Abstract
Our paper presents a novel approach to pattern classification. The general disadvantage of a traditional classifier is in too different behaviour and optimal parameter settings during training on a given pattern set and the following cross-validation. We describe the term critical sensitivity, which means the lowest reached sensitivity for an individual class. This approach ensures a uniform classification quality for individual class classification. Therefore, it prevents outlier classes with terrible results. We focus on the evaluation of critical sensitivity, as a quality criterion. Our proposed classifier eliminates this disadvantage in many cases. Our aim is to present that easily formed hidden classes can significantly contribute to improving the quality of a classifier. Therefore, we decided to propose classifier will have a relatively simple structure. The proposed classifier structure consists of three layers. The first is linear, used for dimensionality reduction. The second layer serves for clustering and forms hidden classes. The third one is the output layer for optimal cluster unioning. For verification of the proposed system results, we use standard datasets. Cross-validation performed on standard datasets showed that our critical sensitivity-based classifier provides comparable sensitivity to reference classifiers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
There exist a lot of different classifiers designed for classification from n-dimensional space. These classifiers are usually used for classification in two or more classes and are based on various principles. For example, linear combination (Orozco-Alzate et al. 2019), use of mutual distances (Shahid and Singh 2019; Liu and Wang 2022; Veneri et al. 2022), variants of gradient boosting (Bentejac et al. 2021; Liang and Sur 2022), adaboost (Hu et al. 2020), and bagging (Medina-Pérez et al. 2017; Jafarzadeh et al. 2021).
Our new approach is based on an assumption that for data with a complex structure, it is beneficial to divide the data into an unspecified number of sub-groups. These artificial groups, so-called hidden classes, can be formed easily using unsupervised or supervised learning techniques. We can use small low-level classifiers with limited validity and focus on data belonging to these classes. The way how we form hidden classes is not crucial, but they could be helpful for the final classification, which will be improved subsequently by their optimal unification. Such an optimal union leads to better classification results than using only the original datasets. This idea is the cornerstone of the proposed classifier. Furthermore, it is necessary to emphasize that we focus on the sensitivity of each, individual class, and not omit any of them.
We propose using simple clustering methods resulting in more clusters than the final classes, which will not be many or few. After forming such clusters, we propose a union of them with the optimal union method (Hrebik et al. 2019). Such unioning enables the formation of a classifier with the highest possible critical sensitivity. Our aim is to introduce a novel classifier suitable for processing data with a higher dimension, so dimension reduction must also be part of our classifier. Dimensionality reduction can be optional, but in our opinion, it generally increases the efficiency of the system and our recommendation is data whitening, thanks to which we get a dimensionless description.
A separate part of our paper considers meaning of critical sensitivity and why we propagate this criterion. The basic idea is that in the case of classification into classes, we have a separately evaluated percentage of correctly classified patterns for each class, which we refer to as the sensitivity of the relevant class. However, most classifiers maximize their accuracy on all data, which, of course, represents an unbalanced evaluation. We focus on the class providing worst results, and this sensitivity we note as critical sensitivity. Even simple clustering methods, such as DBSCAN, can provide high-quality clusters, which by unification will create a classifier in which even the worst class will have sufficient level sensitivity. Therefore, even in the case of an unbalanced size of classes, using our proposed classifier, we do not omit even the smallest one because of a low number of patterns. Our proposed method focuses on generating hidden classes from current datasets. Some authors suggest to generate synthetic samples (Rekha and Madhu 2022).
In this section, we provide a brief basic summary of the current state of research. Pattern recognition (Duda et al. 2012) represents a common aim of artificial intelligence and machine learning. In the case of machine learning, we distinguish between unsupervised and supervised approaches. The typical clustering models (Karlsson 2010) are connectivity, centroid, distribution, density, subspace, graph-based, and neural network ones (Back et al. 2018; Shi et al. 2019; Lin et al. 2020). A well-known unsupervised neural network model is represented by self-organizing maps (SOM). We can include subspace models as Principal Component Analysis or Independent Component Analysis as data processing techniques for dimension reduction (Eldar and Oppenheim 2003; Jolliffe 2011; Nguyen and Holmes 2019) of an original dataset consisting of a large number of interrelated variables. Dimensionality reduction leads to data representation using fewer features. Another approach to dimension reduction represent linear discriminant analysis into two classes (Eldar and Oppenheim 2003; Croux et al. 2008) or more classes (Rao and Toutenburg 1995).
Our proposed approach consists of three steps:
-
Dimensionality reduction and standardization,
-
Data clustering in the space of reduced dimension,
-
Hidden class forming and their optimal union to desired output classes.
Dimensionality reduction is based on data whitening (Eldar and Oppenheim 2003) or multi-class discriminant analysis, as an alternative, in the first step. The following clustering is performed by parameter-driven DBSCAN (Ester et al. 1996) generating several classes, their structure, and outliers. Proposed clustering technique is quick and can be reduced to the SLINK (Sibson 1973) algorithm in many cases. In the last step, we define hidden classes as clusters from the previous step. The relationship between these hidden classes and output classes learned is optimized using a binary programming technique that is focused on the maximization of classifier critical sensitivity.
We summarize several principles of multi-classifiers in the second section focusing on data whitening, multiple discriminant analysis, data clustering, and the concept of hidden classes. We discuss the framework and structure of a novel multi-classifier in the third section. The numerical experiments on basic pattern sets and resulting optimal settings and quality measures are summarized in the next section. The last section concludes our proposed classifier.
2 Multi-classification preliminaries
Basic facts related to the classification into several classes are summarized in this section. The optional methods of preprocessing are discussed first as unsupervised and supervised approaches. Data clustering is then discussed as a method of how to generate hidden classes. Finally, the concept of unioning of hidden classes is presented as a kernel of the novel approach.
