Abstract
Artificial intelligence (AI) has advanced rapidly, but it has limited impact on biomedical text understanding due to a lack of annotated datasets (a.k.a. few-shot learning). Multi-task learning, which uses data from multiple datasets and tasks with related syntax and semantics, has potential to address this issue. However, the effectiveness of this approach heavily relies on the quality of the available data and its transferability between tasks. In this paper, we propose a framework, built upon a state-of-the-art multi-task method (i.e. MT-DNN), that leverages different publicly available biomedical datasets to enhance relation extraction performance. Our model employs a transformer-based architecture with shared encoding layers across multiple tasks, and task-specific classification layers to generate task-specific representations. To further improve performance, we utilize a knowledge distillation technique. In our experiments, we assess the impact of incorporating biomedical datasets in a multi-task learning setting and demonstrate that it consistently outperforms state-of-the-art few-shot learning methods in cases of limited data. This results in significant improvement across most datasets and few-shot scenarios, particularly in terms of recall scores.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Relation extraction (RE) is a subfield of text classification, a natural language processing (NLP) task that aims to automatically associate unstructured text to one (Hosseinalipour et al. 2021a, b) or several (Khataei Maragheh et al. 2022) labels. Specifically, RE aims to identify and extract relationships between entities in unstructured text data. This task is crucial for various applications such as information retrieval (Gharehchopogh and Khalifehlou 2012), question answering (Lee et al. 2007), knowledge graph construction (Wang et al. 2017) and text summarization (Mahalleh and Gharehchopogh 2022.) One of the major challenges in the field of relation extraction is the high variability and complexity of the language used to express relationships. To address this challenge, various methods have been proposed, including rule-based methods (Nebhi 2013; Ben Abacha and Zweigenbaum 2011), machine learning-based methods (Alimova and Tutubalina 2020; Hong et al. 2020), and hybrid approaches (Huang et al. 2006; Zhang et al. 2018) that combine both.
In recent years, there has been a surge in research in the fields of soft computing (Afradi and Ebrahimabadi 2020; Afradi et al. 2021, 2020; Afradi and Ebrahimabadi 2021) and relation extraction and their potential for various NLP applications, especially in biomedical text understanding. Applications include detecting protein–protein interactions (PPIs) (Pyysalo et al. 2006) and extracting information on adverse drug events (ADEs) (Gurulingappa et al. 2012). One major driving factor behind the advancements in relation extraction for biomedical text is the integration of attention mechanisms (Li et al. 2023) into NLP models. These mechanisms enable the models to concentrate on specific parts of the input, which is crucial when dealing with complex biomedical text that contains a high density of specialized terminology. Furthermore, the widespread availability of pre-trained biomedical language models has also been a key factor in enhancing the performance of relation extraction tasks. These models have been trained on vast amounts of biomedical data and can be fine-tuned for specific tasks, resulting in substantial improvements in performance (Lewis et al. 2020). Overall, the recent advancements in NLP and the availability of pre-trained biomedical language models have paved the way for a new generation of relation extraction models with improved performance. These models can extract valuable information from biomedical text with greater accuracy and efficiency, providing benefits for various biomedical applications.
While relation extraction for biomedical text has seen significant progress, the lack of large and high-quality annotated biomedical datasets remains a major challenge. The annotation process is time-consuming and requires extensive domain knowledge, making it expensive to obtain large amounts of annotated data. As a result, this has a significant impact on the performance of relation extraction models in real-world applications. To overcome these limitations, there is a growing need to shift focus from model-centric to data-centric AI, emphasizing the critical role of data in the learning process and the need to extract maximum value from it. Such a shift would enable the development of more effective and robust relation extraction models, addressing the limitations of limited annotated datasets.
Multi-task learning (Caruana 1998) is a technique that aims to address the issue of limited annotated training data by leveraging the similarities between different datasets. This approach involves training a single model on multiple related tasks, using the similarities between the tasks to improve the training process. This technique has been widely adopted in biomedical text understanding and has demonstrated its usefulness in several studies (Peng et al. 2020). However, despite its advantages, multi-task learning can also result in a degradation of performance if the datasets used have different structures and objectives. The size and underlying properties of the datasets can also have an impact on the performance of the model (Alonso and Plank 2017). Thus, careful consideration should be given to the choice of datasets used in multi-task learning to ensure optimal results.
In this study, we present a multi-task framework for biomedical relation extraction (RE) that utilizes a well-established multi-task learning approach (Liu et al. 2019). We use three publicly available multi-class datasets, namely DDI-2013, ChemProt, and I2B2-2010 RE, which are annotated with relationships among drugs, chemical compounds and proteins, medical problems, treatments, and tests. Our framework consists of a transformer-based model with shared layers across the three RE tasks, and separate classification heads for each dataset. To further enhance performance, we adopt a training framework based on knowledge distillation. Our experiments investigate the effectiveness of our multi-task framework in few-shot scenarios, where annotated training data is scarce. Our results surpass the state-of-the-art few-shot learning techniques in the majority of the few-shot scenarios and datasets, with up to 65% improvement in F1 compared to the second-best technique with only 10 training examples. This highlights the potential of multi-task learning techniques in the challenging context of few-shot biomedical text understanding, where collecting large annotated datasets is difficult, but acquiring smaller similar datasets from different clinical organizations is more feasible. The code is available on github.Footnote 1. The contributions of our work can be summarized as follows:
-
We propose a framework that utilizes multi-task learning and knowledge distillation to deal with the few-shot learning issue in the biomedical relation extraction field.
-
We evaluate the quality of our approach with recent baselines, showing that it outperforms the existing state-of-the-art in the majority of few-shot learning scenarios.
