Abstract
In this study, we tested the hypothesis that the growth efficiency of freshwater bacteria is differentially affected by ultraviolet radiation (UVR, 280–400 nm) as mediated through changes in their production and respiration rates. Five bacterial strains affiliated to Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Actinobacteria were isolated from different freshwater habitats and exposed in the laboratory to photosynthetically active radiation (PAR) and PAR + UVR, or kept in the dark for 4 h. Afterward, bacterial carbon production and respiration were assessed by measuring leucine incorporation and oxygen consumption rates, respectively. Ultraviolet radiation decreased significantly the bacterial production of Acidovorax sp., Pseudomonas sp. and Actinobacterium MHWTa3, and the respiration rate of Acidovorax sp. and Acinetobacter lwoffii. Measurements of respiration of a natural bacterial community collected from the same lake where A. lwoffii was isolated resulted in significantly higher rates after exposure to PAR + UVR than in the dark. In the presence of UVR, bacterial growth efficiency significantly decreased in Acidovorax sp., Pseudomonas sp., and Actinobacterium MHWTa3, but it increased in A. lwoffii or it remained unchanged in Sphingomonas sp. Our results indicate that although the outcome was strain-specific, UVR has the potential to alter the efficiency by which dissolved organic matter is transformed into bacterial biomass and thus to affect the biogeochemical carbon cycle.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Heterotrophic bacteria are a key component in the biogeochemical carbon cycle of aquatic ecosystems. The bacterial community is responsible for the remineralization of the dissolved organic matter (DOM) pool, which leads to the production of new bacterial biomass and also to the oxidation of this reduced organic carbon pool to inorganic carbon that is released into the atmosphere through bacterial respiration. The balance between these metabolic processes can be assessed by calculating the bacterial growth efficiency, a parameter that reflects the efficiency of heterotrophic bacteria in converting organic carbon into bacterial cellular carbon (del Giorgio and Cole 1998). The accurate estimation of bacterial growth efficiency in natural communities is far from being trivial although it is the basis to understand ecosystem functioning.
Bacterial production is measured by following the incorporation rates of radiolabeled thymidine (Fuhrman and Azam 1980) or leucine (Kirchman et al. 1985) into bacterial deoxyribonucleic acid and proteins, respectively. A recent study, however, has cautioned on the use of thymidine for the measurements of bacterial production in lakes because a significant fraction of the Betaproteobacteria is not able to taken up this substrate (Pérez et al. 2010). To estimate the produced bacterial biomass, incorporation rates are transformed either by theoretical or by empirical conversion factors, which are however highly variable across aquatic systems (Biddanda et al. 1994; Buesing and Marxsen 2005). Bacterial respiration can be estimated by measuring oxygen consumption or CO2 production rates, and likewise important as for the assessment of bacterial production, incubation times should be kept as short as possible to minimize changes in bacterial community composition (Gattuso et al. 2002). For example, this can be achieved by using fast-responding microelectrodes for oxygen measurements (Warkentin et al. 2007; Pringault et al. 2009) or by estimating oxygen consumption from electron transport system (ETS) activity (del Giorgio 1992). However, in oligotrophic and cold aquatic ecosystems, incubations are necessarily long and in some cases detection of significant oxygen changes can represent a real challenge.