2.1 Multi-classification Task
Basic frame of classification of vector patterns (Duda et al. 2012) into several classes is established first. Let \(n, m, N \in {\mathbb {N}}\) be number of features, patterns, and classes satisfying \(N \ge 2\). Let \({\textbf{x}} \in {\mathbb {R}}^n\) be the feature vector and \(y,y^* \in \lbrace 1,..., N \rbrace\) be a classifier output and its required value, denoting a pattern as \({\textbf{p}} = ({\textbf{x}}, y^*)\), the classifier is defined as a function
and the classifier response is therefore \(y = \textrm{c}({\textbf{x}})\). Denoting \({\textbf{x}}_k \in {\mathbb {R}}^n\), \(y_k^* \in \lbrace 1,..., N \rbrace\) as a feature vector and given output of k-th pattern, we define a pattern set as
The pattern set can be represented by any input matrix \({\textbf{X}} \in {\mathbb {R}}^{m \times n}\) and any output vector \({\textbf{y}}^* \in \lbrace 1,..., N \rbrace ^m\). Any classifier is a complex system that applies various data processing techniques to obtain the final decision. Selected approaches are summarised in the following subsections.
2.2 Data whitening
The first but optional step of any classification is an efficient transformation that decreases the number of features but saves information about pattern differences. The main idea of principal component analysis (PCA) (Jolliffe 2011) is to reduce the dimensionality of the original dataset consisting of a large number of interrelated variables. The reduction retains as much as possible of the variation present in the data set. The aim is achieved by transforming into a new set of variables called principal components. These components are uncorrelated and ordered so that the first few retain most of the variation present in all of the original variables.
Let \(D \in {\mathbb {N}}\) be reduced dimension satisfying \(D<n\). The dimensionality reduction from \({\mathbb {R}}^n\) to \({\mathbb {R}}^D\) using PCA is based on a linear transformation
The PCA is designed to satisfy \(\textrm{E} \,{\textbf{z}} = {\textbf{0}}\) and \(\textrm{var} \,\,{\textbf{z}} = {\textbf{D}}\), where \({\textbf{D}}\) is a diagonal matrix. Resulting parameters of PCA are \({\textbf{W}}_1 \in {\mathbb {R}}^{n \times D}\) and
The transforming matrix \({\textbf{W}}_1\) is calculated as follows. First, we shift the input matrix to obtain \({\textbf{X}}_{\textrm{S}} = {\textbf{X}} - {\textbf{1}}_m {\textbf{x}}_0^{\textrm{T}}\), where \({\textbf{1}}_m\) is m-dimensional vector of units. Then we calculate a covariance matrix \({\textbf{A}} = {\textbf{X}}_{\textrm{S}}^{\textrm{T}} {\textbf{X}}_{\textrm{S}} \ge 0\) and apply Eigen-Value Decomposition (EVD) as finding of eigenvalues \({\textbf{v}} \in {\mathbb {R}}^n\) and eigenvectors \(\lambda \ge 0\) in equation \(({\textbf{A}} - \lambda {\textbf{I}}_n){\textbf{v}} = {\textbf{0}}\) where \({\textbf{I}}_n \in {\mathbb {R}}^{n \times n}\) is identity matrix.
The solutions can be ordered as \(\lambda _{(1)} \ge \lambda _{(2)} \ge ... \ge \lambda _{(D)} \ge 0\) with corresponding normalized eigenvectors \({\textbf{v}}_{(1)}, {\textbf{v}}_{(2)},..., {\textbf{v}}_{(D)}\). Resulting PCA matrix (Jolliffe 2011) is
and the dimensionality reduction generates new feature matrix \({\textbf{Z}} = {\textbf{X}}_{\textrm{S}} {\textbf{W}}_1\).
Data Whitening (DWH) (Eldar and Oppenheim 2003) represents improved process of PCA, which guarantees unit covariance matrix of resulting vector. The transform is defined as
The matrix \({\textbf{W}}_2^{\textrm{T}}\) is designed to satisfy \(\textrm{E} \,{\textbf{z}} = {\textbf{0}}\) and \(\textrm{var} \,{\textbf{z}} = {\mathbb {I}}_n\). Using the result of EVD we directly calculate (Eldar and Oppenheim 2003)
Due to duality, we can perform the data whitening for \(m<n\) in more efficient way. We calculate \({\textbf{B}} = {\textbf{X}}_{\textrm{S}} {\textbf{X}}_{\textrm{S}}^{\textrm{T}} \ge 0\) and perform its EVD. Resulting EVD equation is \(({\textbf{B}} - \lambda {\textbf{I}}_m){\textbf{u}} = {\textbf{0}}\). The solutions can be ordered again as \(\lambda _{(1)} \ge \lambda _{(2)} \ge ... \ge \lambda _{(D)} \ge 0\) with corresponding normalized eigenvectors \({\textbf{u}}_{(1)}, {\textbf{u}}_{(2)},..., {\textbf{u}}_{(D)} \in \textrm{R}^m\). Resulting whitening matrix is
and the data whitening generates new feature matrix \({\textbf{Z}} = {\textbf{X}}_{\textrm{S}} {\textbf{W}}_2\) in both cases. Data whitening in primal or dual form is preferred in this paper for optional data preprocessing.
2.3 Multiple discriminant analysis
Another approach of dimensionality reduction is based on knowledge of class membership. Having information about classes we can also perform linear data transformation to obtain higher data separation. Classical Fisher discriminant analysis (FDA) (Mika et al. 1999; Croux et al. 2008) is designed for two classes but Rao (Rao and Toutenburg 1995; Duda et al. 2012) generalized it for multi-classification task as follows.
The Rao method transforms the data from \({\mathbb {R}}^n\) to \({\mathbb {R}}^{N-1}\) for \(N \ge 2\) using a linear transformation
where \({\textbf{W}}_3 \in {\mathbb {R}}^{n \times (N-1)}\).
