Abstract
Artificial intelligence (AI)-aided general clinical diagnosis is helpful to primary clinicians. Machine learning approaches have problems of generalization, interpretability, etc. Dynamic Uncertain Causality Graph (DUCG) based on uncertain casual knowledge provided by clinical experts does not have these problems. This paper extends DUCG to include the representation and inference algorithm for non-causal classification relationships. As a part of general clinical diagnoses, six knowledge bases corresponding to six chief complaints (arthralgia, dyspnea, cough and expectoration, epistaxis, fever with rash and abdominal pain) were constructed through constructing subgraphs relevant to a chief complaint separately and synthesizing them together as the knowledge base of the chief complaint. A subgraph represents variables and causalities related to a single disease that may cause the chief complaint, regardless of which hospital department the disease belongs to. Verified by two groups of third-party hospitals independently, total diagnostic precisions of the six knowledge bases ranged in 96.5–100%, in which the precision for every disease was no less than 80%.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
AI-aided clinical diagnosis can help clinicians working at primary hospitals and clinics to avoid or reduce misdiagnoses and missing diagnoses. The ML models based on processed big data are well known, e.g., convolutional neural network (CNN), deep neural network (DNN), recurrent neural network (RNN) and Bayesian network (BN) (Fukushima and Neocognitron 1982; Lo et al. 1995; Russakovsky et al. 2015; Szegedy et al. 2015; Brosch et al. 2016; Shin et al. 2016; Duraisamy and Emperumal 2017; Bardou et al. 2018; Christodoulidis et al. 2017; Lin et al. 2018; Er et al. 2016; Ceccon et al. 2014), etc. However, most of them are applied to solve image and speech recognitions. AI-aided general clinical diagnosis is really needed in practice but is relatively rare. References (Wu et al. 2018) and (Liang et al. 2019) report two deep learning models that can perform general clinical diagnoses. However, it is not clear whether or not they have the same precisions when being applied in different application scenarios as being achieved in the testing dataset, which is called the generalization problem, although some comparisons between the models and clinicians have been made. The real world (primary level) applications are not qualified to judge the precisions, because of lacking the medical check measures, professional knowledge and experience. It is reasonable to doubt the generalization ability of the two models described in Wu et al. (2018) and (Liang et al. 2019), because the essence of deep learning is to establish a nonlinear mapping between the input (combinations of variable states including unknown states) and output (diseases) by adjusting the structure and parameters of the neural network. When the actual application scenario is different from the dataset in terms of sample space, which is common, the precision may drop, leading to the generalization problem.
In the general clinical diagnoses, there are at least 10,000 input variables. Each variable has at least 3 states: negative, positive and unknown. Thus, the number of state combinations of input variables are at least 310,000 = 1.6 × 104771, a huge number. The training and testing datasets cover only a small part of these state combinations, which is called the training and testing sample space (TTSS). The real application sample spaces (RASSs) are usually different from TTSS, while different application scenarios may have different RASSs. Thus, the mapping in TTSS may be different from that in RASSs. How the trained model based on TTSS can be applied in different RASSs needs to be verified. In fact, our experience is that the diagnostic precision drops significantly in real applications.
Moreover, how to ensure the model be able to diagnose the rare diseases is another problem, where the common diseases are relatively easy to be diagnosed even by primary clinicians and the rare diseases are really needed to be differentially diagnosed by the AI-aided models, which means that we need not only the high precision in total but also the high precision for each disease including rare diseases. Note that the common diseases are the majority in the training and testing datasets and the rare diseases may be marginalized in ML models, while the high precisions can still be achieved in the testing dataset due to the high proportion of common diseases. For the example of arthralgia shown in Table 4 in this paper, five common diseases (Gout, SLE, Osteoarthritis, RA and Trauma) have 95.8% case records in group 1, which implies that once the five diseases are correctly diagnosed, the total diagnostic precision will be 95.8%, even the diagnoses for the other 18 diseases are all incorrect. In practice, the correct diagnoses for the other 18 diseases are really needed.
Furthermore, because of the black box problem of deep learning models, the two models described in Wu et al. (2018) and (Liang et al. 2019) lack interpretability.
To solve these problems, the model based on the domain knowledge/causality is needed, because domain knowledge has invariance, which is essentially different from ML models basing on big data.
DUCG developed in recent years is such a model (Zhang 2012, 2015a, b; Zhang et al. 2014, 2018; Zhang and Geng 2015; Zhang and Zhang 2016; Zhang and Yao 2018) and has achieved promising application results for fault diagnoses of large, complex industrial systems (Zhang and Yao 2018; Zhang et al. 2018; Dong et al. 2014a, 2018; Qu et al. 2015; Zhao et al. 2014; Geng and Zhang 2014) and general clinical diagnoses (Dong et al. 2014b; Hao et al. 2017; Fan et al. 2018; Jiao et al. 2020; Ning et al. 2020; Zhang et al. 2021).
