique frequently employed to assess the performance of research undertaken by institutions and organizations (Kumar et al. 2021). It is a term derived from the words “biblio,” which refers to evaluation, and “metrics,” which pertains to literature. By scrutinizing variables like article count, citations, and reviews, bibliometrics can provide an accurate assessment of research data (xxx 2021). This type of analysis can be useful in pinpointing areas for future research and providing authors with suggestions on how they can contribute to the field. Performance analysis and scientific mapping (Noyons et al. 1999) are the two basic methods used in bibliometric analysis. Science mapping lays more emphasis on comprehending the links and interconnections between various research features, such as themes and areas, whereas performance analysis concentrates on analysing the specific contributions of research components, such as authors and publications.
4.1 Overview
4.1.1 Basic information of dataset
The Fig. 2 illustared the basic description of 109 studies extracted and filtered from Scopus database and further used in this manuscript. The retrieved information has been categorized using primary data, document contents, authors, authors’ collaborations, and document categories. The selected research includes a total of 1345 references, comprising 65 journal and book sources. On average, each paper cites 13.52 articles and demonstrates an annual growth rate of 42.18%. The papers feature 574 keywords, with 241 unique author keywords from 337 authors. Collaboratively, the data gathering includes 4.33 co-authors and 11.93 foreign co-authors, with three individuals working independently. The publications are classified into different categories, such as book chapters, conference papers, Review papers, Book Chapters, and reviews.
4.1.2 Scientific publications per year
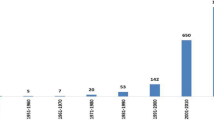
In order to assess changes in apple disease research, the project has collected data on apple leaves covering 11 years, from 2011 to 2022. The study’s accompanying graph demonstrates that the area of apple disease research is continuously evolving, with a noticeable surge in publications beginning in 2018 shown in Fig. 3. It is clear that the topic has gained greater interest recently and is relatively fresh, even though the exact number of publications for 2022 has not yet been calculated (due to the continuing nature of the search).
4.1.3 Three field plot
A Sankey diagram (Riehmann et al. 2005; Koo 2021) was made to graphically show the distribution of research subjects among nations and the recentness of referenced references. The Plot which contains the titles,nation, and Keyword as “TI_TM”,“AU_CO” and “DE” parameters of publication of the cited references, gives a clear picture of the proportion of study subjects for each country as well as the freshness of the papers they mentioned. Figure 4 provides a comprehensive representation of the complex interconnections among three fundamental components of apple leaf disease detection based on machine learning. The left side of the graph displays the 10 most often used title keywords by researchers. The inclusion of specific terms such as “leaf,” “apple,” “disease,” “neural,” “deep,” and “learning” plays a pivotal role in shaping the scholarly literature of this topic, exerting a significant influence on the ongoing discourse and research trajectory. In the intermediate phase, an examination is conducted on the 10 primary nations that contribute the most, including countries such as China, India, and Saudi Arabia, among others. These countries are crucial in the advancement of apple leaf disease diagnostics, highlighting the worldwide scope and importance of this area of study. In this discourse, we examine the foremost 10 methodologies for illness identification, including “Deep Learning,” “Transfer Learning,” “CNN,” “Machine Learning,” “Image Processing,” and “Feature Extraction.” The aforementioned strategies include the array of tools used by researchers to tackle the intricate task of detecting apple leaf diseases via the utilization of sophisticated computer algorithms. The interplay between these three areas is apparent. The choice of title keywords significantly influences the selection of research methodologies, which are often shaped by the research agendas of different nations and areas. Comprehending these relationships is of utmost importance in order to ascertain the direction of future research and devise efficient decision support systems for the control of diseases in apple orchards.