The method is based on pattern index sets \({\mathcal {D}}_i \in \lbrace k \in {\mathbb {N}}: y_k^* = i \rbrace\) for \(i=1,...,N\) and their cardinalities \(m_i = \textrm{card} \, {\mathcal {D}}_i\). After the evaluation of cluster centres
we can calculate within matrix
where
The total and between matrices are calculated as
When the pattern set is non-degenerated then \({\textbf{S}}_{\textrm{W}} > 0\) and we solve generalized EVD problem which is driven by equation
Solutions of generalized EVD can be ordered as \(\lambda _{(1)} \ge \lambda _{(2)} \ge ... \ge \lambda _{(N-1)} \ge 0\) with corresponding eigenvectors \({\textbf{e}}_{(1)}, {\textbf{e}}_{(2)},..., {\textbf{e}}_{(N-1)}\). Finally, the transformation matrix of Rao method is
and the pattern set has new feature matrix \({\textbf{Z}} = {\textbf{X}}_{\textrm{S}} {\textbf{W}}_3\) again.
The Rao method is well informed and the new dimension is fixed to \(N-1\) but the data whitening has the optional dimension of result and the information about class membership is missing. Therefore, these two approaches can generate different matrices \({\textbf{W}}\) and \({\textbf{Z}}\). The Rao method is also useful for data preprocessing.
2.4 DBSCAN technique
There are also various approaches to pattern classification. In this section, we focus on modern sequential clustering algorithms as SLINK, CLINK and finally DBSCAN. The SLINK algorithm (Sibson 1973) carries out single-link cluster analysis on an arbitrary dissimilarity coefficient and provides a representation of the resultant dendrogram which can readily be converted into the usual tree-diagram. There exist also alternative implementation (Goyal et al. 2020) which comes from a reduction in the number of distance calculations required by the standard implementation of SLINK with time complexity O\((m \, \textrm{log} m)\) in the case of m patterns. Hierarchical clustering omitting the initial sorting and consecutive clustering (Schmidt et al. 2017) having a linear time complexity as alternative to single linkage clustering has also been presented.
An algorithm for a complete linkage clustering (Patel et al. 2015) is based, same as SLINK, on a compact representation of a dendrogram. Fast algorithms (Banerjee et al. 2021) for CLINK clustering show that complete linkage clustering of m points can be computed in O\((m \, \textrm{log}^2 m)\) time.
The density-based spatial clustering of applications with noise (DBSCAN) (Ester et al. 1996) represents a non-parametric algorithm with a given set of points in some metric space and groups together points that are closely packed together and marks outliers.
We will study patterns in vector space as \({\textbf{x}}_k \in {\mathbb {R}}^n, k=1,2,...,m\), where m, n are number of patterns and space dimensionality but the DBSCAN is defined in metric space. After application of Euclidean distance we can define mutual distances as \(d_{i,j}=|| {\textbf{x}}_i - {\textbf{x}}_j ||_2\). Various versions of this algorithm (Antony and Deshpande 2016; Bai et al. 2017) differ in the method of distance computation. The inefficient implementations (Shen et al. 2016; Schubert et al. 2017) calculate all mutual distances before data clustering but there are more effective procedures that rapidly decrease the time complexity of DBSCAN to O\((m \, \textrm{log} m)\) as in the case of SLINK.
The DBSCAN is driven by two parameters \(\epsilon > 0\), \(k_{\textrm{min}} \ge 2\) which fully depends on users opinion. We will set them to obtain the best sensitivity of resulting classifier in the process of cross-validation. The DBSCAN generates an undirected graph \({\mathcal {G}}\) with vertex set \({\mathcal {V}} = \{ 1,2,...,m \}\) and edge set \({\mathcal {E}}= \{e_1,e_2,...,e_t \}\) and the pattern \({\textbf{x}}_i\) is placed in vertex i for \(i=1,2,...,m\). There are three types of vertices: a hard member, a soft member and an outlier.
The vertex i is called the hard member when \(\textrm{card} \{ j:d_{i,j} \le \epsilon \} \ge k_{\textrm{min}}\). Every edge \(e= \{ i,j \}\) has to satisfy \(d_{i,j} \le \epsilon\). The edge e is called a hard connection when the vertices i, j are hard members. A soft connection is the edge e where the node i is the hard member but the node j is not. Remaining edges are eliminated. Resulting graph \({\mathcal {G}}\) has several components. The component is declared as a cluster when it has two vertices at least. Remaining discrete components are declared as the outliers.
The main advantage of SLINK, CLINK, and DBSCAN is in the ability of sequential learning with acceptable time complexity. Exactly, the hard members of DBSCAN, number of clusters, and outliers are invariant to pattern order during the learning process.
2.5 Union of hidden classes
Both supervised and unsupervised approaches to the pattern classification can be used to form hidden classes inside the final classifier. The system of hidden classes arises from uncertainty of class membership, imperfectness of classification or any context out approach. We will apply a deterministic approach which is based on the relationship between hidden groups and the output classes (Hrebik et al. 2019) as follows. The aim is to optimize this relationship as the best mapping from the hidden to the output classes. The strict classifier is defined as mapping \({\textrm{c}}: {\mathscr {L}}_H \rightarrow {\mathscr {L}}_N\) from the set \({\mathscr {L}}_H\) of hidden class indices to the set \({\mathscr {L}}_N\) of final class indices, where \({\mathscr {L}}_n = \lbrace 1,..., n \rbrace\). This mapping can be expressed via the matrix \({\mathbb {X}} \in \lbrace 0,1 \rbrace ^{N \times H}\), where \(x_{i,j} = 1\) iff \(d_k \in {\mathscr {H}}_j \Rightarrow d_k \in {\mathscr {C}}_i\). Therefore, \(x_{i,j} = 1\) just when for any pattern belonging to \({\mathscr {H}}_j\) it also belongs to \({\mathscr {C}}_i\). The uniqueness conditions \(\sum _{i=1}^{N} x_{i,j}= 1\) have to be satisfied for \(j=1,...,H\).