It is noted that the existing DUCG model is entirely based on causalities. However, in many practical cases, some non-causal knowledge representations and associated probabilistic reasoning are needed. For example, when representing an uncertain causal relationship between a disease and a blood routing test, it is desirable to use the blood routing test as an inspection type variable, and to use the results of the test as its consequential variables. However, there is no direct causal relationship between the disease and the blood routing test itself, because the blood routing test is not the consequence of the disease. What actually exists is the uncertain causal relationship between the disease and the blood routing test results, i.e. the indicators. On the other hand, such causalities cannot be represented intuitively without the blood routing test variable, where the test is an action to find the consequences/indicators of diseases. In the hierarchical domain knowledge representation, the action is actually a classifier between the disease and the indicators. To solve this problem, the classification type (C-type) variable along with its unit matrix I drawn as its input directed arc is introduced as illustrated in Figs. 1 and 2.
It is proved in Sect. 3 that the DUCG without C-type variables is equivalent to that with them in the sense of inference. The former is resulted from the latter and is really used in the invisible DUCG inference, because the former is obviously easier to compute than the latter, while the latter remains as the visible knowledge base for better DUCG construction and interpretability.
Six DUCG knowledge bases including C-type variables for clinical diagnoses were constructed by clinical experts at Peking Union Medical College Hospital, Beijing Hospital, Xuanwu Hospital and Youan Hospital of Capital Medical University, Beijing, China. The diagnostic precisions were verified by two groups of third-party hospitals. Group 1 was Suining Central Hospital, Sichuan, China, which has a long history of more than 100 years. Group 2 was six hospitals officially organized as a whole by Chongqing Science and Technology Commission: West-South Hospital, Daping Hospital, The Second Affiliated Hospital of Chongqing Medical University, Chongqing Tumor Hospital, Chongqing Traditional Chinese Medicine Hospital (CTCMH) and Wanzhou Central Hospital, Chongqing, China. In which CTCMH was the leading unit. All hospitals are the Grade IIIA (the highest grade in China) hospitals and are located in southwest of China, far from Beijing where the knowledge bases were constructed. The verification results of the two groups are close to each other. Therefore, the generalization ability of DUCG were verified, which means that the DUCG-aided general clinical diagnoses can be applied in any application scenario without generalization problem that usually exists in ML models.
Section 2 introduces DUCG briefly. Section 3 presents the C-type variable methodology. Section 4 applies the C-type variable methodology to the diagnoses of six chief complaints. Two groups of third-party verifications were made. Section 5 summarizes this paper.
2 Brief Introduction to DUCG
DUCG is a newly developed model that can explicitly and graphically represent causalities with uncertainties and perform probabilistic reasoning. In clinical diagnoses, it can easily represent various complex and uncertain causalities between diseases (root causes) and risk factors, symptoms, signs, image findings and laboratory results, etc., namely the observations or evidences. Conditional on the evidences collected for each patient, DUCG calculates the conditional probabilities of the found possible diseases, and thus performs intelligent diagnoses with clear casual and mathematical meanings (Zhang et al. 2021). To have the primary clinicians take responsibilities instead of DUCG, DUCG’s strong interpretability in knowledge bases, diagnostic results and computation process are very important.
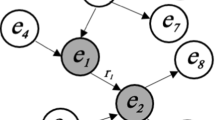
DUCG is composed of two sub-models: single-valued DUCG (S-DUCG) and multivalued DUCG (M-DUCG). The so called single-valued means that only the causes of the true state of a child variable can be specified, while the false state is the complement of the true state. The so-called multivalued means that the causes of every state of a variable can be specified separately (Zhang 2012). In this paper, only M-DUCG is addressed and therefore is abbreviated as DUCG. Figure 3 is an illustrative DUCG. The symbols are described in Table 1. The basic idea of the DUCG model is shown in Fig. 4.