4.2 Sources
4.2.1 Most relevant sources
Sources that are frequently cited and have a high level of impact are considered the most significant in a particular research field. By utilizing these sources, researchers can gain valuable insights and knowledge that can keep them informed about advancements in their field and help them make informed decisions about which sources to prioritize for their own research. The study’s findings highlight the numerous platforms’ most pertinent sources for research publishing. With nine publications included in this study, the analysis shows in Fig. 5 that “Lecture Notes in Networks and Systems” is the most significant source. In addition, “Frontiers in Plant Science,” “Computers and Electronics in Agriculture,” “IEEE Access,” “Sensor,” and “Symmetry” are also considered significant sources for this research field. These findings underscore the importance of utilizing a diverse set of sources to gain a comprehensive understanding of a research topic.
4.2.2 Law of Bradford
Law of Bradford (Thompson and Walker 2015) is a bibliometric principle that describes the distribution of scientific literature among journals. The law posits that, when journals in a particular field are ranked by article productivity, the number of articles published in each journal will be inversely proportional to its rank. This means that the top-ranked journals will have a higher number of published articles than the middle- and lower-ranked journals. The law was named after Samuel C. Bradford, who observed this pattern while examining the distribution of papers in geology journals (Ikpaahindi 1985). The Fig. 6 will illustrate the higher cite source generated by “Lecture Notes in Networks and Systems” with the rank one, Frequency nine, Cumulative Frequency nine and under Zone1 and followed by “Frontiers in Plant Science”, “Computers and Electronics in Agriculture”, “IEEE Access”,“Computers, Materials and Continua”,“Sensors”, “Symmetry” and “2022 IEEE Delhi Selection Conference, Delcon 2022”.
4.2.3 Annual source production
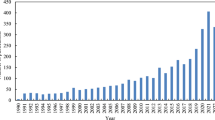
In bibliometric analysis, “source production over time” refers to the number of publications produced within a specific field, by an individual author or group of authors, or by a particular institution during a designated time-frame. This information can be obtained by analyzing bibliographic data from databases such as Scopus or Web of Science, which provide information on the publication date, author, journal, and citation counts of each publication. The presented figure displays the pattern of source production rate over time, with distinct colors indicating different time frames. The graph 7 illustrates a consistent upward trend in the number of articles produced between 2018 to 2022, reaching its maximum value during this period. This indicates a noteworthy level of research activity during these years, possibly influenced by various factors such as technological advancements, increased funding, or emerging research areas (Fig. 7).
4.3 Authors
4.3.1 Most significant authors
The term “most significant author” in bibliometrics refers to an individual who has made a notable and impactful contribution to a specific research field. This determination often relies on bibliographic data and citation analysis, which can reveal an author’s body of work and the extent of their influence. To identify the most significant authors in a field, bibliometric analysis may consider various metrics such as the total number of publications, citation counts, the h-index, and other indicators of scholarly impact. The provided Fig. 8 demonstrates that “LIU B” holds the highest number of publications among all authors featured in the graph, indicating their noteworthy impact in the field of apple leaf disease detection. The remaining authors on the list are arranged in decreasing order of publication count.
4.3.2 Most significant affiliation
To determine the Most Significant Affiliation (MSA) of a research output, bibliometric tools such as the Affiliation Index and the Fractional Count approach are utilized to analyze the affiliations of the authors listed in the publication and estimate the relative contribution of each institution. The Fig. 9 highlights the institution with the maximum affiliation in a particular research area, which is the “Northwestern University of Agricultural and Forestry Science and Technology,” having collaborated on 59 articles. In contrast, the “JILIN AGRICULTURAL UNIVERSITY” and “THIAGARAJAR COLLEGE OF ENGINEERING” had the minimum affiliation, with only 8 articles published in the same research area.
4.3.3 Most significant cited countries
The term “Most Significant Cited Countries” (MSCC) pertains to countries that have made the most substantial contributions to the scholarly literature in a particular field. The identification of MSCC is based on the frequency with which scholars from that country are cited in academic works within that area of study. To determine the MSCC, citation patterns within a specific study area, such as a discipline or subfield, are analyzed using various bibliometric methods, such as Scopus and the Essential Science Indicators (ESI) database.