The relation between the classes and the hidden groups is presented via the contingency table \({\mathbb {F}} \in {\mathbb {N}}_0^{N \times H}\), where \(f_{i,j} = \text {card} \lbrace k:d_k \in {\mathscr {C}}_i \bigcap {\mathscr {H}}_j \rbrace\) is the result of pattern counting. Here, \(f_{i,j}\) is the number of patterns belonging to both class \({\mathscr {C}}_i\) and group \({\mathscr {H}}_j\) as joint frequency, which can be relativized as
where \(i = 1,...,N\), \(j=1,...,H\).
There are two approaches to evaluating the quality of a classifier. One of them is accuracy, which is the success of the classifier as a whole, i.e. the proportion of successfully classified patterns and all patterns. The main disadvantage of such approach is the imbalance in results for individual classes. Against this, there is another concept based on sensitivity, which is very often used in medicine when we evaluate the percentage of success in classifying a sick patient. If, on the other hand, we are interested in the percentage of success in determining whether a patient is healthy, then medicine uses the term specificity. If we do not know in advance how many classes there will be in the task, it is more advantageous to call the classical sensitivity the sensitivity for the first class (\(se_1\)) and the specificity the sensitivity of the second class (\(se_2\)) and generally introduce \(se_N\), which is actually the success rate of the classifier for the given class, i.e. the number of correct individuals in that class relative to the total number of individuals in that class. It is obvious that if we want to have strict requirements for the classifier, it is not a good idea to maximize its accuracy, but to maximize the so-called critical sensitivity, which is nothing but the smallest value of the individual \(se_N\). When we substitute the values \(f_{i,j}\) and \(x_{i,j}\) into those definitions, we receive the following formulas.
The accuracy of given classifier can be expressed as
Using the concept of class sensitivity as a relative frequency of true classification, we can calculate it for \(i = 1,...,N\) as
An average sensitivity can be defined as
A lower estimate of class sensitivity is defined as a critical sensitivity
We prefer critical sensitivity as the strength criterion of classifier efficiency and maximize them via the union of hidden classes. The accuracy criterion is also used as the traditional measure which is frequently used by many authors.
In accordance with Hrebik et al. (2019), we will maximize the critical sensitivity \(se^*\). An adequate mixed binary optimization task is
subject to
with real artificial variable \(\textit{se}^*\). The inequalities Eq. (22) guarantee that \(se^*\) is a lower bound of critical sensitivity during the optimization process.
From the theoretical point of view it is necessary to think whether this task has a solution in general. If we choose \(se^*=0\) then all inequalities Eq. (22) holds regardless of what the values of x are. This means that if the task was too complicated, then in the worst possible scenario it will case that the optimal value of critical sensitivity will be \(se^*=0\), as a symbol that the original task has no solution, but the system of inequalities Eq. (22) has a solution. So, if we obtain \(se^*=0\), it means that the given task cannot be solved by the given method. In the case of degeneration, the task can have more solution. Therefore is important following consideration.
After the specification of \(se^*\), we can yield from the task degeneration and solve additional binary programming task which guarantees the same critical sensitivity and maximize accuracy as
subject to
3 Framework of multi-classification
The novel approach of vector pattern multi-classification is based on a combination of the approaches mentioned above. Basic assumptions and procedures are summarized in this section.
3.1 Assumptions
Let N, M, n be number of output classes, number of patterns, and number of pattern dimensions that are unlimited in general. But there is a threshold value \(n^*\) of pattern dimension which switch between deterministic and random sub-sampling approaches. In both cases, the first step of classification is dimensionality reduction using data whitening or multi-class discriminant analysis which converts the data into the space of dimension \(D \le n\). In the second step, the reduced data are clustered using the DBSCAN technique and the hidden classes are formed. Optimal unions of these hidden classes are performed in the last step of the multiple classifications.
3.2 Classification for \(n \le n^*\)
The learning strategy of classification is based on the pattern dimension. When the patterns are not too large we will proceed with the whole set of patterns. In the case of data whitening we use learning procedure with parameters \(D, k_{\textrm{min}}, \epsilon\). But in the case of multi-class discriminant analysis, we have only two free parameters \(k_{\textrm{min}}, \epsilon\) because of \(D=N-1\). In the first step we transform data matrix \({\textbf{X}} \in {\mathbb {R}}^{M \times n}\) to \({\textbf{Y}} \in {\mathbb {R}}^{M \times D}\) using data whitening Eqs. (1–5) or using Rao method Eqs. (7–14). Then we use the DBSCAN technique for clustering in \({\mathbb {R}}^D\) with parameters \(k_{\textrm{min}}, \epsilon\). Every cluster forms a new hidden class of patterns and the outliers which are also localized by the DBSCAN are ignored. Finally, the optimal union of hidden classes Eqs. 20–24) is performed.
There are no problems with large pattern number M because both data whitening, Rao method, and DBSCAN are designed for a large amount of data patterns. But the parameters of DBSCAN must be selected to generate not too large number H of hidden classes.
3.3 Approximated classification for \(n > n^*\)
When the pattern vector length n is too large its reduction is necessary preprocessing step which is a kind of context out data whitening. We select \(n_{\textrm{red}} \le n^*\) first and create a random sub-sample of m patterns which are supposed to be representatives of the given pattern set. When \(m \le n^*\), the dual form of learning Eq. (6) is performed as an alternative data whitening which produces the weight matrix \({{\textbf {W}}} \in {\mathbb {R}}^{n \times n_{\textrm{red}}}\). Using this matrix we transform the original data to obtain a matrix \({\textbf{X}}_{\textrm{red}} \in {\mathbb {R}}^{M \times n_\textrm{red}}\). This matrix of reduced patterns is used instead of the original pattern set using the learning strategy (Sect. 3.2). This process is of a stochastic nature and \(n_{\textrm{red}}, m\) are two additive parameters that control the preliminary dimensionality reduction. Therefore, the novel classification algorithm is also applicable to long pattern vectors but with context out imperfectness related to the random sampling of patterns.