Besides variations caused by methodological factors, numerous environmental variables could affect the bacterial growth efficiency in natural aquatic ecosystems (del Giorgio and Williams 2005). Among those environmental variables, sunlight is known, for example, to stimulate bacterial production in humic lakes due to the phototransformation of DOM and the consequent increase in the availability of nutrients (Reche et al. 1998). However, this indirect stimulatory effect of sunlight on production is usually not observed in transparent aquatic systems where UV radiation (UVR, 280–400 nm) has usually a negative effect on bacteria (i.e., Chatila et al. 2001; Buma et al. 2003 and references therein). In fact, in UV transparent lakes such as alpine ones, bacterial production in the upper meters of the water column is strongly reduced by UVR (Sommaruga et al. 1997). Solar UVR is known to induce oxidative stress in aquatic bacteria through the production of reactive oxygen species, which in turn damage cell components (Jeffrey et al. 2000; Maranger et al. 2002). Direct negative effects of UVR have been observed on bacterial growth (Sieracki and Sieburth 1986), viability (Helbling et al. 1995), and enzymes production (Herndl et al. 1993; Garde and Gustavson 1999). However, there are large differences in sensitivity to solar UVR among marine bacterial taxa when growth patterns (Agogué et al. 2005), viability (Joux et al. 1999), and activity (Alonso-Sáez et al. 2006) are assessed. Thus, generalizations on the response of bacterial groups or communities to UVR is difficult, because bacteria have evolved diverse protection mechanisms against UV stress such as the synthesis of photoprotective substances (Müller et al. 2005) or have efficient repair processes to minimize UV-induced DNA damage (Boelen et al. 2001; Buma et al. 2003).
Currently, information on how light and particularly UVR affects bacterial growth efficiency is scarce (Pakulski et al. 1998; Pringault et al. 2009), and the studies available have addressed the whole bacterial community of marine ecosystems. Equivalent information on bacterial isolates or in freshwater ecosystems is not available. Further, in some studies bacterial respiration has been estimated indirectly (Hernandez et al. 2007), thus making difficult the direct comparison of results. In this study, we assessed the production and respiration rates of freshwater bacterial isolates kept in darkness or exposed to photosynthetically active radiation (PAR, 400–700 nm) and PAR + UVR and calculated their growth efficiencies. Most of the isolates included in the study are members of different bacterial groups found in alpine lakes (i.e., lakes located above the treeline), which are very oligotrophic and UV transparent (Laurion et al. 2000) or they were included because of its uniqueness regarding cultured members (Hahn et al. 2003). We hypothesized that the effect of UVR on bacterial growth efficiency is strain-specific among freshwater bacteria. In addition, we also tested the effect of PAR + UVR on respiration rates of the natural bacterial community from one of the lakes where the isolates were obtained.
Materials and methods
Sources of bacterial strains
The bacterial strains used in this study include GKS10 and GKS12b both isolated from the plankton of the alpine lake Gossenköllesee (47°13′N, 11°01′E, 2,417 m above sea level); SOS1 that was isolated from the plankton of the alpine lake Schwarzsee ob Sölden (46°57′N, 10°56′E, 2,799 m above sea level); and RC1 that was isolated from freshly collected rainwater in the catchment area of Gossenköllesee (Table 1). All lake samples were collected with a Schindler-Patalas sampler at 1 m depth. Rainwater was collected in combusted (450°C; 2 h) glass bottles of 2 l. Finally, the strain MHWTa3 isolated from the shallow eutrophic lake Tai-Hu, China, and kindly provided by M. Hahn (Hahn et al. 2003) was selected to include a representative of Actinobacteria, as we did not succeed to isolate a member of this group from alpine lakes.
Media used for isolation and cultivation of bacterial strains
For the isolation of bacteria, 0.25 ml of unfiltered water was plated onto solid nutrient broth-soyotone-yeast extract (NSY) medium (Hahn et al. 2003). Isolated single colonies were recultured on solid NSY medium and transferred to liquid NSY several times before being recultured in inorganic basal medium (IBM) (Hahn et al. 2003) supplemented with 2.5 mg glucose l−1 (IBMG). The concentration of dissolved organic carbon (DOC) in IBMG was 1.0 mg l−1. This value was selected as a compromise between being able to detect growth and resembling those DOC concentrations found in the habitats (0.3–0.7 mg l−1, Laurion et al. 2000) from which bacteria were isolated. In addition, glucose has also the advantage that is not photodegraded under the radiation conditions we used in the experiment (i.e., absence of UVC where glucose absorbs) and that bacteria are usually able to transform this sugar into all the necessary amino acids, vitamins, and nucleotides that make up cells (Gottschalk 1986). For the strain MHWTa3 that originates from a very eutrophic lake, NSY medium was used for cultivation and determination of growth curves. Cultivation took place in a walk-in room set at 15 ± 1°C. During cultivation, the isolates were not exposed to UVR, but except for isolate MHWTa3, all the others originate from high UV environments. For example at the depth where bacteria were isolated in Gossenköllesee and Schwarzsee ob Sölden (i.e., 1 m), UVR at 320 nm is still ca. 85 and 70%, respectively, of that measured at the surface (Laurion et al. 2000).