Based on the citation breakdown by country in the provided Fig. 10, “China” emerged as the leading country with the highest number of citations, having contributed to 323 published articles. “India” came in second place with 248 citations, followed by “Pakistan”, “ Turkey”, “Spain”, “Indonesia”, “Norway”, “Saudi Arabia” and “Mexico”.
4.4 Document
4.4.1 Most frequent words
In bibliometric analysis, stopwords and punctuation are often removed from the text data like in other text analysis methods in order to determine the most frequently occurring words. After counting the remaining words, the most frequent words are determined. Numerous textual pieces of information, including publication abstracts or full-text articles, are frequently analysed as part of bibliometric study. The most popular themes, keywords, or study areas may be found within this data by using the most frequent terms. Deep leaning, apple leaf disease detection, malus, precision agriculture, and feature extraction were among the terms used in this study. The phrases “Deep Learning” and “Apple Leaf Disease Detection” with occurrence numbers of 64 and 41, respectively, have the highest frequency, per the results shown in Fig. 11.
4.4.2 Word cloud
A word cloud (Heimerl et al. 2014) is a graphic depiction of the words that appear most frequently in a corpus, dataset, or collection of texts. Word clouds (Heimerl et al. 2014) can be used in bibliometrics to examine the main themes, subjects, and areas of study within a certain field or discipline. The most popular terms are graphically represented as word clouds, with the size of each word corresponding to its frequency in the text corpus. Use software tools like Wordle, TagCrowd, or Voyant Tools to do this.
Figure 12 uses two alternative word clouds created by selecting 20 keywords based on the title and author’s keywords.
4.4.3 Word tree map
A rectangular form of hierarchical data may be shown using a treemap (Scheibel et al. 2020), a visualisation tool. The rectangles that make up the treemap each indicate a particular value or amount by virtue of their size. The rectangles are arranged in a hierarchy, with each rectangle broken into smaller rectangles to symbolise each subcategory under the parent category. The illustration showed a treemap of several keywords, each specified by a rectangle-shaped image in a distinct colour. The Fig. 13 included terms like leaf (which occurred 84 times), apple (which occurred 78 times), and soon.
4.5 Conceptual structure
4.5.1 Co-occurennce all keywords analysis
The underlying themes and subjects within a certain text corpus or dataset can be discovered by analysing the co-occurrence (Hosseini et al. 2021) of keywords or phrases. Additionally, it can be useful to find connections between various phrases or keywords, such as synonyms or associated ideas, which can assist to hone search terms and increase the precision of keyword-based searches (Fig. 14, Table 2).
The effective method of grouping related objects or entities based on shared traits or qualities is known as cluster analysis. Cluster analysis is employed in network visualisation maps to locate node clusters that are tightly related to one another and to show these nodes as separate clusters. According to the research, just 43 of the total 1466 keywords had at least 10 occurrences, which was the cutoff point. The links and correlations among these 43 keywords were highlighted by further grouping them into 3 groups. Three primary clusters are depicted in the illustration 14 as “Red,” “Green,” and “Blue” bubbles. There are ten things in the red colour bubble, including “Convolutional Neural Network,” “Deep Neural Network,” “Leaf Disease,” and “Convolution.” Twelve components, including “Segmentation,” “Extraction,” “Feature Extraction,” “CNN,” “Textures,” and “Recognition Accuracy,” are included in the green colour bubble. Nine objects, including “Crops,” “Machine Learning,” etc., are contained in the blue colour bubble.
4.5.2 Co-occurrence all index keywords analysis
A technique used in computational linguistics to find and quantify the frequency of co-occurrence of words (Pourhatami et al. 2021) or phrases within a particular corpus of text is called co-occurrence index keywords analysis. The co-occurrence index is derived by dividing the total co-occurrences of two words within a given word frame by the product of the individual frequencies of the two words. The Fig. 15 depict the maximum occurrence of deep learning, features extraction, plants disease and many more.