For simplicity, the subscript ji in Fig. 3 is abbreviated as j. The rectangular node Bn is the basic or root cause event variable, without any input, and Bnj is state j of Bn. The circular node Xn is the result event variable, Xnj is state j of Xn, and Xn can be both the cause/input and the consequence/output of other nodes. The pentagonal node Dn is the default or unknown cause event of Xn or Xnj, without any input, and its occurrence probability is defined as 1. The hexagonal node SXn is a special X-type event variable, and SXnj is state j of SXn. When SXnj occurs, where j ≠ 0 and 0 indicates normal state, a particular disease or variable state must be true with a certain confidence θ, and therefore SXnj is called gold-criterion in clinical diagnosis. The double-circle node BXn is a B&X-type variable with both B and X properties. Its state division and definition are exactly the same as Bn, and only the state probability distribution of BXn may be different from Bn (affected by the associated risk factors). The logic gate variable Gn represents the various state combinations of the input variable and its input is connected with a directed arc  . The double line logic gate SGn represents various state combinations of the associated risk factors (such as age, gender, etc., represented as X-type variables), changing the state probability distribution of Bn as that of BXn. The output of SGn is BXn, through a double-line directed arc
. The double line logic gate SGn represents various state combinations of the associated risk factors (such as age, gender, etc., represented as X-type variables), changing the state probability distribution of Bn as that of BXn. The output of SGn is BXn, through a double-line directed arc  that zooms in or zooms out the state probabilities of Bn as that of BXn according to the combinations of risk factors. The reversal logic gate RGn drawn as
that zooms in or zooms out the state probabilities of Bn as that of BXn according to the combinations of risk factors. The reversal logic gate RGn drawn as  represents that the input of RGn may cause some combinations of output. The single-line directed arc
represents that the input of RGn may cause some combinations of output. The single-line directed arc  represents the causality matrix Fn;i = (rn;i/rn)An;i, where Ank;ij is the element in the matrix An;i, Ank;ij is the virtual random event that the parent event Vij (V ∈ {B, X, D, G, BX, SX}) causes the child event Xnk (including SXnk) directly. rn;i > 0 is the strength of the causal relationship between Vi and Xn, \(r_{n} \equiv \sum\limits_{i} {r_{n;i} }\). The dashed directed arcs
represents the causality matrix Fn;i = (rn;i/rn)An;i, where Ank;ij is the element in the matrix An;i, Ank;ij is the virtual random event that the parent event Vij (V ∈ {B, X, D, G, BX, SX}) causes the child event Xnk (including SXnk) directly. rn;i > 0 is the strength of the causal relationship between Vi and Xn, \(r_{n} \equiv \sum\limits_{i} {r_{n;i} }\). The dashed directed arcs  or
or  is conditional
is conditional  or
or  respectively, conditional on condition event Zn;i, where n indexes the child/output and i indexes the parent/input. When Zn;i is true,
respectively, conditional on condition event Zn;i, where n indexes the child/output and i indexes the parent/input. When Zn;i is true,  or
or  becomes
becomes  or
or  respectively; otherwise,
respectively; otherwise,  or
or  is eliminated.
is eliminated.
In DUCG, the upper-case letter represents event or event variable and the corresponding lower-case letter represents the probability, i.e., bnj = Pr{Bnj}, bxnj = Pr{BXnj}, xnj = Pr{Xnj}, sxnj = Pr{SXnj}, gnj = Pr{Gnj}, rgnj = Pr{RGnj}, dn = Pr{Dn}≡1, zn;i = Pr{Zn;i}, fnk;ij = Pr{Fnk;ij} = (rn;i/rn)ank;ij, ank;ij = Pr{Ank;ij}, fn;i = Pr{Fn;i}, an;i = Pr{An;i}, etc. The indices before “;” are for the child and the indices after “;” are for the parent. The {a-, b-, r-}-type parameters are usually given by domain experts based on statistics or their experience. Note that the main formulas of DUCG are in the form of numerator divided by denominator (see (Zhang et al. 2021) for details). Therefore, only the relative values of parameters are sensitive, not the absolute values, which means that the parameters are easy to be given by clinical experts.
The variable index is inside the symbol without the letter of the variable type. The symbol shape represents the variable type. State index 0 denotes the normal/negative state, while the other states indicate abnormal/positive states. Moreover, Vnj ∈ {Xnj, SXnj, RGnj}, j ≠ 0, is assigned with attention parameter εnj ≥ 1 that quantifies the attention of domain experts to explain the cause of Vnj. If no cause can be found, a virtual Dn drawn as dashed pentagon will be assigned as the default cause of Vnj according to the DUCG simplification rule 10 listed in the Appendix of Zhang et al. (2021), and anj;nD between Vnj and the virtual Dn is defined as anj;nD = 1/εnj, in which the index D indicates the invariable state of Dn. In this case, Vnj is called the isolated evidence. Also, 0 < θnj ≤ 1 is assigned to SXnj to quantify the confidence that the specific disease does exist given SXnj, where j ≠ 0. Ref. (Zhang et al. 2021) gives more details.
As shown in Fig. 4, the above events and probabilities satisfy Eqs. (1) and (2) respectively:
In which, Fn;i≡(rn;i/rn)An;i and fn;i≡(rn;i/rn)an;i. Fnk;ij≡(rn;i/rn)Ank;ij, fnk;ij≡(rn;i/rn)ank;ij and ank;ij = Pr{Ank;ij}, where Fnk;ij, fnk;ij, Ank;ij and ank;ij are members of Fn;i, fn;i, An;i and an;i respectively. In the case of only one input to Xn, Fn;i = An;i and fn;i = an;i.
Equation (1) can be repeatedly applied until the expression becomes the sum-of-products composed of {BX-, D-, A-, r-}-type events and parameters, which is the event expanding process, and then the probability of the expression can be calculated by replacing the upper-case letters with the corresponding lower-case letters as illustrated in Eqs. (1) and (2). The state probability distribution of BXk can be calculated from \(bx_{km} = sa_{km;kj} b_{km}\), where sakm;kj is the zoom factor transforming bkm to bxkm (see (7) in Zhang et al. (2021) for details). Then, BX-type variables can be treated as root causes/diseases.