Genotypic analysis
For the isolation of bacterial DNA, 2 ml of bacterial culture was filtered onto polycarbonate white filters (Millipore GTTP, 0.22 μm pore size) and the dried filters were frozen (−20°C) until further processing. DNA was isolated from the filters with a customary DNA isolation kit (PowerSoil Isolation Kit, MO BIO laboratories, CA, USA). PCR amplification of the bacterial 16S rRNA gene was done with the primers 27 forward (5′-AGA GTT TGA TCM TGG CTC AG-3′) (Lane et al. 1985) and 1,492 reverse (5′-TAC GGY TAC CTT GTT ACG ACT T-3′) (Kane et al. 1993) using a HotStarTaq® Plus Master Mix Kit (QIAGEN GmbH, Hilden, Germany) following the manufacturer’s instructions. The thermocycling program consisted of the following steps: initial activation at 95°C for 5 min, 30 cycles of denaturation at 94°C for 1 min, annealing at 52°C for 1 min, extension at 72°C for 2 min, and a final extension step for 10 min at 72°C. Sequencing reactions were done using external facilities (http://www.macrogene.com). To search for related 16S rRNA sequences with high similarity values, the obtained sequences were submitted to the Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi.nlm.nih.gov/Blast.cgi; Altschul et al. 1997) for preliminary identification (Table 1).
Nucleotide sequence accession numbers
Sequence data were deposited in the GenBank under accession numbers GQ451825—GQ451827 (Table 1).
Bacterial growth curves
Before the experiments, growth curves of the bacterial strains were established to characterize the different growth phases. Bacteria (except for strain MHWTa3) were inoculated into a fresh IBMG medium, and 1.5 ml samples were collected every 24 h for several days and fixed with formaldehyde (2% final concentration). Bacterial growth rates (μ) were calculated for the exponential phase according to the equation: μ = (ln N 2 − ln N 1)/(t 2 − t 1), where N 2 and N 1 are the number of cells at two different times. Generation times (g) were calculated as g = ln 2/μ.
Experimental design
For the experiments, three sets of muffled (450°C; 2 h) quartz tubes (n = 3) were filled with 100 ml of bacterial culture growing in IBMG and in the middle of the exponential phase. The first set of tubes was wrapped with a double layer of aluminum foil and served as dark control (DARK). The second set was wrapped with two layers of URUV foil (Digefra, Germany) that had 50% transmittance at 380 nm and excludes most of the UVR (PAR treatment), whereas the third set of tubes was exposed without further manipulation (PAR + UVR treatment). Simulated solar UVR was provided by four aged (100 h) fluorescent lamps (UVA-340, Q-Panel Co., Cleveland, OH, USA) with a maximum emission at 340 nm. The integrated irradiance between 280 and 320 nm (i.e., UV-B) was 1.4 W m−2 corresponding to a final dose of 20.2 kJ m−2, which is equivalent to a typical daily integrated value for summer at mid latitudes. PAR was provided by two white fluorescent tubes (cool white L36/W20, Osram) emitting 80 μmol quanta m−2 s−1. This PAR intensity is low compared to the natural solar spectrum and we have previously not observed negative effects on heterotrophic organisms, but it is efficient in promoting photorepair. A spectrum of the combination of lamps is found in Sommaruga et al. (1996).