4.5.3 Thematic map
The term “thematic map” (Yu and Muñoz 2020) refers to a sort of map that presents geographic data about a particular theme or topic rather than merely physical or political elements. Thematic maps are useful for displaying a variety of facts on a certain topic, such as population density, economic activity, or climatic trends. A range of visual components, including colours, symbols, and shading, are frequently used in the representation of data in thematic maps (Forrest 2015). To illustrate varying amounts of a variable, such as income or temperature, over several geographic locations, a choropleth map can, for instance, utilise various colour tones.
The Fig. 16 explore the concept of four themes such as “Basic”, “Motor”, “Nache” and “Emerging or Declined”. The each instance identified with round shaped different color bubbles. The Basic theme contains “Leaf Apple Disease” and “Detection Learning classification”. The Motor theme contains “Approach”, “Agriculture” and “Artificial”. The Niche theme described “Rust”,“Detecting”, “Images”, “Category”, “Multi” and “Rapid”. The Emerging theme descrubed about “Leaves”, “Data” and “Learning Based”.
5 Discussion and future direction
Finally, this work performed a bibliometric analysis to investigate the trend of applying machine learning approaches for apple leaf disease diagnosis. According to the survey, China and India are major contributors in this field, using machine learning, deep learning, and convolutional neural network (CNN) methodologies. “Lecture Notes in Networks and Systems” emerged as a key source, as did journals such as “Frontiers in Plant Science,” “Computers and Electronics in Agriculture,” “IEEE Access,” “Sensor,” and “Symmetry.” The study also identified key terms for precise keyword searches, such as “Convolutional Neural Network,” “Deep Neural Network,” “Leaf Disease,” “Convolution,” “Segmentation,” “Extraction,” “Feature Extraction,” “CNN,” “Textures,” “Recognition Accuracy,” “Crops,” and “Machine Learning.” This research lays the groundwork for future research into machine learning-based apple leaf disease identification. The integration of cutting-edge algorithms like RNNs and GANs, the use of multispectral imaging and sensor fusion techniques, the development of real-time monitoring systems, the promotion of collaboration and data sharing, and field validation for practical deployment are key areas that require more investigation. By addressing these issues, disease detection accuracy will increase, allowing for prompt intervention and increasing agricultural yield. In the foreseeable future, our objective is to create a decision support system that prioritizes user-friendliness, utilizing machine learning and deep learning techniques in its underlying framework. The proposed system aims to facilitate the uploading of leaf photos for analysis by various users, including farmers and researchers. Subsequently, the system will furnish categorizations, so denoting whether the leaf exhibits signs of sound health or manifests symptoms of a certain ailment in apple trees. The primary objective of this automated system is to enhance efficiency and save expenses, therefore facilitating users in making well-informed determinations regarding the health of their apple trees.
Data availability
Not applicable.
Code availability
Not applicable.
References
Abbasi P, Ali S, Braun G, Bevis E, Fillmore SAE (2019) Reducing apple scab and frogeye or black rot infections with salicylic acid or its analogue on field-established apple trees. Can J Plant Pathol 41(3):345–354. https://doi.org/10.1080/07060661.2019.1610070
Alruwaili M, Alanazi S, Abd El-Ghany S, Shehab A (2019) An efficient deep learning model for olive diseases detection. Int J Adv Comput Sci Appl 10(8)
Aria M, Cuccurullo C (2017) Bibliometrix: an r-tool for comprehensive science mapping analysis. J Informet 11(4):959–975