The evidences can be written as \(E = \bigcap\limits_{i} {X_{ij} }\). The diagnostic inference is to calculate the conditional probability Pr{BXkj|E} = Pr{BXkjE}/Pr{E}, BXkj ∈ SH, SH is the possible disease set conditional on E. We need to expand E as the sum-of-products composed of {BX-, D-, A-, r-}-type events and parameters. In which, logic computations such as absorption and exclusion and the r-type parameter calculation are applied.
In general, Eq. (3) is satisfied, in which “1” denotes complete set.
Based on Eq. (3), we have the following theorem expressed as Eq. (4).
Theorem 1
Which means that the causality chains in DUCG are self-relied. Therefore, we do not need to specify all parameters in an;i. For example, we may have Eq. (5).
Which means that we can specify only the parameters in concern. In other words, for a variable whose state is normal (indexed by 0), we do not care about the causality and probability related to this state. What we are interested in is the causality between abnormal states. For example, a certain disease Bij (j ≠ 0) causes a certain abnormal state Xnk (k ≠ 0), where Xn may represent a medical check result. We also do not care about the unconditional probability bi0 (i.e. without disease). That is to say, bi0, an0;ij and ank;i0 in {a-, b-}-type matrices do not need to be given. Usually, we express bi0, an0;ij and ank;i0 as “ − ” or blank, which is equivalent to null set in expanding E.
The DUCG diagnostic inference is to calculate the probability distribution of BXi affected by risk factors observed for a patient and calculate Pr{BXkj|E} = Pr{BXkjE}/Pr{E}, j ≠ 0, in which BXkj ∈ SH is composed of the abnormal states of BX-type variables. SH is the set of possible diseases conditional on E, and is found by the logical expanding and simplification of DUCG. The appendix in Zhang et al. (2021) lists the DUCG simplification rules. The detailed inference algorithm can be found in Zhang (2012)-(Zhang and Zhang 2016; Zhang et al. 2021).
3 Introducing C-type Variables to Extend DUCG to Include Classification Relationship
3.1 The basic idea
Consider Fig. 5, where B1 represents pituitary prolactin adenoma, X2 indicates whether thyroid function is normal, X3 indicates whether TSH (Thyroid Stimulating Hormone) is low, and X4 indicates whether FT3 (free triiodothyronine) is low.
In Fig. 5, the hierarchy and relationships are clearly represented. It also embodies the medical knowledge of the disease, that is, pituitary prolactinoma (B1) may cause thyroid function abnormal (X2), and these abnormalities are manifested as TSH (X3) and FT3 (X4). However, problems are exposed when assigning values to the a-type matrices for each directed arc. Since A2;1 is a causal event matrix between pituitary prolactinoma B1 and thyroid function X2, a2,1;1,1 should be the probability of thyroid dysfunction caused by pituitary prolactinoma. Since A3;2 is a causal event matrix representing the causality from thyroid function X2 to TSH (X3), a3,1;2,1 should be the probability that thyroid dysfunction (X2,1) triggers low TSH (X3,1). Similarly, a4,1;2,1 should be the probability of thyroid dysfunction (X2,1) triggering low FT3 (X4,1). But this is obviously wrong, because the real causal relationship is: X3,1 and X4,1 are the causes of X2,1, not the opposite. At the same time, there is no direct causal relationship between B1 and X2. It is an indirect causal relationship with X2 through X3 and X4, and the direction is opposite. According to the expression in Fig. 5, the inference results of DUCG and the diagnosis results of clinical experts will be inconsistent, because the knowledge of the clinical experts is actually as shown in Fig. 6. In other words, Fig. 5 is incorrect. This example illustrates how easy the mistake may occur without classification variables.
To solve this problem, we introduce C-type variable along with I matrix as follows:
Definition 1
The state partition of the classification variable Cn drawn as  is identical to its parent variable i, Fn;i is fixed as a unit matrix \(I_{n;i} = \left( {\begin{array}{*{20}c} 1 & 0 & \cdots & 0 \\ 0 & 1 & \cdots & \vdots \\ \vdots & \vdots & \ddots & 0 \\ 0 & \cdots & 0 & 1 \\ \end{array} } \right)\), and Fm;n is actually the causality between cause variable i and consequence variable m.
is identical to its parent variable i, Fn;i is fixed as a unit matrix \(I_{n;i} = \left( {\begin{array}{*{20}c} 1 & 0 & \cdots & 0 \\ 0 & 1 & \cdots & \vdots \\ \vdots & \vdots & \ddots & 0 \\ 0 & \cdots & 0 & 1 \\ \end{array} } \right)\), and Fm;n is actually the causality between cause variable i and consequence variable m.
Equivalently, fn;i = In;i, because fn;i = Pr{Fn;i} = Pr{In;i} = In;i. Note that “1” in DUCG stands for both numerical one and complete set. With this definition, Fig. 6 can be better represented as Fig. 7.
In Fig. 7, according to Definition 1, f2;1 = I2;1, and f3;2 and f4;2 equal to f3;1 and f4;1 in Fig. 6 respectively.
Theorem 2
In the sense of inference, the DUCG with C-type variable along with its corresponding I matrix is equivalent to the DUCG without C-type variables.