-
We provide an in-depth data-driven analysis of the main factors influencing multi-task learning when integrating the knowledge of heterogeneous biomedical datasets.
-
We show how the performance of the proposed approach varies in scenarios with varying degrees of data scarcity (i.e. 1, 10, 50, 100 and 1000 training samples).
The remainder of this paper is organized as follows. Section 2 reviews the related studies on relation extraction and its applications in the biomedical field and in few-shot scenarios. Section 3 presents the datasets and the proposed method in details. In Sect. 4, experiments about the method and its application in few-shot scenarios are provided, and implications about results are discussed. Finally, this paper is concluded in Sect. 5.
2 Related work
Relation extraction (RE) has been thoroughly investigated in the realm of NLP and Information Extraction (IE). One of the most widely adopted rule-based methods is the use of regular expressions and lexical patterns to identify relationships, which rely on the manual creation of patterns that are specific to the target relationships and the domain of the text (Nebhi 2013; Ben Abacha and Zweigenbaum 2011). While this approach has demonstrated good results, it is heavily dependent on the quality and coverage of the patterns. In contrast, machine learning-based approaches (Alimova and Tutubalina 2020; Hong et al. 2020) leverage supervised learning techniques to train models on annotated text data, enabling the models to learn to identify relationships based on context and features of the entities and their interactions. This approach is more robust and adaptable to new domains and relationships, but requires a substantial amount of annotated text data, which can be costly and time-consuming to obtain. Hybrid approaches (Huang et al. 2006; Zhang et al. 2018) combine the advantages of both rule-based and machine learning-based methods by using rule-based methods to pre-process the text and extract candidate relationships, which are then fed to a machine learning model for final classification. This approach can enhance performance and reduce the need for annotated data.
Relation extraction in biomedical applications presents a unique set of challenges compared to traditional NLP tasks. One of the key difficulties is the complexity of the domain-specific medical language, which often includes technical terms, acronyms, and abbreviations that are not found in general English text. Additionally, the relationships between entities in biomedical texts can be highly nuanced, with subtle differences in meaning that require a deep understanding of the biological and medical context. Despite these challenges, relation extraction has a wide range of potential applications in biomedical research, including the discovery of biological pathway (Kim et al. 2018) and associations between genes and diseases (Marchesin and Silvello 2022).
However, another important challenge is that annotated training data for relation extraction in the biomedical domain is limited, making it difficult to train machine learning models to accurately recognize relationships. While a vast amount of works on few-shot learning exist on image data (Tang et al. 2020; Sung et al. 2018), these scenarios in RE are relatively under-studied. Hong et al. (2020) propose a method based on distant supervision that automatically extract biomedical relations from large-scale literature repositories. Li et al. (2017) propose a joint model for named entity recognition and relation extraction based on a CNN for charactel-level representations and BiLSTMs. Chen et al. (2020) introduce transformers as encoding layers of joint models to improve the performance in identifying patients suitable for clinical trials. Li et al. (2018) explores the relatedness among multiple tasks by applying simple multi-task learning approaches.
Despite its advantages, when learning from multiple tasks it is possible that the performance of the resulting model may decrease compared to training a separate model for each task (Alonso and Plank 2017). This can occur because the model may struggle to balance the optimization of multiple tasks, leading to sub-optimal performance on one or more tasks. Additionally, the tasks may have conflicting objectives or requirements, which can result in poor performance on some tasks. Furthermore, the model may over-generalize or over-fit to the training data, making it less effective at making predictions on unseen data. Therefore, it is important to carefully evaluate the trade-off between the potential benefits of multi-task learning and the potential risks to performance before choosing this approach for a given problem. In contrast to prior studies, this work goes beyond the evaluation of multi-task biomedical relation extraction models in few-shot scenarios and provides a comprehensive examination of the inter-task influences, both positive and negative, in our multi-task models.
3 Materials and methods
In this section, we describe data, models and algorithms used to perform our experiments.
3.1 Datasets
The biomedical datasets used in this study are described in this section. We focus on three publicly available multi-class datasets for relation extraction: DDI-2013 (Herrero-Zazo et al. 2013), ChemProt (Kringelum et al. 2016), I2B2-2010 RE (Uzuner et al. 2011). We use the same pre-processing procedure as in Lewis et al. (2020).
3.1.1 DDI-2013
This corpus consists in documents from the DrugBank databaseFootnote 2 and MedLineFootnote 3 abstracts annotated with pharmacological substances and their interactions. It is the first dataset highlighting (1) pharmacodynamic (PD), i.e. the changes in pharmacological effects of a drug caused by the presence of another drug, and (2) pharmacokinectic (PK), which occurs in presence of interference in the intake of one drug (i.e. the distribution or elimination of one drug from another).
The annotated relations are described as follows:
-
Mechanism describes the PK interference mechanism
-
Effect describes the effect of the intake of a drug or the PD mechanism
-
Advice highlights a recommendation or advice which regards interactions between drugs
-
Int indicates a drug–drug interaction without any additional information, explanations or advice
Size of training, development and test sets is: \(\mid {\mathcal {D}}_{train} \mid = 29,334\), \(\mid {\mathcal {D}}_{dev} \mid = 7245\), \(\mid {\mathcal {D}}_{test} \mid = 5762\).
3.1.2 ChemProt
This corpus contains data from open source databases (e.g. CheMBL, BindingDB, PDSP Ki, DrugBank) annotated with chemical compounds, proteins and their interactions. We will consider the following groups of chemical–proteins relations (CPRs) in our study:
-
CPR 3 indicates upregulation relations (activation, promotion, increased activity)
-
CPR 4 indicates downregulation (inhibition, block, decreased activity)
-
CPR 5, CPR 6 are related to interactions of type “agonist” and “antagonist”, respectively.