Bashir T, Poudyal K, Bhattacharya T, Sambandam S (2011) Bashir et al 2011 Ottawa abstract
Belete T, Boyraz N (2017) Critical review on apple scab (Venturia inaequalis) biology, epidemiology, economic importance, management and defense mechanisms to the causal agent. J Plant Physiol Pathol 5(2):2
Bhardwaj R, Jaroli P, Singla C, Bhardwaj V, Mohapatra S (2022) Deep learning model based novel semantic analysis. In: 2022 2nd international conference on advance computing and innovative technologies in engineering (ICACITE), pp 1454–1458. https://doi.org/10.1109/ICACITE53722.2022.9823741
Bonkra A, Dhiman P (2021) Iot security challenges in cloud environment. In: 2021 2nd international conference on computational methods in science & technology (ICCMST), pp 30–34. IEEE
Bonkra A, Dhiman P, Goyal S, Gupta N (2022a) A systematic study: implication of deep learning in plant disease detection. In: 2022 IEEE international conference on current development in engineering and technology (CCET), pp 1–6. IEEE
Bonkra A, Noonia A, Kaur A (2022b) Apple leaf diseases detection system: a review of the different segmentation and deep learning methods. In: Artificial intelligence and data science: first international conference, ICAIDS 2021, Hyderabad, India, December 17–18, 2021, Revised Selected Papers. Springer, pp 263–278
Bonkra A, Bhatt P, Rosak-Szyrocka J, Muduli K, Pilař L, Kaur A, Chahal N, Rana A (2023a) Apple leave disease detection using collaborative ml/dl and artificial intelligence methods: scientometric analysis. Int J Environ Res Public Health 20(4):3222. https://doi.org/10.3390/ijerph20043222
Bonkra A, Bhatt PK, Kaur A, Kamboj S (2023b) Scientific landscape and the road ahead for deep learning: apple leaves disease detection. In: 2023 international conference on artificial intelligence and smart communication (AISC), pp 869–873. IEEE
Börner K (2011) Science of science studies: Sci2 tool. Commun ACM 54(3):60–69
Bracino AA, Concepcion RS, Bedruz RAR, Dadios EP, Vicerra RRP (2020) Development of a hybrid machine learning model for apple (Malus domestica) health detection and disease classification. In: 2020 IEEE 12th international conference on humanoid, nanotechnology, information technology, communication and control, environment, and management (HNICEM), pp 1–6. IEEE
Chalabi NE, Attia A, Akrouf S (2021) Machine learning and deep learning: recent overview in medical care. In: Internet of Things, infrastructures and mobile applications: proceedings of the 13th IMCL conference 13. Springer, New York, pp 223–231
Chowdhury ME, Rahman T, Khandakar A, Ayari MA, Khan AU, Khan MS, Al-Emadi N, Reaz MBI, Islam MT, Ali SHM (2021) Automatic and reliable leaf disease detection using deep learning techniques. AgriEngineering 3(2):294–312
Chuanlei Z, Shanwen Z, Jucheng Y, Yancui S, Jia C (2017) Apple leaf disease identification using genetic algorithm and correlation based feature selection method. Int J Agric Biol Eng 10(2):74–83
Cohen S (2021) The basics of machine learning: strategies and techniques. In: Artificial intelligence and deep learning in pathology. Elsevier, Amsterdam, pp 13–40
Cui X, Li S, Yu M, Kim H, Kwack K-S, Min B-H (2013) Automatic cartilage segmentation and measurement for diagnosis of OA using 3d box and gaussian filters. Osteoarthr Cartil 21:232–233
Dhiman P, Kaur A, Iwendi C, Mohan SK (2023a) A scientometric analysis of deep learning approaches for detecting fake news. Electronics 12(4):948
Dhiman P, Kaur A, Bonkra A (2023b) Fake information detection using deep learning methods: a survey. In: 2023 International conference on artificial intelligence and smart communication (AISC), pp 858–863. IEEE
Di J, Li Q (2022) A method of detecting apple leaf diseases based on improved convolutional neural network. PLoS ONE 17(2):0262629