Exposure took place in a walk-in room at 15°C for 4 h. During exposure, quartz tubes were kept horizontal at 25 cm distance from the lamps in a water bath to maintain constant temperature. Before (T0) and immediately after exposure (T4), samples were removed to estimate bacterial abundance, bacterial carbon production, and respiration rates. Bacterial abundance was checked again at the end of the respiration incubations (Tend) to control for potential changes. The incubations for respiration ranged between 8 and 14 h depending on the isolate (see bacterial respiration).
Bacterial abundance
Samples for bacterial abundance (n = 3) were filtered onto black polycarbonate filters (Millipore GBTP, 0.22 μm pore size) and stained with DAPI (4′,6′-diamidino-2-phenylindole; Molecular Probes, Eugene, OR, USA) according to the method described by Porter and Feig (1980). At least 15 monochromatic pictures were taken using a charge coupled device (CCD) camera installed on an epifluorescence microscope (Zeiss Z1 Imager) equipped with a filter set for DAPI (Zeiss Nr. 1) at an overall magnification of 1,000×. More than 600 DAPI-positive cells were counted semi-automatically using the image analysis software Scoreedo (http://scoreedo.sengaro.net).
Bacterial carbon production (BCP)
Leucine incorporation rates were measured using [4,5-3H]-l-leucine (Amersham, specific activity = 17.7 GBq mmol−1) for strain MHWTa3 and [U-14C]-l-leucine (Amersham, specific activity = 11.3 GBq mmol−1) for the other strains at the final concentration of 20 nmol l−1 (Kirchman et al. 1985). One formaldehyde-fixed control and triplicate samples (5 ml) were incubated for 1 h at 15°C in the dark. Incubations were stopped by adding formaldehyde (2% final concentration). Then, samples were filtered onto 0.22-μm pore size polycarbonate filters (Millipore GTTP). The filters were rinsed twice with 5 ml of cold trichloroacetic acid (5%) for 5 min before being dissolved by adding 6 ml of scintillation cocktail (Ready-Safe, Beckman Coulter). Radioactivity was measured after 15 h in a scintillation counter (Beckman LS 6000IC). The conversion of bulk leucine incorporation rates (mol l−1 h−1) into bacterial carbon production (μg C l−1 h−1) was done using a conversion factor of 1.44 × Leuinc (Leuinc = leucine incorporation in mol) as recommended by Buesing and Marxsen (2005).
Bacterial respiration and bacterial growth efficiency
Bacterial respiration in the control and treatments was estimated from rates of oxygen consumption before and after exposure. Triplicate respiration glass microchambers (4 ml; Unisense, Denmark) were filled with 4 ml of culture and incubated in the dark. Oxygen concentration was measured immediately after filling the respiration chambers and at regular times using an oxygen microsensor OX-MR (Unisense, Denmark). This microsensor is designed with an exterior guard cathode (Revsbech 1989), which results in extremely low oxygen consumption by the electrode itself (1.5–15 × 10−8 mg O2 h−1, http://www.unisense.com). The microsensor and the gas tight microchambers allow highly precise repeated measurements to be done in every chamber without affecting the oxygen concentration. The microsensor has a response time shorter than 1 s and a precision of 0.05%, which is equivalent to the Winkler technique (Briand et al. 2004). All the measurements were done under temperature-controlled conditions, and temperature was kept constant (±0.1°C) during the whole incubation. Previous to the experiments, the oxygen consumption of the different bacterial strains was monitored during 16 h to define an appropriate incubation period, i.e., to detect a significant decrease in oxygen concentration. Accordingly, the following incubation times were chosen: 8 h (GKS12b, SOS1, and RC1), 11 h (MHWTa3), and 14 h (GKS10). The different times used, however, do not affect the comparisons because oxygen decrease was linear for all strains. Oxygen consumption was computed from the slope of oxygen concentration versus time and was converted into μg carbon respired per cell (DAPI abundance) assuming a respiratory quotient of 1 (del Giorgio and Cole 1998) and using the mean cell number of the beginning and the end of the respiration measurement. Bacterial growth efficiency (BGE) was computed as BGE (%) = Bacterial carbon production/(Bacterial carbon production + Bacterial respiration) × 100.