3.4 Classification verification
We suppose the parameters of classification are set on the complete pattern set to obtain the classifier with maximum possible critical sensitivity \(se^*\). The role of parameter \(\epsilon\) for fixed \(D, k_{\textrm{min}}\) is crucial and can rapidly change the class sensitivities but the critical sensitivity peaceful-wise continuous function of \(\epsilon\) and therefore there is an interval of \(\epsilon\) which maximizes \(se^*\). After this preliminary parameter setting we have to perform the cross-validation. When the number of patterns M is small, we prefer Leave-One-Out (Wong 2015; Gronau and Wagenmakers 2019) cross-validation technique but for large M we can use 10-fold (Xu et al. 2018; Steyerberg 2019) cross-validation scheme as generally recommended. When we map the role of parameter \(\epsilon\) in the case of cross-validation the values of \(se^*\) are not so high in many cases. The interval of an optimal \(\epsilon\) can be also different in the case of cross-validation. There are no general rules and this phenomenon will be studied experimentally in the next section.
3.5 Classifier structure
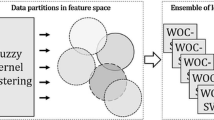
The new classifier serves to ensure that for all patterns \({\textbf{x}}_k \in {\mathbb {R}}^n, k=1,2,...,m\), where m, n are number of patterns and space dimensionality, recognized to which class \({\mathscr {C}}_i, i=1,2,...,N\) belongs. The new classifier consists of three parts, and the proposed structure is captured in Fig. 1. The first part of the system, is a linear transformation that displays all patterns from dimension n to a lower dimension D. Either multiple discriminant analysis (Sect. 2.3) or data whitening (Sect. 2.2) is used for this transformation. The second part of the system uses the standard DBSCAN tool with parameters \(\epsilon\) and \(k_{\textrm{min}}\) (Sect. 2.4). Depending on these parameters, individual patterns are classified unsupervised into H classes, representing hidden classes. The third part of the system is optimal union using binary programming techniques (Sect. 2.5). This creates a system that unambiguously assigns each x on the input to which class it belongs.
4 Experimental part
To demonstrate the results of proposed approach we have selected ten basic classification tasks (Dua 2020). This approach allow clear comparison with other classification methods. All datasets analysed during the current study are available in the repositories referred in Table 1, mainly in UC Irvine Machine Learning Repository (Dua 2020). The table includes also the number of patterns, their dimensionality, and the number of classes. For comparison of our results, we work with relevant papers presenting classification results. The main aim is to confirm that our method is comparable with other ones. As our approach based on critical sensitivity is not commonly used we compare the basic criteria of classification accuracy. We present and compare both cases’ optimal setting on the training set as well as the leave-one-out cross-validation.
4.1 Case study: iris flower classification task
As the primal research dataset, we decided to use the well-known and widely used iris dataset (Swain et al. 2012). Iris dataset contains three classes of fifty instances each, where each class refers to a type of iris plant. One class is linearly separable from the other two, the latter is not linearly separable from each other. Every iris pattern is a four-dimensional real positive vector.
First of all, we have to prepare the data for our model. We can use the data whitening or Rao method. Our aim is to demonstrate using of DBSCAN. We test different values of \(\epsilon\) and investigate results for \(k_{\textrm{min}}\) from 2 to 5. The \(\epsilon\) values in the testing examples we set from 0 to 1.26. The optimal settings with the highest value of \(se^*\) are summarized in Table 2 for training set and in Table 3 for Leave-One-Out cross-validation.
We can confirm the best results using \(k_{\textrm{min}} = 2\), Rao method and \(\epsilon\) from \(\epsilon _{\textrm{min}} = 0.08\) to \(\epsilon _{\textrm{max}} = 0.09\) in naive case. Leave-One-Out cross-validation led to the wider interval of \(\epsilon\) reaching 0.04 to 0.14 for the same setting. The value of critical sensitivity is above ninety percent in both cases. The reached maximum accuracy value is fully comparable to other presented results (Kulluk et al. 2012; Ozyildirim and Avci 2014; Asafuddoula et al. 2017; Yin and Gelenbe 2018) summarized in Tables 4 and 5 along with reached minimum and maximal accuracy together with placement among benchmark techniques.
4.2 Application to other pattern sets
We have applied our novel method also to nine other datasets: Wine, Glass, Cancer (Wisconsin), Haberman, Liver, Ionosphere, Cancer (Coimbra), Transfusion, and Cryotherapy described in Table 1. Most of the tasks are classification into two classes. In the case of the Glass dataset, there is known also an alternative task to seven classes. The results of training are collected in Table 2 as optimal values of classification parameters and adequate values of accuracy and sensitivities. The proposed method has been also treated by leave-one-out cross-validation. Obtained results are summarized in Table 3 in a similar way. There are no dramatic changes in parameter setting and resulting accuracy and sensitivities in Tables 2 and 3. The SLINK algorithm as DBSCAN for \(k_{\textrm{min}} = 2\) is a good choice in a majority of cases. But the type dimension of primal coordinate reduction is task sensitive. The Rao technique multi-class discriminant analysis is useful in many cases.
As seen in Tables 4 and 5 our method is comparable with standard techniques of classification. To see how competitive our results are, we went through several papers on this topic (Kulluk et al. 2012; Ozyildirim and Avci 2014; Rani and Ganesh 2014; Abdar et al. 2017; Asafuddoula et al. 2017; Kahramanli 2017; Aslan et al. 2018; Li and Chen 2018; Talabni and Engin 2018; Yin and Gelenbe 2018; Austria et al. 2019; Chan and Chin 2019; Kraipeerapun and Amornsamankul 2019; Rahman et al. 2020). The rank of our novel method has been evaluated for every dataset and during both training and cross-validation processes. Comparison results including the rank among others is in Tables 4 and 5. The proposed method is in the second quartile related to the involved referential methods in most cases. As the use of datasets varies, also the number of used benchmark techniques for comparison is variant and summarized in the following paragraphs.