Donthu N, Kumar S, Mukherjee D, Pandey N, Lim WM (2021) How to conduct a bibliometric analysis: an overview and guidelines. J Bus Res 133:285–296
Dubey SR, Jalal AS (2012) Detection and classification of apple fruit diseases using complete local binary patterns. In: 2012 third international conference on computer and communication technology, pp 346–351. IEEE
Dubey SR, Jalal AS (2016) Apple disease classification using color, texture and shape features from images. SIViP 10:819–826
Dursunoglu S, Ertunc F (2006) Distribution of Apple mosaic Ilarvirus (ApMV) in Turkey. In: XX international symposium on virus and virus-like diseases of temperate fruit crops-fruit tree diseases, vol 781, pp 131–134
Evgeniou T, Pontil M (2001) Support vector machines: theory and applications. Adv Course Artif Intell 2049:249–257. https://doi.org/10.1007/3-540-44673-7_12
Forrest D (2015) Thematic maps in geography. Int Encycl Soc Behav Sci. https://doi.org/10.1016/B978-0-08-097086-8.72069-1
Gawade A (2021) Early-stage apple leaf disease prediction using deep learning. Biosci Biotechnol Res Commun 14(5):40–43
Goyal S, Bhushan S, Kumar Y, Rana AuHS, Bhutta MR, Ijaz MF, Son Y (2021) An optimized framework for energy-resource allocation in a cloud environment based on the whale optimization algorithm. Sensors 21(5):1583
Hardcastle J (2011) Scopus: a quick introduction. Editors’ Bull 7(3):90–91
Hasan S, Jahan S, Islam MI (2022) Disease detection of apple leaf with combination of color segmentation and modified dwt. J King Saud Univ-Comput Inf Sci 34(9):7212–7224
Heimerl F, Lohmann S, Lange S, Ertl T (2014) Word cloud explorer: text analytics based on word clouds. In: 2014 47th Hawaii international conference on system sciences, pp 1833–1842. https://doi.org/10.1109/HICSS.2014.231
Heryawan L, Febriansyah F, Bukhori A (2022) Deep learning and machine learning model comparison for diagnosis detection from medical records. In: The 2022 international conference on computer, control, informatics and its applications, pp 306–310
Hosseini S, Baziyad H, Norouzi R, Jabbedari Khiabani S, Gidófalvi G, Albadvi A, Alimohammadi A, Seyedabrishami S (2021) Mapping the intellectual structure of gis-t field (2008–2019): a dynamic co-word analysis. Scientometrics 126:2667–2688
Hough WS, Mason AF, et al (1951) Spraying, dusting and fumigating of plants. Principles and applications. In: Spraying, dusting and fumigating of plants. Principles and applications
Ikpaahindi L (1985) An overview of bibliometrics: its measurements, laws and their applications. Libri 35:163
Islam MA, Islam MS, Hossen MS, Emon MU, Keya MS, Habib A (2020) Machine learning based image classification of papaya disease recognition. In: 2020 4th international conference on electronics, communication and aerospace technology (ICECA), pp 1353–1360. IEEE
Jiang P, Chen Y, Liu B, He D, Liang C (2019) Real-time detection of apple leaf diseases using deep learning approach based on improved convolutional neural networks. IEEE Access 7:59069–59080
Kalwad PD, Kanakaraddi SG, Preeti T, Ichalakaranji S, Salimath S, Nayak S (2022) Apple leaf disease detection and analysis using deep learning technique. In: Information and communication technology for competitive strategies (ICTCS 2020) ICT: applications and social interfaces. Springer, New York, pp 803–814
Kaur A, Kaur B (2022) Load balancing optimization based on hybrid heuristic-metaheuristic techniques in cloud environment. J King Saud Univ-Comput Inf Sci 34(3):813–824
Kaur G, Kaur A, Khurana M (2022a) A review of opinion mining techniques. ECS Trans 107(1):10125
Kaur G, Kaur A, Khurana M, et al (2022b) A stem to stern sentiment analysis emotion detection. In: 2022 10th international conference on reliability, infocom technologies and optimization (trends and future directions)(ICRITO), pp 1–5. IEEE