Effect of PAR + UVR on respiration of a natural bacterial community
On October 30 2007, a water sample was collected from Schwarzsee ob Sölden at 1 m depth with a Schindler-Patalas sampler (3 l). In the laboratory, the sample was filtered through a 0.8-μm polycarbonate membrane (ATTP, Millipore) to exclude organisms larger than bacteria and then was exposed as described above to PAR + UVR for 4 h or kept in the dark. Incubation and respiration measurements were done at 14°C.
Statistical analysis
One-way analysis of variance (ANOVA) was done on the dataset to test for significant differences between the control (DARK) and the treatments. To test for significant changes in bacterial abundance during exposure and respiration measurements (i.e., T0—T4 and T4—Tend) and in the different treatments, a one-way repeated measures ANOVA was used. The post hoc multiple comparisons of all ANOVAs were made pairwise by the Holm-Sidak method with an overall significance level of 0.05. Significant differences in the experiment with the natural bacterial assemblage were tested with a Student t-test. Statistical tests were carried out using SigmaStat (Systat, Software Inc., San Jose, CA).
Results
Growth conditions and bacterial generation time
After an overall lag phase of about 24 h, all strains entered in exponential growth for a minimum of 73 h (RC1) to a maximum of 141 h (GKS12b) before they reached the stationary phase (Fig. 1). These results indicated that the isolates from alpine lakes and rain were able to grow on glucose as the sole source of carbon. GKS10 was the fastest growing strain (μ = 0.050 h−1) and had the shortest generation time (g = 13.9 h), while GKS12b was the slowest growing strain (μ = 0.019 h−1) with the longest generation time (g = 37.0 h) (Table 1). The highest bacterial abundance at the end of the exponential phase was found for MHWTa3 (2.3 × 109 cells ml−1; Fig. 1), whereas the lowest one was observed in RC1 (4.3 × 106 cells ml−1; Fig. 1).
Changes in bacterial abundance during and after exposure
As revealed by the one-way repeated measures ANOVA and post hoc comparisons, bacterial abundance in GKS10 significantly decreased between T0 and T4 in the PAR + UVR treatment, but also in the DARK control, whereas between T4 and Tend it decreased in all treatments (Fig. 2). Similarly, in RC1 bacterial numbers decreased between T0 and T4 in all treatments, but remained constant afterward. In GKS 12b, bacterial numbers significantly increased between T0 and T4 in the DARK and under PAR, but not under PAR + UVR (Fig. 2). After exposure, numbers remained unchanged. By contrast, bacterial abundance of strain SOS1 increased during exposure in all treatments and also after exposure, except for the PAR + UVR treatment, where changes between T4 and Tend were not significant. Strain SOS1 was the only isolate where differences in bacterial numbers between the control and treatments were significant at Tend (Holm-Sidak post hoc analysis). Finally, bacterial abundance in MHWTa3 increased in all treatments between T0 and T4 and remained unchanged afterward (Fig. 2).