Theorem 2 constitutes the inference algorithm of the DUCG with C-type variables, i.e. we can use the C-type variables along with I matrices to construct the DUCG with C-type variables, while the corresponding DUCG without C-type variables is really used in the DUCG inference. The latter is resulted from the former by (1) the elimination of C-type variables along with I directed arcs and (2) the connections between the cause and consequences of the C-type variable in the former. i.e., simplify Fig. 7 as Fig. 6. The inference equivalence is proved in follows:
Proof
First, we prove a simple case, i.e. Figures 6 and 7 are equivalent in inference. For this, we only need to prove that Pr{B1X3X4} in Fig. 6 and in Fig. 7 are equal. According to Fig. 6 and Eq. (1), we have.
In which the operator “*” indicates to multiply the corresponding elements in the two matrices as defined in Corollary 15Footnote 1 in Zhang et al. (2014). According to Fig. 7 and Eq. (1), we have
The last step in Eq. (7) is because f3;2 in Fig. 7 equals to f3;1 in Fig. 6, and f4;2 in Fig. 7 equals to f4;1 in Fig. 6. Thus, we have Eq. (7) equals to Eq. (6).
Obviously, the above proof can be applied in the case when the child variables of B1 in Fig. 6 and C2 in Fig. 7 are increased, which covers all cases of theorem 2. ■
According to Theorem 2, we can use Fig. 7 to express the medical hierarchical knowledge in the DUCG editor, automatically change Fig. 7 as Fig. 6 in the invisible inference, and perform the inference according to Fig. 6.
More details are addressed in follows.
3.2 Single parent
In Fig. 8, C3 has more than one parent, where the real causalities that we want to represent are as shown in Fig. 9. However, Fig. 8 may cause some trouble.
The real causalities behind Fig. 8
Suppose evidence E = X5,1X6,2, and f5;3 and f6;3 are given as follows:
\(f_{5;3} = \left( {\begin{array}{*{20}c} - & - \\ - & {f_{5,1;3,1} } \\ \end{array} } \right)\), \(f_{6;3} = \left( {\begin{array}{*{20}c} - & - \\ - & {f_{6,1;3,1} } \\ \end{array} } \right)\).
Based on Fig. 8, we have f3;1 = I3;1 and f3;2 = I3;2 as defined. According to Eq. (1), we have
However, based on Fig. 9, we have
Equation (9) is not equal to Eq. (8). To solve this problem, we have the following definition:
Definition 2
Each C-type variable can have only one parent variable, while different C-type variables may be the same in content.
Thus, Fig. 8 is changed as Fig. 10, in which C3 = C4. As defined, f5;3, f5;4, f6;3 and f6;4 in Fig. 10 equal to f5;1, f5;2, f6;1 and f6;2 in Fig. 9 respectively.
Based on Fig. 10, we have Eq. (10).
It is seen that Eq. (10) equals to Eq. (9), which means that Fig. 10 is equivalent to Fig. 9 in the sense of inference. In conclusion, Fig. 8 is not allowed and Fig. 10 should be used.
Figure 11 shows another case that cannot be represented by one C-type variable. According to Definition 2, the corresponding DUCG with C-type variables should be as shown in Fig. 12. It is easy to prove that Figs. 11 and 12 are equivalent to each other in inference.
3.3 Normalizing paths
In practice, the repeated paths shown in Fig. 13 are possible. These repeated paths can be merged, that is, Fig. 13 can be calculated according to Fig. 14.
Normalized C-type path corresponding to Fig. 13
Figure 14 merges C8 and C9 in Fig. 13 into C10. The calculation of the merged parameters is as follows:
First, I8;4 and I9;4 in Fig. 13 are merged as I10;4 in Fig. 14. Next, F5;10 in Fig. 14 is equal to the sum of F5;8 = (r5;8/r5)A5;8 and F5;9 = (r5;9/r5)A5;9 in Fig. 13 as shown in Eq. (11).
Theorem 3
Once a group of C-type variables share a same child variable and a same parent variable, this group of C-type variables can be merged as a single C-type variable along with its single I matrix. The merged F-type variable as the only output of the merged C-type variable is the sum of the group of F-type variables as the outputs of the group of C-type variables.
Proof
Suppose the group of C-type variables are Ci, i ∈ SC. They share a child variable Xn and a parent variable Vm. Let Cj be the merged C-type variable, j ∉ SC, and Fn;j be the merged F-type variable that is the single output directed arc of the merged C-type variable. According to Eq. (1) and based on the original group of C-type variables, we have.
Also, according to Eq. (1) but based on the merged C-type variable, we have
Let Eq. (12) equal to Eq. (13), we have
Of course, the merged DUCG with C-type variable can be replaced in inference by the one without C-type variable.
4 The Third-Party Verifications
To verify the diagnostic precisions and generalization ability of DUCG, we constructed six DUCG knowledge bases according to six chief complaints respectively, in which the C-type variables were used.
4.1 Construction of DUCG with C-type variables
The construction steps are as follows.
Step 1 Determine the diseases that may cause the chief complaints across hospital departments, which means that the diseases are not limited in a specific hospital department and the triage may not be necessary, although the DUCG triage methodology has been presented in Bu et al. (2020).