Kaushal C, Kaushal K, Singla A (2020) Firefly optimization based segmentation technique to analyze medical images of breast cancer. Int J Comput Math 98:1–19. https://doi.org/10.1080/00207160.2020.1817411
Kaushal C, Islam MK, Althubiti S, Alenezi F, Mansour R (2022) A framework for interactive medical image segmentation using optimized swarm intelligence with convolutional neural networks. Comput Intell Neurosci. https://doi.org/10.1155/2022/7935346
Khan MA, Lali MIU, Sharif M, Javed K, Aurangzeb K, Haider SI, Altamrah AS, Akram T (2019) An optimized method for segmentation and classification of apple diseases based on strong correlation and genetic algorithm based feature selection. IEEE Access 7:46261–46277
Khiabani SJ, Batani A, Khanmohammadi E (2022) A hybrid decision support system for heart failure diagnosis using neural networks and statistical process control. Healthc Anal 2:100110
Koo M (2021) Systemic lupus erythematosus research: a bibliometric analysis over a 50-year period. Int J Environ Res Public Health 18(13):7095
Kumar S, Patil RR, Kumawat V, Rai Y, Krishnan N, Singh S (2021) A bibliometric analysis of plant disease classification with artificial intelligence using convolutional neural network. Library Philos Pract 5777:1–14
Lages FS, Douglas-de Oliveira DW, Costa FO (2018) Relationship between implant stability measurements obtained by insertion torque and resonance frequency analysis: A systematic review. Clin Implant Dent Relat Res 20(1):26–33
Lefebvre M, Langrell SR, Gomez-y-Paloma S (2015) Incentives and policies for integrated pest management in Europe: a review. Agron Sustain Dev 35:27–45
Liu B, Zhang Y, He D, Li Y (2017) Identification of apple leaf diseases based on deep convolutional neural networks. Symmetry 10(1):11
Madhu G, Nabi SU, Mir JI, Raja WH, Sheikh MA, Sharma OC, Singh DB (2020) Alternaria leaf and fruit spot in apple: symptoms, cause and management. Eur J Biotechnol Biosci 8:24–26
Mohapatra S, Sarangi PK, Sarangi P, Sahu P, Sahoo B (2022) Text classification using NLP based machine learning approach. In: AIP conference proceedings, vol 2463, p 020006. https://doi.org/10.1063/5.0080301
Noyons E, Moed H, Van Raan A (1999) Integrating research performance analysis and science mapping. Scientometrics 46(3):591–604
Omrani E, Khoshnevisan B, Shamshirband S, Saboohi H, Anuar NB, Nasir MHNM (2014) Potential of radial basis function-based support vector regression for apple disease detection. Measurement 55:512–519
Ouhami M, Hafiane A, Es-Saady Y, El Hajji M, Canals R (2021) Computer vision, IoT and data fusion for crop disease detection using machine learning: a survey and ongoing research. Remote Sens 13(13):2486
Pourhatami A, Kaviyani-Charati M, Kargar B, Baziyad H, Kargar M, Olmeda-Gómez C (2021) Mapping the intellectual structure of the coronavirus field (2000–2020): a co-word analysis. Scientometrics 126(8):6625–6657
Pradhan P (2017) Science mapping and visualization tools used in bibliometric & scientometric studies: an overview
Prasanna DL, Tripathi SL (2022) Machine and deep-learning techniques for text and speech processing. In: Machine learning algorithms for signal and image processing, pp 115–128
Qiu H, Song Y, Zhao T (2012) The EM algorithm and its packages in r project: a literature review. In: Proceedings of the 2012 second international conference on electric information and control engineering, vol 1, pp 1464–1467
Rehman ZU, Khan MA, Ahmed F, Damaševičius R, Naqvi SR, Nisar W, Javed K (2021) Recognizing apple leaf diseases using a novel parallel real-time processing framework based on mask rcnn and transfer learning: An application for smart agriculture. IET Image Proc 15(10):2157–2168
Riehmann P, Hanfler M, Froehlich B (2005) Interactive sankey diagrams. In: IEEE symposium on information visualization. INFOVIS 2005, pp 233–240. IEEE