-
CPR 9 is related to substrate or part of relations. Therefore, this relation does not have particularly relevant features and is thus difficult to extract.
Size of training, development and test sets is: \(\mid {\mathcal {D}}_{train} \mid = 19,461\), \(\mid {\mathcal {D}}_{dev} \mid = 11,821\), \(\mid {\mathcal {D}}_{test} \mid = 16,944\).
3.1.3 I2B2-2010 RE
This corpus focuses on relationships between medical concepts such as tests and treatments. The relation extraction task has 8 classes divided into 3 categories depending on the entities involves. We describes these categories as follows:
-
Medical problem-treatment relations
-
TrIP the treatment improves or cures the medical problem
-
TrWP the treatment worsens the medical problem
-
TrCP the treatment causes the medical problem
-
TrAP the treatment is administered for the medical problem (the result is not mentioned in the sentence)
-
TrNAP the tratment is not provided or is intermittently administered due to the medical problem
-
-
Medical problem-test relations
-
TeRP the test reveals the medical problem
-
TeCP the test is conducted to investigate the medical problem (the sentence does not indicate the result but the reason for the test)
-
-
Medical problem-medical problem relations
-
PIP medical problem indicates medical problem
-
Size of training, development and test sets is: \(\mid {\mathcal {D}}_{train} \mid = 21,385\), \(\mid {\mathcal {D}}_{dev} \mid = 873\), \(\mid {\mathcal {D}}_{test} \mid = 43,001\).
3.2 Method
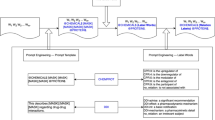
In this section, we outline the methodology employed in our study. Specifically, we utilize a multi-task learning framework, MT-DNN (Liu et al. 2019), on three biomedical datasets for the purpose of Relation Extraction, as detailed in Sect. 3.1. As depicted in Fig. 1, an Encoder based on a transformer architecture is shared among the tasks, and specialized classification heads are fine-tuned for each of the datasets. Subsequently, a knowledge distillation process is employed to enhance performance, as illustrated in Fig. 2: multiple multi-task models are trained, with their predictions constituting the knowledge that is distilled by a single multi-task model.
3.2.1 Multi-task learning architecture: MT-DNN
We use a Multi-Task Deep Neural Network (MT-DNN) (Liu et al. 2019) as the multi-task framework for our experiments. The overall architecture is shown in Fig. 1. The input \(X=\{[CLS], x_2, \dots , x_m\}\) is a word sequence of length m from one of the three analyzed datasets. The Lexicon Encoder maps each token \(x_i\) to its input embedding vector \(l_i\) obtained by summing the corresponding word, segment and positional embeddings. The pre-trained Transformer Encoder maps input embedding vectors into a sequence of contextual embedding vectors thus forming a shared representation across the different tasks. In this work, we use one of the pre-trained models made available by (Lewis et al. 2020) as the backbone of the multi-task framework. Task specific layers are defined as sentence classification models: the first token [CLS] of each sentence X is a semantic representation of the sentence and the probability that X contains a relation between medical entities is predicted by a logistic regression with softmax:
where \({\textbf{W}}^T_t\) is the parameter matrix for the task t.
Overview of the multi-task architecture applied to our study. The Lexicon Encoder and Transformer Encoder are shared across the different tasks and maps the input first to a sequence of embedding vectors (one for each token) and then to shared contextual embedding vectors which take count of contextual information. A task-specific layer is then used for each dataset to generate dataset-specific representations
3.2.2 Knowledge distillation
The knowledge distillation (KD) method has been successfully used with multi-task learning to enjoy the advantages of ensemble learning while not needing to keep the entire ensemble of models but just one single model (Liu et al. 2019), our KD methodology is shown in Fig. 2: we start by training three MT-DNN networks with three dropout values \(p=\{0.1, 0.15, 0.2\}\) and each of them is then used as the backbone for a single-task network fine-tuned on each task dataset. Soft labels produced by teachers for each training example are then averaged to produce the dark knowledge to be distilled. We studied the effects of two types of KD loss: (1) Mean Squared Error (MSE) and (2) a hybrid loss based on Kullback Leibler divergence. MSE minimizes the mean squared discrepancy between the soft labels of the teacher and values estimated by the student network:
The hybrid loss is based on two contribution: the first is given by the Kullback Leibler loss which minimizes the divergence between two probability distributions, i.e. the soft labels of the teacher and the predictions of the student: the second contribution assumes that the teacher is not perfect and thus takes into account the ground truth by means of the cross-entropy loss:
where \({\mathcal {L}}_{CE}(y, {\hat{y}})\) denotes the cross-entropy loss, \(y=y_\tau ^i\) being the ground truth label for the i-th sample at time step \(\tau\) and \({\hat{y}}=f_\tau (x_\tau ^i, \theta )\) representing the predicted output for the i-th sample at time step \(\tau\), given the model parameters \(\theta\); \({\mathcal {L}}_{KL}\) denotes the Kullback–Leibler divergence between the output probability distribution from the model with parameters \(\theta\) and the teacher with parameters \(\theta _T\); the parameter \(\lambda\) controls the weighting of the contribution of the knowledge distillation and ensures that the student also learns from the actual ground truth.
Overview of the knowledge distillation process applied in our study. First, MT-DNN networks are trained with different dropout values \(p=\{0.1, 0.15, 0.2\}\). Each MT-DNN network is then fine-tuned on each dataset and all the soft-labels produced by teachers are averaged to produce the dark knowledge to be distilled. A single MT-DNN student is trained with a knowledge distillation loss which takes count of the knowledge acquired by teachers
4 Experiments
Our analysis will be focused on answering the questions reported as follows.