Step 2 Construct the subgraph for every disease determined in step 1 as illustrated in Figs. 15 and 16 in which the symbols are described in Table 2. In subgraphs, the interpretability of DUCG knowledge bases is well demonstrated.
Step 3 Synthesize the subgraphs under a same chief complaint as a DUCG by fusing the same variables in different subgraphs. For example, the synthesized arthralgia DUCG is as shown in Fig. 17.
4.2 Verifications, precisions and comparisons
After the DUCG construction, we tested its correctness carefully by using the case records in the hospital information system (HIS) of the knowledge base constructor’s hospitals as illustrated in Ref. (Zhang et al. 2021). Then, two groups of third-party verifications for six DUCG knowledge bases were performed independently to verify the generalization ability and diagnostic precisions of DUCG. The verifications done by Group 1 contain more diseases than Group 2, because Group 2 did verifications earlier than Group 1 when less diseases were considered. However, the diseases in Group 2 are all included in Group 1, so that we can compare the results of them in a comparable scale. The verifications were performed as follows:
-
1.
Under each chief complaint, search the cases recorded in the HISs of the third-party hospitals for each disease.
-
2.
For the total cases searched for each disease, randomly select no more than 10 cases for test.
-
3.
Check the selected case record to ensure that it is in high quality, otherwise give up the case and make a new selection.
-
4.
Manually input the evidences found in the tested case record into the DUCG cloud platform developed to implement the DUCG methodology.
-
5.
Click the DUCG diagnosis function on the platform to find the possible diseases and rank them according to their conditional probabilities.
-
6.
Compare the diagnosed diseases with the tested case record. If the diagnosed diseases with significant conditional probabilities cover the diseases in the record, and the clinical experts confirm that the diseases not in the record (if any) are also reasonable, label this tested case as “correct,” otherwise label it as “incorrect.” In fact, because of the uncertain quality, norm and format in the records, it was not easy to judge the correctness. In the confusing cases, discussions with clinical experts were the final means to make judgements.
-
7.
Calculate the precision for each disease by the correct case number divided by the total tested case number of the disease.
-
8.
Calculate the total precision for the DUCG of the chief complaint by the total correct case number divided by the total tested case number under the chief complaint.
As an example, the arthralgia DUCG verified in Group 1 is as shown in Fig. 17. Total 23 diseases are listed in Table 3, in which the 16 diseases in Group 2 are included. The verification results are shown in Tables 4, 5. The results for the other five chief complaints are in Tables 6, 7, 8, 9, 10 respectively in the Appendix. The total precisions from the two groups are listed and compared in Table 5. Note that the precisions from Group 2 are all 100%.
It is seen that the total precisions of the six DUCGs from the two groups respectively are very close to each other and no less than 96.5%, in which the lowest precision for all diseases was no less than 80%. The precision difference of the two groups is no more than |96.53 − 100|% = 3.47%. The mean precision difference of the six chief complaints is:
4.3 Verification discussions
For some relatively rare diseases, the case records were less than 10. In such cases, all the qualified records were selected. If there was no case found, the precision of this disease could not be calculated and was not considered in the precision calculations.
We believe that it is enough to test no more than 10 randomly selected cases for a disease in verifications, because 10 cases can cover most knowledge points related to the disease. If the knowledge base is correct, the test results will be correct, regardless of how many cases are tested. Given the total number of cases, if we increase the tested cases for every disease, only the tested cases of common diseases will be increased and the results will likely be correct, while the tested cases of rare diseases will not be increased due to the lack of cases, leading to an improper higher precision in total. The scientific way to perform the verification is to have the numbers of tested cases as equal as possible for all diseases. As a balance, we chosen to have no more than 10 tested cases.
The so-called “rare” disease means that it is rare under the chief complaint. A disease is rare under a chief complaint does not mean that it is also rare under other chief complaints.
It is easy to understand that only the discharged patient case records meet the high-quality requirement (the recorded information was sufficient and diagnosis was correct) for the third-party verifications. We did not use the outpatient case records for verifications, because it was hard to judge whether the outpatient diagnoses were correct or not. In general, the case record for a discharged patient contains more medical information than the case record of an outpatient. How to verify the diagnostic precision of DUCG conditional on less information for an outpatient is another issue and will be addressed elsewhere.
5 Summery and discussions
The C-type variables are used only in the DUCG construction. Without C-type variables, the DUCG knowledge base is hard to be well organized and interpreted, and mistakes occur easily. The inference is based on the DUCG without C-type variables, which is automatically generated from the DUCG with C-type variables and is invisible.
Two groups of independent verifications for the six DUCG knowledge bases corresponding to six chief complaints verify that DUCG has strong generalization ability, which means that DUCG can be applied in any real application scenarios with almost the same precisions. This is because of the knowledge invariance.
The diagnostic interpretability of DUCG is provided by the generated sub-DUCG for each possible disease. A sub-DUCG is for a possible disease, in which all the evidences and causalities including the connected state-known variables and the isolated state-abnormal variables to this possible disease are displayed to the users in a graphical manner with text. More details can be found in Zhang et al. (2021).