According to Kulluk et al. (2012) for training and cross-validation comparison we used five different variants of harmony search algorithms and standard backpropagation algorithm. For training, we compared the results with approximator using a spiking random neural network using five other benchmarks (Yin and Gelenbe 2018). For cross-validation results comparison we used generalized classifier neural network and its logarithmic learning implementation, probabilistic neural network and standard multilayer perceptron as presented in Ozyildirim and Avci (2014). The results of an incremental ensemble classifier method (Asafuddoula et al. 2017) were also used as a benchmark.
Comparison of training and testing for Haberman and Liver datasets is included for product-unit neural networks as a special class of feed-forward neural network, together with results for backpropagation and Levenberg–Marquardt algorithms, as presented in Kahramanli (2017).
For Ionosphere and Transfusion datasets, we used also method proposed in Chan and Chin (2019) to tackle the problem of imbalanced data based on cosine similarity together with results for synthetic minority oversampling technique and adaptive synthetic sampling approach. The Liver dataset is compared to the results of two proposed methods Boosted C5.0 and CHAID based on the decision trees and five other methods (backpropagation, NB tree, decision tree, C5.0, support vector machine, and basic neural network) presented in Abdar et al. (2017). The training results for Transfusion dataset were obtained by the naive Bayesian classifier, implementation of algorithm iterative Dichotomiser 3, and random tree, all presented in Rani and Ganesh (2014).
Both breast cancer datasets, Cancer Wisconsin and Coimbra, were compared with five different classification models including decision tree, random forest, support vector machine, neural network and logistics regression as presented in Li and Chen (2018). Coimbra dataset is compared with results of an artificial neural network, standard extreme learning machine, support vector machine and K-nearest neighbour presented in Aslan et al. (2018), and additionally to ten classification algorithms and their variations including logistic regression, k-nearest neighbour, support vector machine, decision tree, random forest, gradient boosting method, and naive Bayes presented in Austria et al. (2019).
In the case of Cryotherapy dataset, we used comparison with four methods using kernel functions for improving the learning capacity of support vector machine presented in Talabni and Engin (2018). Seven methods additional methods for Cryotherapy, two methods based on the combination of cascade generalization and complementary neural network, and five existing methods (neural network, stacked generalization, cascade generalization, complementary neural network, the combination of stacked generalization as presented in Kraipeerapun and Amornsamankul (2019). Cryotherapy results were also compared with proposed using of support vector machine and nine standard methods k-nearest neighbours, binary logistic regression, linear discriminant analysis, quadratic discriminant analysis, classification and regression trees, random forest, adaptive boosting, gradient boosting, and bagging, presented and summarized in Rahman et al. (2020).
Results for training using Iris, Wine, Glass, Haberman, Liver and Ionosphere datasets are compared also with 1-NN classifier and two its variants, namely the Hypersphere Classifier and the Adaptive Nearest Neighbor Rule, as presented in Orozco-Alzate et al. (2019). Results of Fuzzy Pattern Trees using Grammatical Evolution, called Fuzzy Grammatical Evolution (Murphy et al. 2022), is used for comparison on Iris, Wine, Haberman and Transfusion datasets.
For additional comparison we have included also average accuracy results for cross-validation of scalable ensemble technique XGBoost, random forest, gradient boosting, LightGBM using selective sampling of high gradient instances and Ordered CatBoost modifiing the computation of gradients to avoid the prediction shift as presented in Bentejac et al. (2021) for Iris, Wine, Cancer (Wisconsin), Liver, and, Ionosphere. Cross-validation results in case of hybrid classification model named HyCASTLE (Veneri et al. 2022) is also included for Iris, Glass and Haberman.
Based on all previous experiments we see our method as quite robust. Robustness means a similar parameter setting in the training and cross-validation processes in this case. The novel method is therefore comparable with current methods and its learning is very simple and robust related to the parameter value. Moreover, we present additionally the values of critical sensitivities, which we believe are relevant for the classifier quality.
5 Conclusion
The proposed type of pattern classifier with embedded dimensionality reduction and hidden class forming has been tested. Based on training and cross-validation using ten standard datasets, the new type of classifier has several advantages evaluated below.
Using a training materialized for direct verification or leave-one-out cross-validation, we set DBSCAN parameter \(k_{\textrm{min}}=2\) in most cases to obtain the best value of critical sensitivity of a given system. Therefore, there is no need to use general DBSCAN approach because its reduced version the clustering algorithm SLINK produces less compact hidden clusters. It is usually the main weakness of the SLINK approach, but in our case, the optimal unioning of hidden classes eliminates this disadvantage. The future realization of this classifier can use only SLINK instead.
The most important property of this classifier is similar behavior during learning on the training set and the cross-validation. We suppose that selected method of dimensionality reduction and length of resulting vectors depend only on the dataset but are independent of the verification strategy.
There is also a similarity in the range of parameter \(\epsilon [ \epsilon _{\textrm{min}}, \epsilon _{\textrm{max}} ]\). Moreover, the optimal \(\epsilon\) is included in the optimum range of cross-validation. Therefore the optimal set of parameter \(\epsilon\) can be directly used as relevant estimate of \(\epsilon\) for the leave-one-out cross-validation processes.
The proposed method aims the critical sensitivity maximization, which produces non-trivial unions of hidden classes. But the authors of referential techniques are oriented mainly to the accuracy evaluation of classifiction, which complicates the method comparison.
There are several general recommendations for the setting of a classifier. The user decides first whether to apply multi-class discriminant analysis or whether he prefers data whitening of given dimension D. The SLINK method with parameter \(\epsilon\) is suggested for hidden class forming. The optimal values of D and \(\epsilon\) can be estimated using only the training set for verification. Finally, the leave-one-out cross-validation is a simple process focused only on parameter \(\epsilon\) improvement, which remains unchanged in many cases. The proposed classifier, represents a comparable alternative to other pattern classification techniques focusing on critical sensitivity.