-
Q1: Comparison with few-shot baselines. How does few-shot MT-DNN perform as compared to few-shot learning baselines? We use three few-shot learning baselines to perform a comparison with the multi-task architecture leveraged in this work: a Siamese network (Koch et al. 2015), ProtoNET Snell et al. (2017), BioBERT Lee et al. (2020), ClinicalBERT Alsentzer et al. (2019) and PET Schick and Schütze (2021).
-
Q2: Effects of multi-task learning. Can it improve the performance w.r.t. single-task models? We select one of the publicly available biomedical pre-trained transformer architectures as the base for our multi-task MT-DNN model, which is then enhanced with Knowledge Distillation and compared with single-task performance over the entire training-sets. Furthermore, we study how knowledge distillation impacts the overall performance by analyzing the effects of different values assigned to the loss weight \(\lambda\)
-
Q3: Tasks influence analysis. What are the main influencing factors in multi-task learning? Different datasets can have a different impact over the multi-task performance. We will analyze similarities and differences between datasets to understand their effects on positive and negative transfer when training the multi-task model. On the basis of the above, we will analyze the mutual influence between different tasks by pairwise training, i.e. selectively excluding datasets from the training procedure to analyze their overall effects over the multi-task performance.
-
Q4: Few-shot scenarios. How does the performance vary in few-shot scenarios? We are interested in the understanding of the value of multi-task learning when only a few set of data is available for each dataset, and how its effects vary when the training dataset increases. More in detail, we will train our multi-task models by simulating few-shot scenarios in where only k training examples are available for each dataset (with k varying from 1 to 1000) and we will test their performance over the entire test set.
4.1 Training parameters
In this section we report the training parameters used in our experiments. We fixed the input sequence length to 512 and the batch size to 8. We used the training parameters suggested in Liu et al. (2019) for both the multi-task and single-task experiments. In particular, we conducted experiments by setting various hyperparameters such as learning rate, weight decay and optimizer using an initial random search and subsequently performing a greedy search focusing on the neighborhood of the default values on a subset of the training data, as commonly done in the literature. These preliminary experiments confirmed the suggested parameter values. Thus, we used an Adamax optimizer with learning rate set to 5e−5 and weight decay to 0.01 with adam eps to 1e−7. To avoid gradient explosion, the grad clipping parameter is set to 1.0. Additionally, we provide an empirical study on the value of the loss weighting parameter \(\lambda\) used in the knowledge distillation process.
When the training procedure involves the entire training dataset or at least 1000 examples, we set the number of epochs to 10 (in both the single-task and multi-task cases), while we set it to 20 in 1-, 10-, 50- and 100-shot scenarios.
The loss functions vary according to the type of approach: in single-task and simple multi-task learning, we use the cross-entropy loss; when using knowledge distillation, we experimented with MSE and a hybrid loss formed by cross-entropy and Kullback Leibler divergence.
The training parameters used for few-shot baselines are reported as follows:
-
Siamese Network (Koch et al. 2015): we use GloVe embeddings (embedding size = 100)
-
ProtoNET (Snell et al. 2017): learning rate is set to 1e−5, Euclidean loss is used and the support set varies depending on the number of shots. In 1-shot training, a support set equal to 1 is necessarily chosen; in 10-shot training we select a support set equal to 5 and this value remains the same in all the other scenarios due to RAM availability constraints.
-
BioBERT (Lee et al. 2020) and ClinicalBERT (Alsentzer et al. 2019): same parameters used to train our multi-task networks.
-
PET (Schick and Schütze 2021): 5 epochs with 250 steps, learning rate set to 1e−4, batch size to 8, weight decay to 0.01. Furthermore, we initialize the weights of the transformer architecture with the biomedical checkpoint publicly made available in Lewis et al. (2020), which is the same we use for our MT-DNN models.
Note that the number of epochs and the learning rate were selected based on the complexity of the model and the amount of data available, and were determined through appropriate tuning to avoid overfitting, obtaining the best possible model on the validation set. It was observed that as the amount of data increased in few-shot tasks, fewer training epochs were required. To maintain fairness in comparing the results between the different tasks, common evaluation metrics such as F1, recall, and precision were used. The dependence on the number of shots and the initialization of the various networks was mitigated by sampling with 5 different seeds for each shot of training for each task, and initializing the network with these seeds during different trainings. This helps to increase the reliability and generalizability of the results and ensure a fair comparison between the different tasks.
4.2 Results
4.2.1 Q1: Comparison with few-shot baselines
Tables 1, 2 and 3 report the comparison between our framework and state-of-the-art baselines in terms of precision, recall and F1 scores, respectively.
The results presented in Table 1 indicate that ProtoNET yields the highest precision in scenarios with extremely limited training data (1-shot and 10-shot). This method is based on a prototypical network that emphasizes on the representation of each relation type and the calculation of prototypes for each relation type, which enhances precision in relation identification when the training samples are relevant. However, when a slightly larger number of training samples are available, the multi-task learning approach demonstrates superior performance. This is due to the information shared among the three relation extraction tasks and the increased robustness and generalization capability of the model resulting from the larger number of training samples.
Despite its precision in identifying relations, ProtoNET exhibits a low recall as evidenced by the results presented in Table 2. The utilization of language models pre-trained with biomedical data as BioBERT and ClinicalBERT, the implementation of prompts in PET, which effectively leverages the knowledge gained by language models, and multi-task approaches that incorporate information from additional tasks may enhance recall and thus make these approaches more suitable for identifying a greater number of relevant relationships. Among these methods, our multi-task learning approach guarantees the highest results in terms of recall scores.