DUCG does not deal with AI-aided medical image examination and medical sound recognition. They could be done by ML models. Hence, the relationship between DUCG and ML is cooperation.
In real applications, the AI-aided system should be able to recommend next medical checks based on the known information to collect further information for more accurate diagnosis. This will be discussed in another paper.
Notes
Corollary 15: \(A_{{nk_{n} ;i}} V_{i} A_{{mk_{m} ;i}} V_{i} = \left( {A_{{nk_{n} ;i}} *A_{{mk_{m} ;i}} } \right)V_{i}\), in which \(\left( {A_{{nk_{n} ;i}} *A_{{mk_{m} ;i}} } \right) \equiv \left( {\begin{array}{*{20}c} {A_{{nk_{n} ;i1}} A_{{mk_{m} ;i1}} } & {A_{{nk_{n} ;i2}} A_{{mk_{m} ;i2}} } & \ldots & {A_{{nk_{n} ;ij}} A_{{mk_{m} ;ij}} } & \ldots & {A_{{nk_{n} ;iJ}} A_{{mk_{m} ;iJ}} } \\ \end{array} } \right)\)
Correspondingly,
\(a_{{nk_{n} ;i}} *a_{{mk_{m} ;i}} \equiv \left( {\begin{array}{*{20}c} {a_{{nk_{n} ;i1}} a_{{mk_{m} ;i1}} } & {a_{{nk_{n} ;i2}} a_{{mk_{m} ;i2}} } & \ldots & {a_{{nk_{n} ;ij}} a_{{mk_{m} ;ij}} } & \ldots & {a_{{nk_{n} ;iJ}} a_{{mk_{m} ;iJ}} } \\ \end{array} } \right)\)
where, “*” is an AND/multiplication matrix operator specially defined in DUCG.
In format, the * operator is similar to Hadamard product.
References
Danal Bardou, Kun Zhang, Sayed Mohammad Ahmad. Classification of breast cancer based on histology images using convolutional neural networks. IEEE Access, vol. 6, pp. 24680–24693. 2018.
Brosch T et al (2016) Deep 3D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. IEEE Trans Medical Imaging 35(5):1229–1239
Bu X, Lu L, Zhang Z, Zhang Q, Yan Z (2020) A general outpatient triage system based on dynamic uncertain causality graph. IEEE Access. https://doi.org/10.1109/ACCESS.2020.2995087
Ceccon S, Garwayheath DF, Crabb DP et al (2014) Exploring early Glaucoma and the visual field test: classification and clustering using bayesian networks. IEEE J Biomed Health Infom 18(3):1008–1014
Christodoulidis S, Anthimopoulos M, Ebner L, Chresti A, Mougiakakou S (2017) Multisource transfer learning with convolutional neural networks for lung pattern analysis. IEEE J Biomed Health Inform 21(1):76–84
Dong C, Zhang Q, Geng S (2014a) A modeling and probabilistic reasoning method of dynamic uncertain causality graph for industrial fault diagnosis. Int J Autom Comput 11(3):288–298
Dong C, Wang Y, Zhang Q, Wang N (2014b) The methodology of dynamic uncertain causality graph for intelligent diagnosis of vertigo. Comput Methods Programs Biomed 113:62–174
Dong C, Zhao Y, Zhang Q (2018) Cubic causality modeling and uncertain inference method for dynamic fault diagnosis. J Tsinghua Univ (Sci Technol) 58(7):614–622
Duraisamy Sawaswathi, Emperumal Srinivasan (2017) Computer-aided mammogram diagnosis system using deep learing convolutional fully complex-valued relaxation neural network classifier. IET Computer Vision 11(8):656–662
Er O, Cetin O, Bascil MS, Temurtas F (2016) A comparitive study on Parkinson’s disease diagnosis using neural networks and artifial immune system. J Med Imaging Health Inf 1:264–268
Fan Y, Zhang Z, Jing Z, Wang Y, Liu Z, Guo M, Wang R, Feng M (2018) Diagnostic value of dynamic uncertain causality graph DUCG in sellar region disease. Chinese J Minimal Invasive Neurosurg 06:249–253
Fukushima K, Miyake S (1982) Neocognitron: A self-organizing neural network model for a mechanism of visual pattern recognition. Competition and Cooperation in Neural Nets. Springer, Berlin Heidelberg
Geng S and Zhang Q (2014) Calculation method to diagnose intigrated causes of faults in process systems by means of dynamic uncertain causality graph. In: proceeding of 2014 Aisa-Pasific computer science and application confreence (CSAC 2014), Shanghai, China, pp 306–311
Hao S, Geng S, Fan L, Chen J, Zhang Q, Li L (2017) Intelligent diagnosis of jaundice with dynamic uncertain causality graph model. J Zhejiang Univ-Sci B (Biomed Biotechnol) 18(5):393–401
Jiao Y, Zhang Z, Zhang T, Shi W, Zhu Y, Hu J, Zhang Q (2020) Development of an artificial intelligence diagnostic model based on dynamic uncertain causality graph for the differential diagnosis of dyspnea. Front Med 14:488–497
Liang H, Tsui BY, Ni H, Calentim CCS, Baxter SL, Liu G et al (2019) Evaluation and accurate diagnoses of pdiatric diseases using artificial intelligence. Nat Med. https://doi.org/10.1038/s41591-018-0335-9