Rodriguez-Galiano V, Sanchez-Castillo M, Chica-Olmo M, Chica-Rivas M (2015) Machine learning predictive models for mineral prospectivity: an evaluation of neural networks, random forest, regression trees and support vector machines. Ore Geol Rev 71:804–818
Salomão BdCM, Muller C, Amparo HCd, Aragão GMFd (2014) Survey of molds, yeast and Alicyclobacillus spp. from a concentrated apple juice productive process. Braz J Microbiol 45:49–58
Sanal MG, Paul K, Kumar S, Ganguly NK (2019) Artificial intelligence and deep learning: the future of medicine and medical practice. J Assoc Phys India 67(4):71–73
Scheibel W, Trapp M, Limberger D, Döllner J (2020). A taxonomy of treemap visualization techniques. https://doi.org/10.5220/0009153902730280
Shaffer WH, Wrather JA (1996) Cedar apple rust
Stefanis C (2014) Global food security: an agricultural perspective. J Agric Sustain 6:69–87
Sumithra M, Saranya N (2021) Particle swarm optimization (PSO) with fuzzy c means (PSO-FCM)-based segmentation and machine learning classifier for leaf diseases prediction. Concurr Comput 33(3):5312
Synnestvedt MB, Chen C, Holmes JH (2005) Citespace II: visualization and knowledge discovery in bibliographic databases. In: AMIA annual symposium proceedings, vol 2005. American Medical Informatics Association, p 724
Thompson DF, Walker CK (2015) A descriptive and historical review of bibliometrics with applications to medical sciences. Pharmacotherapy 35(6):551–559
Tian L, Xue B, Wang Z, Li D, Yao X, Cao Q, Zhu Y, Cao W, Cheng T (2021) Spectroscopic detection of rice leaf blast infection from asymptomatic to mild stages with integrated machine learning and feature selection. Remote Sens Environ 257:112350
Trivedi NK, Gautam V, Anand A, Aljahdali HM, Villar SG, Anand D, Goyal N, Kadry S (2021) Early detection and classification of tomato leaf disease using high-performance deep neural network. Sensors 21(23):7987
Van Eck N, Waltman L (2010) Software survey: Vosviewer, a computer program for bibliometric mapping. Scientometrics 84(2):523–538
Van Eck NJ, Waltman L (2011) Vosviewer manual. Manual for VOSviewer version 1(0)
Verma K, Bhardwaj S, Arya R, Islam U, Bhushan M, Kumar A, Samant P (2019) Latest tools for data mining and machine learning. Int J Innov Technol Explor Eng 8(9S):18–23
Wang Y, Wu J, Li H (2020) Human detection based on improved mask r-cnn. In: Journal of Physics: Conference Series, vol 1575. IOP Publishing, p 012067
Wöhner T, Emeriewen OF (2019) Apple blotch disease (Marssonina coronaria (Ellis & Davis) Davis)-review and research prospects. Eur J Plant Pathol 153(3):657–669
Yadav D, Yadav AK et al (2020) A novel convolutional neural network based model for recognition and classification of apple leaf diseases. Traitement du Signal 37(6)
Yu J, Muñoz J (2020) A bibliometric overview of twitter-related studies indexed in web of science. Future Internet 12:91. https://doi.org/10.3390/fi12050091
Zhang S, Wang H, Huang W, You Z (2018) Plant diseased leaf segmentation and recognition by fusion of superpixel, k-means and phog. Optik 157:866–872
Funding
There was no particular grant for this research from any funding organisation in the public, private, or nonprofit sectors.
Author information
Authors and Affiliations
Contributions
AB contributed the statistics and tables in addition to writing the initial drafting. The data sources and editing were done by SP. AK and MAS oversaw and validated the work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical Approval
Yes.
Consent for participation
Yes.
Consent to publication
Yes.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bonkra, A., Pathak, S., Kaur, A. et al. Exploring the trend of recognizing apple leaf disease detection through machine learning: a comprehensive analysis using bibliometric techniques. Artif Intell Rev 57, 21 (2024). https://doi.org/10.1007/s10462-023-10628-8
Accepted:
Published:
DOI: https://doi.org/10.1007/s10462-023-10628-8