To sum up, our approach consistently produced the best results in 50-shot contexts with regard to precision, recall, and F1. In 10-shot contexts, our approach still achieved the best F1, as shown in Table 3, although precision was comparable or slightly lower compared to other baselines. However, our approach excelled in terms of recall, significantly outperforming other methods. This is attributed to the use of data from other tasks, which allowed us to identify a larger number of relevant relationships.
4.2.2 Q2: Effects of multi-task learning
The results of utilizing MT-DNN and its extension through knowledge distillation are presented in Table 4. It is evident from the table that multi-task learning provides a significant improvement for the inference task on the ChemProt and I2B2-2010 datasets. However, it results in a decrease in performance when applied to the DDI-2013 dataset. The application of knowledge distillation is advantageous for all downstream tasks but fails to outperform the single-task performance on the DDI-2013 dataset. This phenomenon, referred to as negative transfer, will be thoroughly analyzed in research question Q3.
Furthermore, we analyzed the impact of knowledge distillation on the overall performance. In particular, we have performed hyper-parameter tuning on the weighting parameter \(\lambda\) which controls the contribution of ground truth to the knowlege distillation loss as in Eq. 3. Specifically, the tuning was conducted using shots 1, 10, and 50, while fixing the network initialization and shot extraction seeds to be the same across experiments with different \(\lambda\) values. The parameters used in these experiments are the same as those used in our multi-task few-shot experiments. The \(\lambda\) values used for tuning are: 0, 0.2, 0.4, 0.6, 0.8, and 1. Results in Fig. 3 show that the best F1 score is achieved with \(\lambda\) values that imply considering both the ground truth and teachers. In particular, the optimal value obtained in every few-shot scenario and with all the datasets—with the only exception of DDI-2013 (10-shot)—is \(\lambda = 0.4\), slightly biased towards the teacher’s additional knowledge. Hence, the student network can learn from the teacher how to capture more subtle and complex patterns in the data such as uncertainties and correlations between different classes and the nuances and complexities of the language. However, results degrade when the student network relies too heavily on the teachers’ predictions.
4.2.3 Q3: Tasks influence analysis
We first analyze the three tasks based on their similarities, and then study their mutual influence and effects in the multi-task learning framework used.
Differences in syntax Initially, a vocabulary was derived from each dataset that encompasses the occurring words. The number of shared words between the tasks is depicted in the pie chart of Fig. 4. It can be observed that the tasks of DDI-2013 and ChemProt exhibit the highest number of shared words, which is 42.9% of the total vocabulary. Conversely, the words in the I2B2-2010 dataset are distinct from those in the other two datasets, with a similarity of 30.8% and 26.3% compared to ChemProt and DDI-2013, respectively.
In Fig. 5, the distributions of sentence lengths are presented, where the sentences are represented as a sequence of words. It is evident that, despite the similarities in median values across the various tasks, DDI-2013 exhibits a substantial quantity of lengthy sentences, with approximately 1000 instances surpassing 600 words. Conversely, sentences in I2B2-2010 tend to be comparatively shorter in comparison to those in other tasks.
Differences in semantics The semantic similarity between various tasks was determined by computing the similarity between sentence embeddings generated with SentenceBERT (Reimers and Gurevych 2019). This was achieved by utilizing BlueBERT (Peng et al. 2019) as the primary encoder. The method involved calculating the cosine similarity score between each sentence from each dataset and all the examples in each dataset, and then averaging the scores to obtain the similarity score between the target sentence and the three datasets. To obtain the similarity scores between datasets \(D_1\) and \(D_2\), the average similarity scores between sentences \(s\in D_1\) and dataset \(D_2\) were calculated.
The results presented in Fig. 6 indicate that I2B2-2010 is the most heterogeneous dataset, as evidenced by the low similarity score with itself. This is likely due to the fact that the data was collected from eight distinct hospitals. Conversely, ChemProt and DDI-2013 demonstrate a high degree of semantic similarity to each other.
We are interested in understanding the impact of semantic similarity and dissimilarity on performance when considering pairs of tasks. This investigation was conducted through the use of pairwise training (Standley et al. 2020). The results presented in Table 5 show the scores obtained when multi-task training was performed with the task indexed in the row and the task indexed in the column (single-task performance is reported on the diagonal). The table reveals that while the performance of the other tasks is improved through multi-task training, DDI-2013 experiences a negative transfer, probably due to the absence of long sentences in other datasets, resulting in a decrease in performance compared to the single-task scenario. Additionally, the contributions made by DDI-2013 to the performance improvement of the other tasks are generally inferior compared to those made by the other tasks. On the other hand, the I2B2-2010 task, despite its inherent high variability, benefits the most from multi-task training.
4.2.4 Q4: Few-shot scenarios
We examined the impact of multi-task learning on performance in scenarios with varying degrees of data scarcity. To accomplish this, we measured the performance of multi-task models as the number of samples (k) increased (\(k \in {1, 10, 50, 100, 1000}\)), and the results are presented in Fig. 7 in terms of precision, recall, and F1 scores. In contrast to the results obtained in the pairwise experiments as described in Question Q3, we observed a generally positive transfer in performance. Specifically, while the DDI-2013 dataset experienced negative transfer when utilizing the complete training data, we noted a benefit from multi-task learning in low-resource scenarios for all datasets, with relative improvements ranging from 18.3 to 32.4% in F1 scores.