The main advantage of the proposed classifier is that it successfully avoids the curse of dimensionality and includes automatic reduction and standardization of the input dataset. All cluster analysis is carried out in dimensionless coordinates and thus offers a wide range of uses for a whole range of applications. It is a relatively simple classifier having comparable properties comapred to more complicated ones. As one of the other applications, we can recommend, for example, the recognition of unstructured data such as strings, trees, and graphs. In such a case, it is necessary to set correctly a suitable feature description, which precedes the reduction layer of our classifier.
Data availability
The datasets generated during and/or analysed during the current study are available in the UC Irvine Machine Learning Repository (Dua 2020).
Abbreviations
- \({\mathscr {S}}\) :
-
Pattern set
- \({\mathbb {N}}\) :
-
Natural numbers
- \({\mathbb {R}}\) :
-
Real numbers
- \({\mathscr {C}}\) :
-
Output class
- \({\mathscr {H}}\) :
-
Hidden class
- n :
-
Number of features
- m :
-
Number of patterns
- H :
-
Number of hidden classes
- N :
-
Number of classes
- D :
-
Reduced dimension
- \({\textbf{p}}\) :
-
Pattern vector
- \({\textbf{X}}\) :
-
Input matrix
- \({\textbf{y}}\) :
-
Classifier output vector
- \({\textbf{y}}^*\) :
-
Desired output vector
- y :
-
Classifier output/response
- \(y^*\) :
-
Desired output value
- \({\textbf{w}}\) :
-
Weight vector
- \(w_0\) :
-
Bias
- \(\textrm{c}\) :
-
Classifier
References
Abdar M, Zomorodi-Moghadam M, Das R, Ting IH (2017) Performance analysis of classification algorithms on early detection of liver disease. Exp Syst Appl 67:239–251
Antony N, Deshpande A (2016) Domain-driven density based clustering algorithm. Proceedings of international conference on ICT for sustainable development. Springer, pp 705–714
Asafuddoula M, Verma B, Zhang M (2017) An incremental ensemble classifier learning by means of a rule-based accuracy and diversity comparison. International joint conference on neural networks. IEEE, pp 1924–1931
Aslan MF, Celik Y, Sabanci K, Durdu A (2018) Breast cancer diagnosis by different machine learning methods using blood analysis data. Int J Intell Syst Appl Eng 6(4):289–293
Austria YD, Lalata JAP, Maria LB Jr, Goh JEE, Goh MLI, Vicente HN (2019) Comparison of machine learning algorithms in breast cancer prediction using the coimbra dataset. Int J Simul Syst Sci Technol 20:23
Back T, Fogel DB, Michalewicz Z (2018) Evolutionary computation 1: basic algorithms and operators. CRC Press
Bai L, Cheng X, Liang J, Shen H, Guo Y (2017) Fast density clustering strategies based on the k-means algorithm. Pattern Recognit 71:375–386
Banerjee P, Chakrabarti A, Ballabh TK (2021) An efficient algorithm for complete linkage clustering with a merging threshold. Data management, analytics and innovation. Springer, pp 163–178
Basavegowda HS, Dagnew G (2020) Deep learning approach for microarray cancer data classification. CAAI Trans Intell Technol 5(1):22–33
Bentejac C, Csorgo A, Martinez-Munoz G (2021) A comparative analysis of gradient boosting algorithms. Artif Intell Rev 54(3):1937–1967
Chan TK, Chin CS (2019) Health stages diagnostics of underwater thruster using sound features with imbalanced dataset. Neural Comput Appl 31(10):5767–5782
Croux C, Filzmoser P, Joossens K (2008) Classification efficiencies for robust linear discriminant analysis. Stat Sin 2008:581–599
Dua D, Graff C (2020) UCI machine learning repository (2020). http://archive.ics.uci.edu/ml
Duda RO, Hart PE, Stork DG (2012) Pattern classification. Wiley
Eldar YC, Oppenheim AV (2003) Mmse whitening and subspace whitening. IEEE Trans Info Theory 49(7):1846–1851
Ester M, Kriegel HP, Sander J, Xu X et al (1996) A density-based algorithm for discovering clusters in large spatial databases with noise. Kdd 96:226–231
Goyal P, Kumari S, Sharma S, Balasubramaniam S, Goyal N (2020) Parallel slink for big data. Int J Data Sci Anal 9(3):339–359
Gronau QF, Wagenmakers E-J (2019) Limitations of bayesian leave-one-out cross-validation for model selection. Comput Brain Behav 2(1):1–11
Hrebik R, Kukal J, Jablonsky J (2019) Optimal unions of hidden classes. Cent Euro J Op Res 27(1):161–177
Hu G, Yin C, Wan M, Zhang Y, Fang Y (2020) Recognition of diseased pinus trees in uav images using deep learning and adaboost classifier. Biosyst Eng 194:138–151
Jafarzadeh H, Mahdianpari M, Gill E, Mohammadimanesh F, Homayouni S (2021) Bagging and boosting ensemble classifiers for classification of multispectral, hyperspectral and polsar data: a comparative evaluation. Remote Sens 13(21):4405
Jolliffe I (2011) Principal component analysis. Springer
Kahramanli H (2017) Training product-unit neural networks with cuckoo optimization algorithm for classification. Int J Intell Syst Appl Eng 5(4):252–255
Karlsson C (2010) Handbook of research on cluster theory. Edward Elgar Publishing
Khozeimeh F, Alizadehsani R, Roshanzamir M, Khosravi A, Layegh P, Nahavandi S (2017) An expert system for selecting wart treatment method. Comput Biol Med 81:167–175
Khozeimeh F, Jabbari Azad F, Mahboubi Oskouei Y, Jafari M, Tehranian S, Alizadehsani R, Layegh P (2017) Intralesional immunotherapy compared to cryotherapy in the treatment of warts. Int J Dermatol 56(4):474–478
Kraipeerapun P, Amornsamankul S (2019) Using cascade generalization and neural networks to select cryotherapy method for warts. 2019 International conference on engineering, science, and industrial applications (ICESI). IEEE, pp 1–5