Changes in the bacterial abundance of the five strains during and after exposure to PAR, PAR + UVR, or kept in the dark. The different times after exposure depended on achieving a significant change in oxygen consumption. Cell numbers are expressed as the mean of three replicates ±1 SD. In most measurements, error bars are smaller than the symbols. Solid symbols represent the dark control, gray symbols the PAR treatment, and open symbols the PAR + UVR treatment. The vertical dashed line represents the end of the exposure to the different treatments
Bacterial respiration rate
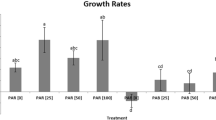
In the DARK control, SOS1 showed the highest bulk oxygen consumption rate, whereas RC1 had the lowest one (Fig. 3). SOS1 had also the highest cell-specific respiration rate (1.8 × 10−8 μg C cell−1 h−1) followed by GKS10 (9.1 × 10−9 μg C cell−1 h−1), GKS12b (8.0 × 10−9 μg C cell−1 h−1), RC1 (6.8 × 10−9 μg C cell−1 h−1), and MHWTa3 (4.2 × 10−10 μg C cell−1 h−1). For all strains, no significant differences in cell-specific respiration rates were found between the DARK control and the PAR treatment (Fig. 4). By contrast, exposure to PAR + UVR significantly decreased cell-specific respiration rates by 74.0 ± 2.2% (Holm-Sidak, P < 0.05) in the isolate SOS1, but not in the others (Fig. 4). In GKS12b, bacterial respiration in the presence of PAR + UVR was not detectable within the measurement period (Fig. 5). Respiration rates of the three other strains decreased (MHWTa3), increased (RC1), or were similar (GKS10) to the control, but changes were not significant (Fig. 4).
a Cell-specific bacterial carbon production (BCP), b cell-specific bacterial respiration (BR), and c bacterial growth efficiency (BGE) of strains GKS10, GKS12b, SOS1, RC1, and MHWTa3 after exposure to photosynthetically active radiation (PAR) and PAR + UVR. Mean values (n = 3) are expressed as percentage of the DARK control (horizontal line reference) ±1 SD. The asterisks above the bar summarize the outcome of the post hoc Holm-Sidak test and indicate a significant difference between the DARK control and the respective treatment. Significance level: * P < 0.05, ** P < 0.01, and *** P < 0.001. nd not detectable
Bacterial carbon production
The highest cell-specific bacterial carbon production in the DARK control corresponded to the RC1 strain (4.83 × 10−9 μg C cell−1 h−1), followed by GKS10 (3.31 × 10−9 μg C cell−1 h−1), GKS12b (2.21 × 10−9 μg C cell−1 h−1), SOS1 (1.03 × 10−10 μg C cell−1 h−1), and MHWTa3 (9.12 × 10−13 μg C cell−1 h−1). After exposure to PAR, no significant changes in bacterial carbon production were observed in all strains (Fig. 4). However, after exposure to PAR + UVR, bacterial carbon production significantly decreased in three out of five strains (Holm-Sidak, P < 0.05) showing a mean reduction of 97.8 ± 1.6% (GKS12b), 87.2 ± 0.9% (RC1), and 91.8 ± 3.0% (MHWTa3) when compared to the DARK control (Fig. 4). The decrease in bacterial carbon production of GKS10 (mean reduction of 36.0%) and SOS1 (mean reduction of 30.3%) after exposure to PAR + UVR was not significantly different from the DARK control (Fig. 4).
Bacterial growth efficiency
The highest BGE in the DARK was found in RC1 (42.3 ± 8.8%) followed by GKS10 (26.8 ± 7.0%) and SOS1 (0.6% ± 0.1), while the lowest value was detected in MHWTa3 (0.2% ± 0.08). Exposure to PAR did not have a significant effect on bacterial growth efficiency, except for strain GKS10 where it slightly increased (Fig. 4), whereas the effect of UVR was strain-specific. Thus, significantly lower bacterial growth efficiency values were found for RC1 (12.4 ± 1.0%) and for MHWTa3 (20.7 ± 11.5%) after exposure to PAR + UVR when compared to the DARK control (Fig. 4). The BGE in GKS10 decreased to 73.6 ± 4.8% in the presence of UVR, but the change was not significantly different when compared to the DARK (Fig. 4). By contrast, a significantly higher BGE value was found in SOS1 (Fig. 4).