Few-shot comparison between single-task and multi-task networks. Performance on the three datasets under analysis (rows) is reported in terms of precision (first column), recall (second column) and F1 (third column). The improvement percentage of the multi-task network w.r.t. the single task network is reported for each k-shot setting
Furthermore, the improvement percentage typically increased as the amount of training data decreased, reaching a maximum of 77.4% in F1 scores on the ChemProt data in the 1-shot scenario. This aligns with previous research (Worsham and Kalita 2020; Standley et al. 2020) that emphasizes the potential benefits of multi-task learning in few-shot learning contexts. Although the improvement in precision scores either remained constant or increased as the number of samples increased, there was a notable decrease in recall scores. This suggests that the advantage of multi-task learning in the few-shot scenarios investigated is mainly due to the improved ability of the trained model to differentiate between true positives and false negatives.
Pair-wise experiment in few-shot scenarios. For each dataset (rows), multi-task performance obtained with by using all the dataset is compared with multi-task performance obtained by using only one other dataset. Performance is reported in terms of precision (first column), recall (second column) and F1 (third column)
We conducted the pairwise experiment in few-shot learning scenarios to gain a deeper understanding of positive and negative transfer in few-shot scenarios. The results displayed in Fig. 8 demonstrate that models trained in a pairwise manner have comparable scores to the multi-task models examined in Fig. 7. The small differences across the pairwise results can be only observed in recall scores, where we can observe small decreases in performance when pairing ChemProt with other datasets. Additionally, the performance of the pairwise models is consistently higher than that of single-task models.
5 Conclusion and future work
In this study, we propose a novel framework for few-shot biomedical relation extraction, which is based on a transformer-based network and multi-task learning method (Liu et al. 2019). Our approach uses a shared layer across biomedical RE tasks and trains a classification head for each task separately. To enhance the model’s performance, we adopt a training framework based on knowledge distillation.
Our evaluation of the factors contributing to positive and negative transfer in biomedical relation extraction demonstrates that our framework achieves positive transfer in all low-resource scenarios, where labeled data is limited for the primary task. Moreover, our approach surpasses state-of-the-art few-shot learning baselines in most tasks and scenarios, especially in recall scores, reaching up to 84% with only 50 training samples. This suggests that our system correctly identifies a large portion of true positive relations in the data.
However, our method’s low precision scores indicate that there is still room for improvement, especially in applications where high-stakes decision-making is involved. To enhance the precision of multi-task models, we suggest incorporating additional features such as dictionaries and medical ontologies, which provide a structured vocabulary and semantic rules for relation identification.
Lastly, it is important to note that our assessment of the system’s performance was based on publicly available data, which may not accurately depict its performance on real-world clinical data. Therefore, further investigations are necessary to examine the system’s performance with real-world clinical data and determine its practical applicability.
References
Afradi A, Ebrahimabadi A (2020) Comparison of artificial neural networks (ANN), support vector machine (SVM) and gene expression programming (gep) approaches for predicting tbm penetration rate. SN Appl Sci 2:1–16
Afradi A, Ebrahimabadi A (2021) Prediction of TBM penetration rate using the imperialist competitive algorithm (ICA) and quantum fuzzy logic. Innov Infrastruct Solut 6(2):103
Afradi A, Ebrahimabadi A, Hallajian T (2020) Prediction of tunnel boring machine penetration rate using ant colony optimization, bee colony optimization and the particle swarm optimization, case study: Sabzkooh water conveyance tunnel. Mining Miner Depos 14(2):75–84
Afradi A, Ebrahimabadi A, Hallajian T (2021) Prediction of TBM penetration rate using fuzzy logic, particle swarm optimization and harmony search algorithm. Geotech Geol Eng 8:1–24
Alimova I, Tutubalina E (2020) Multiple features for clinical relation extraction: a machine learning approach. J Biomed Inform 103:103382. https://doi.org/10.1016/j.jbi.2020.103382
Alonso HM, Plank B (2017) When is multitask learning effective? semantic sequence prediction under varying data conditions. In: Lapata M, Blunsom P, Koller A (eds) Proceedings of the 15th conference of the european chapter of the association for computational linguistics, EACL 2017, Valencia, Spain, April 3–7, 2017, vol 1: Long Papers, pp 44–53. Association for Computational Linguistics. https://doi.org/10.18653/v1/e17-1005
Alsentzer E, Murphy J, Boag W, Weng W-H, Jindi D, Naumann T, McDermott M (2019) Publicly available clinical BERT embeddings. In: Proceedings of the 2nd clinical natural language processing workshop, pp 72–78. Association for Computational Linguistics, Minneapolis, Minnesota, USA. https://doi.org/10.18653/v1/W19-1909
Ben Abacha A, Zweigenbaum P (2011) Automatic extraction of semantic relations between medical entities: a rule based approach. J Biomed Semant 2(5):1–11. https://doi.org/10.1186/2041-1480-2-S5-S4