Kulluk S, Ozbakir L, Baykasoglu A (2012) Training neural networks with harmony search algorithms for classification problems. Eng Appl Artif Intell 25(1):11–19
Li Y, Chen Z (2018) Performance evaluation of machine learning methods for breast cancer prediction. Appl. Comput. Math 7(4):212–216
Liang T, Sur P (2022) A precise high-dimensional asymptotic theory for boosting and minimum-l1-norm interpolated classifiers. Ann Stat 50(3):1669–1695
Lin H, Zhao B, Liu D, Alippi C (2020) Data-based fault tolerant control for affine nonlinear systems through particle swarm optimized neural networks. CAA J Autom Sin 7(4):954–964
Liu F, Wang J (2022) An accurate method of determining attribute weights in distance-based classification algorithms. Math Probl Eng. https://doi.org/10.1155/2022/6936335
Medina-Pérez MA, Monroy R, Camiña JB, García-Borroto M (2017) Bagging-tpminer: a classifier ensemble for masquerader detection based on typical objects. Soft Comput 21(3):557–569
Mika S, Ratsch G, Weston J, Scholkopf B, Mullers KR (1999) Fisher discriminant analysis with kernels. Neural networks for signal processing. IEEE, pp 41–48
Murphy A, Ali MS, Mota Dias D, Amaral J, Naredo E, Ryan C (2022) Fuzzy pattern tree evolution using grammatical evolution. SN Comput Sci 3(6):1–13
Nguyen LH, Holmes S (2019) Ten quick tips for effective dimensionality reduction. PLoS Comput Biol 15(6):1006907
Orozco-Alzate M, Baldo S, Bicego M (2019) Relation, transition and comparison between the adaptive nearest neighbor rule and the hypersphere classifier. International conference on image analysis and processing. Springer, pp 141–151
Ozyildirim BM, Avci M (2014) Logarithmic learning for generalized classifier neural network. Neural Netw 60:133–140
Patel S, Sihmar S, Jatain A (2015) A study of hierarchical clustering algorithms. 2nd International conference on computing for sustainable global development. IEEE, pp 537–541
Patrício M, Pereira J, Crisóstomo J, Matafome P, Gomes M, Seicca R, Caramelo F (2018) Using resistin, glucose, age and bmi to predict the presence of breast cancer. BMC Cancer 18(1):29
Rahman M, Zhou Y, Wang S, Rogers J et al (2020) Wart treatment decision support using support vector machine. University of Texas
Rani SA, Ganesh SH (2014) A comparative study of classification algorithm on blood transfusion. J Adv Res Technol 3:57–60
Rao CR, Toutenburg H (1995) Linear models. Springer, pp 3–18
Rekha G, Madhu S (2022) An hybrid approach based on clustering and synthetic sample generation for imbalance data classification: clustsyn. Proceedings of data analytics and management. Springer, pp 775–784
Schmidt M, Kutzner A, Heese K (2017) A novel specialized single-linkage clustering algorithm for taxonomically ordered data. J Theor Biol 427:1–7
Schubert E, Sander J, Ester M, Kriegel HP, Xu X (2017) Dbscan revisited, revisited: why and how you should (still) use dbscan. ACM Trans Database Syst (TODS) 42(3):19
Shahid AH, Singh M (2019) Computational intelligence techniques for medical diagnosis and prognosis: problems and current developments. Biocybern Biomed Eng 39(3):638–672
Shen J, Hao X, Liang Z, Liu Y, Wang W, Shao L (2016) Real-time superpixel segmentation by dbscan clustering algorithm. IEEE Trans Image Processing 25(12):5933–5942
Shi G, Zhao B, Li C, Wei Q, Liu D (2019) An echo state network based approach to room classification of office buildings. Neurocomputing 333:319–328
Sibson R (1973) Slink: an optimally efficient algorithm for the single-link cluster method. Comput J 16(1):30–34
Steyerberg EW (2019) Validation of prediction models. Springer, pp 329–344
Swain M, Dash SK, Dash S, Mohapatra A (2012) An approach for iris plant classification using neural network. Int J Soft Comput 3(1):79
Talabni H, Engin A (2018) Impact of various kernels on support vector machine classification performance for treating wart disease. International conference on artificial intelligence and data processing. IEEE, pp 1–6
Veneri MD, Cavuoti S, Abbruzzese R, Brescia M, Sperlì G, Moscato V, Longo G (2022) Hycastle: A hybrid classification system based on typicality, labels and entropy. Knowl Based Syst 244:108566
Wong TT (2015) Performance evaluation of classification algorithms by k-fold and leave-one-out cross validation. Pattern Recognit 48(9):2839–2846
Xu L, Fu HY, Goodarzi M, Cai CB, Yin QB, Wu Y, Tang BC, She YB (2018) Stochastic cross validation. Chemom Intell Lab Syst 175:74–81
Yeh IC, Yang KJ, Ting TM (2009) Knowledge discovery on rfm model using bernoulli sequence. Exp Syst Appl 36(3):5866–5871
Yin Y, Gelenbe E (2018) A classifier based on spiking random neural network function approximator. Preprint available in ResearchGate. net
Acknowledgements
The authors would like to acknowledge the support of the research grant SGS20/190/OHK4/3T/14. The second author also acknowledges the support of the OP VVV MEYS funded project CZ.02.1.01/0.0/0.0/16_019/0000765 Research Center for Informatics.
Funding
Open access publishing supported by the National Technical Library in Prague.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hrebik, R., Kukal, J. Concept of hidden classes in pattern classification. Artif Intell Rev 56, 10327–10344 (2023). https://doi.org/10.1007/s10462-023-10430-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10462-023-10430-6