Effect of PAR + UVR on respiration of a natural bacterial community
Exposure of the natural bacterial assemblage from Schwarzsee ob Sölden to PAR + UVR increased the respiration rate significantly by 1.8-fold (t-test, P < 0.05) when compared to the DARK control (Fig. 6).
Discussion
Our results showed that bacteria isolated from different freshwater habitats such as alpine lakes and rain, but grown under the same conditions, significantly differ in their growth efficiency even in the absence of UVR. Different growth efficiencies can result from the fact that even during unconstrained growth, bacteria use different amounts of energy for maintenance of metabolic processes instead of allocating it into new biomass (Russell and Cook 1995). Furthermore, variations in bacterial growth efficiency are often found when growth is limited by substrate availability (del Giorgio and Cole 1998). However, the latter source of variation can be neglected for the bacterial isolates we tested because they were in the exponential phase of growth. Whereas information on how efficiently aquatic bacteria convert DOM into bacterial biomass is essential to understand distribution patterns within bacterial communities, our experiment was not designed to resemble bacterial growth under natural conditions, where a complex mixture of different organic substrates is available. In fact, the growth efficiency values we estimated might be very different when bacteria grow in a more complex and organic-rich medium or in their original water (del Giorgio and Cole 1998). Our aim, however, was to be able to compare the effect of UVR on bacterial growth efficiency under the same conditions for all strains. Thus, whereas we cannot exclude probable physiological changes caused by the medium used, all isolates were able to reach exponential growth though at different rates (Fig. 1).
In general, exposure to UVR had an inhibitory effect on carbon production in all bacterial strains, although only in three strains a significant decrease was observed (Fig. 4a). This negative effect was also observed when changes in bacterial numbers were accounted for during the experiment (Figs. 2 and 4). Solar UVR and in particular the high energetic UV-B radiation (280–315 nm) is known to cause harmful effects in aquatic bacteria (Herndl and Obernosterer 2002; Buma et al. 2003), such as decrease in viability (Joux et al. 1999; Davidson and van der Heijden 2000; Agogué et al. 2005) or inhibition of secondary production (Herndl et al. 1993; Sommaruga et al. 1997; Hernandez et al. 2007). However, the sensitivity to UVR and the degree of damage are highly strain-specific (Arrieta et al. 2000; Zenoff et al. 2006b) and are affected among others factors by the existence of UVR protection mechanisms (Buma et al. 2003), previous exposure to increased levels of UVR (Gustavson et al. 2000), and DNA repair efficiency (Matallana-Surget et al. 2009).
Among the isolates tested in our study, the Gram-negative strain SOS1 (Acinetobacter lwoffii) had the highest rate of cell-specific bacterial production (Fig. 4a) and though there was a reduction (non significant) after exposure to UVR, this bacterium seems to be relatively tolerant against the stress imposed. In fact, strain SOS1 together with strain MHWTa3 were the only ones where a significant increase in bacterial abundance was observed during exposure to UVR, though afterward, numbers remained unchanged (Fig. 2). Strain SOS1 belongs to the Gammaproteobacteria and was isolated from the high-altitude lake Schwarzsee ob Sölden where this group comprises a very small fraction of the total bacterial community. Members of this group seem to be well adapted to the high UVR intensities present in this type of environment. In a study by Zenoff et al. (2006a), the unpigmented strain Acinetobacter johnsonii A6, isolated from an oligotrophic lake in the Antarctic, was similarly resistant to UVR as other Gram-positive pigmented bacteria tested in the same study. Members of the Acinetobacter group are unique regarding their high resistance to desiccation, H2O2 exposure, and even gamma radiation (La Duc et al. 2003). These findings indicate that despite weaker cell wall characteristics than Gram-positive bacteria and lack of pigmentation, representatives of this group are highly tolerant to UVR.