Caruana R (1998) Multitask learning. In: Thrun S, Pratt LY (eds) Learning to learn. Springer, New York, pp 95–133
Chen M, Lan G, Du F, Lobanov VS (2020) Joint learning with pre-trained transformer on named entity recognition and relation extraction tasks for clinical analytics. In: Rumshisky A, Roberts K, Bethard S, Naumann T (eds) Proceedings of the 3rd clinical natural language processing workshop, ClinicalNLP@EMNLP 2020, Online, November 19, 2020, pp. 234–242. Association for Computational Linguistics, Online. https://doi.org/10.18653/v1/2020.clinicalnlp-1.26
Gharehchopogh FS, Khalifehlou Z (2012) Study on information extraction methods from text mining and natural language processing perspectives. AWER Proc Inf Technol Comput Sci 1:1321–1327
Gurulingappa H, Rajput AM, Roberts A, Fluck J, Hofmann-Apitius M, Toldo L (2012) Development of a benchmark corpus to support the automatic extraction of drug-related adverse effects from medical case reports. J Biomed Inform 45(5):885–892. https://doi.org/10.1016/j.jbi.2012.04.008
Herrero-Zazo M, Segura-Bedmar I, Martínez P, Declerck T (2013) The DDI corpus: an annotated corpus with pharmacological substances and drug–drug interactions. J Biomed Inform 46(5):914–920. https://doi.org/10.1016/j.jbi.2013.07.011
Hong L, Lin J, Li S, Wan F, Yang H, Jiang T, Zhao D, Zeng J (2020) A novel machine learning framework for automated biomedical relation extraction from large-scale literature repositories. Nat Mach Intell 2(6):347–355. https://doi.org/10.1038/s42256-020-0189-y
Hosseinalipour A, Gharehchopogh FS, Masdari M, Khademi A (2021) Toward text psychology analysis using social spider optimization algorithm. Concurr Comput 33(17):6325. https://doi.org/10.1002/cpe.6325
Hosseinalipour A, Gharehchopogh FS, Masdari M, Khademi A (2021) A novel binary farmland fertility algorithm for feature selection in analysis of the text psychology. Appl Intell 51(7):4824–4859. https://doi.org/10.1007/s10489-020-02038-y
Huang M, Zhu X, Li M (2006) A hybrid method for relation extraction from biomedical literature. Int J Med Inf 75(6):443–455. https://doi.org/10.1016/j.ijmedinf.2005.06.010
Khataei Maragheh H, Gharehchopogh FS, Majidzadeh K, Sangar AB (2022) A new hybrid based on long short-term memory network with spotted hyena optimization algorithm for multi-label text classification. Mathematics 10(3):488. https://doi.org/10.3390/math10030488
Kim M, Baek SH, Song M (2018) Relation extraction for biological pathway construction using node2vec. BMC Bioinform 19(8):75–84. https://doi.org/10.1186/s12859-018-2200-8
Koch G, Zemel R, Salakhutdinov R, et al (2015) Siamese neural networks for one-shot image recognition. In: ICML Deep Learning Workshop, vol 2, p.0.Lille. https://www.cs.cmu.edu/rsalakhu/papers/oneshot1.pdf
Kringelum J, Kjærulff SK, Brunak S, Lund O, Oprea TI, Taboureau O (2016) Chemprot-3.0: a global chemical biology diseases mapping. Database J Biol Databases Curation. https://doi.org/10.1093/database/bav123
Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, Kang J (2020) Biobert: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 36(4):1234–1240. https://doi.org/10.1093/bioinformatics/btz682
Lee C, Hwang Y, Jang M (2007) Fine-grained named entity recognition and relation extraction for question answering. In: Kraaij W, de Vries AP, Clarke CLA, Fuhr N, Kando N (eds) SIGIR 2007: Proceedings of the 30th annual international ACM SIGIR conference on research and development in information retrieval, Amsterdam, The Netherlands, July 23–27, 2007, pp 799–800. ACM. https://doi.org/10.1145/1277741.1277915
Lewis P, Ott M, Du J, Stoyanov V (2020) Pretrained language models for biomedical and clinical tasks: Understanding and extending the state-of-the-art. In: Proceedings of the 3rd clinical natural language processing workshop, pp 146–157. Association for Computational Linguistics. https://doi.org/10.18653/v1/2020.clinicalnlp-1.17
Li F, Zhang M, Fu G, Ji D (2017) A neural joint model for entity and relation extraction from biomedical text. BMC Bioinform 18(1):198–119811. https://doi.org/10.1186/s12859-017-1609-9
Li Q, Yang Z, Luo L, Wang L, Zhang Y, Lin H, Wang J, Yang L, Xu K, Zhang Y (2018) A multi-task learning based approach to biomedical entity relation extraction. In: Zheng HJ, Callejas Z, Griol D, Wang H, Hu X, Schmidt HHHW, Baumbach J, Dickerson J, Zhang L (eds) IEEE international conference on bioinformatics and biomedicine, BIBM 2018, Madrid, Spain, December 3–6, 2018, pp 680–682. IEEE Computer Society. https://doi.org/10.1109/BIBM.2018.8621284
Li C, Li S, Wang H, Gu F, Ball AD (2023) Attention-based deep meta-transfer learning for few-shot fine-grained fault diagnosis. Knowl-Based Syst. https://doi.org/10.1016/j.knosys.2023.110345
Liu X, He P, Chen W, Gao J (2019) Improving multi-task deep neural networks via knowledge distillation for natural language understanding. CoRR arXiv:1904.09482
Liu X, He P, Chen W, Gao J (2019) Multi-task deep neural networks for natural language understanding. Association for Computational Linguistics, Florence, Italy. https://doi.org/10.18653/v1/P19-1441
Liu X, He P, Chen W, Gao J (2019) Multi-task deep neural networks for natural language understanding. In: Proceedings of the 57th annual meeting of the association for computational linguistics, pp. 4487–4496. Association for Computational Linguistics, Florence, Italy. https://www.aclweb.org/anthology/P19-1441
Mahalleh ER, Gharehchopogh FS (2022) An automatic text summarization based on valuable sentences selection. Int J Inf Technol 14(6):2963–2969. https://doi.org/10.1007/s41870-022-01049-x