Lin Z, Huang Y, Wang J (2018) RNN-SM fast steganalysis of VoIP streams using recurrent neural network. IEEE Trans Inf Forensics Secur 13(7):1854–1868
Lo SB, Lou SA, Lin JS et al (1995) Artificial convolution neural network techniques and applications for lung nodule detection. IEEE Trans Med Imaging 14(4):711
Ning D, Zhang Z, Qiu K, Lu L, Zhang Q, Zhu Y, Wang R (2020) Efficacy of intelligent diagnosis with a dynamic uncertain causality graph model for rare disorders of sex development. Frontiers of Medicine 14:498–505
Qu Y, Zhang Q, Zhu X (2015) Application of dynamic uncertain causality graph to dynamic fault diagnosis in chemical processes. CAAI Trans Intell Syst 10(3):354–361
Russakovsky O, Deng J, Su H et al (2015) Imagenet large scale visual recognition challenge. Int J Comput Vision 115(3):211–252
Shin H-C et al (2016) Deep comvolutional neural networks for computer-aided detection: CNN architectures, dataset charicteristics and transfer learning. IEEE Trans Med Imaging 35(5):1285–1294
Szegedy C, Vanhoucke V, Ioffe S, et al. Rethinking the Inception Architecture for Computer Vision. Computer Sci, 2015: 2818–2826
Wu J, Liu X, Zhang X, He Z, Lv P (2018) Mastrer clinical medical knowledge at certified-doctor-level with deep learning model. Nacture Commun. https://doi.org/10.1038/s41467-018-06799-6
Yao Q, Zhang Q, Liu P, Yang P (2017) Application of dynamic uncertain causality graph in spacecraft fault diagnosis: prediction. Int Core J Eng 3(1):113–119
Zhang Q (2012) Dynamic uncertain causality graph for knowledge representation and reasoning: discrete DAG cases J. Comput Sci Technol 27(1):1–23
Zhang Q (2015) Dynamic uncertain causality graph for knowledge representation and probabilistic reasoning: directed cyclic graph and joint probability distribution. IEEE Trans Neural Netw Learn Syste 26(7):1503–1517
Zhang Q (2018) Increase safety and availability of nuclear power plants by means of DUCG. China Nuclear Power 11(1):59–68
Zhang Q, Geng S (2015) Dynamic uncertain causality graph applied to dynamic fault diagnosis of large and complex systems. IEEE Trans Rel 64(3):910–927
Zhang Q, Yao Q (2018) Dynamic uncertain causality graph for knowledge representation and reasoning: utilization of statistical data and domain knowledge in complex cases. IEEE Trans Neural Netw Learn Syst 29(5):1637–1651
Zhang Q, Zhang Z (2016) Dynamic uncertain causality graph applied to dynamic fault diagnoses and predictions with negative feedbacks. IEEE Trans Rel 65(2):1030–1044
Zhang Q, Dong C, Cui Y, Yang Z (2014) Dynamic uncertain causality graph for knowledge representation and probabilistic reasoning: statistics base, matrix and fault diagnosis. IEEE Trans Neural Netw Learn Syst 25(4):645–663
Zhang Q, Qiu K, Zhang Z (2018) Calculate joint probability distribution of steady directed cyclic graph with local data and domain casual knowledge. China Comun 15(7):146–155
Zhang Q, Bu X, Zhang Z, Zhang M, Hu J (2021) Dynamic uncertain causality graph for computer-aided general clinical diagnoses with nasal obstruction as illustration. Artif Intell Rev 54:27–61
Zhang Q (2015) Dynamic uncertain causality graph for knowledge representation and probabilistic reasoning: continuous variable, uncertain evidence and failure forecast. IEEE Trans Syst, Man Cybern,. 45, 7, pp 990–1003
Zhao Y, Zhang Q, Dong C (2014) Application of DUCG in fault diagnosis of nuclear power plant secondary loop. Autom Sci Technol 48:496–501
Acknowledgements
This research was supported by Institute for Guo Qiang, Tsinghua University (project number: 2020QG0001), Chongqing Science and Technology Commission (project number: cstc2018jscx-mszdx0106), and The Rockefeller-Endowed China Medical Board (Open Competition Program, grant number: 20-384).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
The diagnostic results of the five chief complaints (dyspnea, cough and expectoration, epistaxis, fever with rash, abdominal pain) are shown in following.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, Z., Jiao, Y., Zhang, M. et al. AI-aided general clinical diagnoses verified by third-parties with dynamic uncertain causality graph extended to also include classification. Artif Intell Rev 55, 4485–4521 (2022). https://doi.org/10.1007/s10462-021-10109-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10462-021-10109-w