Marchesin S, Silvello G (2022) TBGA: a large-scale gene-disease association dataset for biomedical relation extraction. BMC Bioinform 23(1):111. https://doi.org/10.1186/s12859-022-04646-6
Nebhi K (2013) A rule-based relation extraction system using dbpedia and syntactic parsing. In: Hellmann S, Filipowska A, Barrière C, Mendes PN, Kontokostas D (eds) Proceedings of the NLP & dbpedia workshop co-located with the 12th international semantic web conference (ISWC 2013), Sydney, Australia, October 22, 2013. CEUR Workshop Proceedings, vol 1064. CEUR-WS.org, Online. http://ceur-ws.org/Vol-1064/Nebhi_Rule-Based.pdf
Peng Y, Yan S, Lu Z (2019) Transfer learning in biomedical natural language processing: An evaluation of BERT and elmo on ten benchmarking datasets. In: Demner-Fushman, D., Cohen, K.B., Ananiadou, S., Tsujii, J. (eds.) Proceedings of the 18th BioNLP Workshop and Shared Task, BioNLP@ACL 2019, Florence, Italy, August 1, 2019, pp. 58–65. Association for Computational Linguistics, Online. https://doi.org/10.18653/v1/w19-5006
Peng Y, Chen Q, Lu Z (2020) An empirical study of multi-task learning on BERT for biomedical text mining. In: Proceedings of the 19th SIGBioMed workshop on biomedical language processing. Association for Computational Linguistics. pp 205–214. https://doi.org/10.18653/v1/2020.bionlp-1.22
Pyysalo S, Ginter F, Heimonen J, Björne J, Boberg J, Järvinen J, Salakoski T (2006) Bioinfer: a corpus for information extraction in the biomedical domain. BMC Bioinform 8:50–50. https://doi.org/10.1186/1471-2105-8-50
Reimers N, Gurevych I (2019) Sentence-bert: Sentence embeddings using siamese bert-networks. In: Inui, K., Jiang, J., Ng, V., Wan, X. (eds.) Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing, EMNLP-IJCNLP 2019, Hong Kong, China, November 3-7, 2019, pp. 3980–3990. Association for Computational Linguistics, Online. https://doi.org/10.18653/v1/D19-1410
Schick T, Schütze H (2021) Exploiting cloze-questions for few-shot text classification and natural language inference. In: Merlo, P., Tiedemann, J., Tsarfaty, R. (eds.) Proceedings of the 16th Conference of the European Chapter of the Association for Computational Linguistics: Main Volume, EACL 2021, Online, April 19 - 23, 2021, pp. 255–269. Association for Computational Linguistics, Online. https://doi.org/10.18653/v1/2021.eacl-main.20
Snell J, Swersky K, Zemel RS (2017) Prototypical networks for few-shot learning. In: Guyon I, von Luxburg U, Bengio S, Wallach HM, Fergus R, Vishwanathan SVN, Garnett R (eds) Advances in neural information processing systems 30: annual conference on neural information processing systems 2017, December 4–9, 2017, Long Beach, CA, USA, pp 4077–4087. https://proceedings.neurips.cc/paper/2017/hash/cb8da6767461f2812ae4290eac7cbc42-Abstract.html
Standley T, Zamir A, Chen D, Guibas LJ, Malik J, Savarese S (2020) Which tasks should be learned together in multi-task learning? In: Proceedings of the 37th International Conference on Machine Learning, ICML 2020, 13-18 July 2020, Virtual Event. Proceedings of Machine Learning Research, vol. 119, pp. 9120–9132. PMLR, Online. http://proceedings.mlr.press/v119/standley20a.html
Sung F, Yang Y, Zhang L, Xiang T, Torr PHS, Hospedales TM (2018) Learning to compare: relation network for few-shot learning. In: 2018 IEEE conference on computer vision and pattern recognition, CVPR 2018, Salt Lake City, UT, USA, June 18–22, 2018. Computer vision foundation/IEEE Computer Society. pp 1199–1208 https://doi.org/10.1109/CVPR.2018.00131
Tang H, Li Z, Peng Z, Tang J (2020) Blockmix: meta regularization and self-calibrated inference for metric-based meta-learning. In: Chen CW, Cucchiara R, Hua X, Qi G, Ricci E, Zhang Z, Zimmermann R (eds) MM ’20: The 28th ACM international conference on multimedia, virtual event/Seattle, WA, USA, October 12–16, 2020, pp. 610–618. ACM. https://doi.org/10.1145/3394171.3413884
Uzuner Ö, South BR, Shen S, DuVall SL (2011) 2010 i2b2/va challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 18(5):552–556. https://doi.org/10.1136/amiajnl-2011-000203
Wang Q, Mao Z, Wang B, Guo L (2017) Knowledge graph embedding: a survey of approaches and applications. IEEE Trans Knowl Data Eng 29(12):2724–2743. https://doi.org/10.1109/TKDE.2017.2754499
Worsham J, Kalita J (2020) Multi-task learning for natural language processing in the 2020s: Where are we going? Pattern Recognit. Lett. 136:120–126. https://doi.org/10.1016/j.patrec.2020.05.031
Zhang Y, Lin H, Yang Z, Wang J, Zhang S, Sun Y, Yang L (2018) A hybrid model based on neural networks for biomedical relation extraction. J Biomed Inform 81:83–92. https://doi.org/10.1016/j.jbi.2018.03.011
Acknowledgements
We acknowledge financial support from the PNRR MUR project PE0000013-FAIR.
Funding
Open access funding provided by Università degli Studi di Napoli Federico II within the CRUI-CARE Agreement.
Author information
Authors and Affiliations
Contributions
Conceptualization: VM, GN, MP and GS Methodology: VM, GN, MP and GS Formal analysis: VM, GN, MP and GS Writing—Original Draft: VM, GN, MP and GS.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Vincenzo Moscato, Giuseppe Napolano, Marco Postiglione and Giancarlo Sperlì have contributed equally to this work.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Moscato, V., Napolano, G., Postiglione, M. et al. Multi-task learning for few-shot biomedical relation extraction. Artif Intell Rev 56, 13743–13763 (2023). https://doi.org/10.1007/s10462-023-10484-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10462-023-10484-6