Similar to SOS1, GKS10 (Sphingomonas sp. B14) showed no significant reduction in bacterial carbon production after exposure to UVR (Fig. 4a), suggesting a high tolerance. Members of the genus Sphingomonas seem to be highly resistant to UV-B radiation due to their low accumulation of cyclobutane pyrimidine dimers (Joux et al. 1999). Additionally, many strains of Sphingomonas have pigments (also strain GKS10, Table 1) that can probably minimize the negative effects of UVR (Buma et al. 2003). By contrast, strains GKS12b (Acidovorax sp.), RC1 (Pseudomonas sp.), and MHWTa3 (Actinobacterium MHWTa3 showed a high UV sensitivity (Fig. 4a). Whereas information on UV sensitivity of members of the genus Acidovorax or Actinobacterium is not available, that on Pseudomonas suggests that members of this genus can be very sensitive to UVR (Fernández and Pizarro 1996; Zenoff et al. 2006b) and may lack DNA repair mechanisms (Simonson et al. 1990; Kidambi et al. 1996). Interestingly, despite their presumable high sensitivity to UVR, members of the genus Pseudomonas are known to be transported through the atmosphere (Amato et al. 2007), where a general stress tolerance is probably necessary.
Regarding bacterial respiration, no general trend was observed after exposure to UVR. Although bacterial respiration decreased in three strains, only SOS1 showed a significant decrease (Fig. 4b). The undetectable bacterial respiration values in strain GKS12b suggest also a severe inhibition (Fig. 5). By contrast, the increase in bacterial respiration found in RC1 (Fig. 4b and 5) could be an indication of a high sensitivity to UVR because metabolic reactions and associated biochemical oxygen demand tend to increase under physiological stress (Aertsen and Michiels 2004; Hecker et al. 2007).
Antagonistic results of UVR effects on respiratory activity have been found for several bacterial freshwater communities. For example, Rae and Vincent (1998) found a significant UV inhibition of actively respiring bacteria, while Ferreyra et al. (1997) detected significantly higher electron transport system activities and consequently higher bacterial respiration in a natural plankton community after exposure to UV-B radiation. Probable reasons for those contrasting results can be differences in bacterial community composition (Reinthaler et al. 2005), as well as changes in bacterial community structure during respiration measurements (Gattuso et al. 2002). Our own results with the natural bacterial community of Schwarzsee ob Sölden indicate that UVR resulted in higher oxygen consumption rates (Fig. 6). These results are interesting because they illustrate the contrasting response of the whole bacterial community (i.e., ‘black box’ approach) and strain SOS1 isolated from the same lake. Though we do not know what the representation of strain SOS1 was in the natural bacterial community of Schwarzsee ob Sölden at the time of the experiment, these results suggest that it may be difficult to interpret bulk respiration rates in natural bacterial communities exposed to solar UVR.
The tested isolates showed contrasting changes in their bacterial growth efficiency after exposure to UVR (Fig. 4c). Different bacterial isolates are known to vary in their sensitivity to UVR (Fernández and Pizarro 1996; Arrieta et al. 2000), but hardly any information is available on changes of strain-specific bacterial growth efficiency after UV exposure. Although the strong inhibition of bacterial growth efficiency by UVR was not totally unexpected, we did not anticipate to observe a negative effect for widespread bacterial taxa such as Acidovorax and Pseudomonas (Fig. 4C). Members of those groups have been found in various freshwater systems and are present even in extreme habitats with high UVR conditions (Amato et al. 2007; Hervàs et al. 2009). Generally, bacterial isolates display large interspecific differences in their recovery efficiencies after UV exposure, as shown for marine bacterioplankton (Kaiser and Herndl 1997) and for marine bacterial isolates (Joux et al. 1999; Arrieta et al. 2000). Strains of Acinetobacter johnsonii were found to endure and even to recover very rapidly from UV damage, despite high accumulation of cyclobutane pyrimidine dimers (Zenoff et al. 2006b). By contrast, strains of Pseudomonas spp. do not recover (Zenoff et al. 2006b). In our study, SOS1 (Acinetobacter lwoffii, Fig. 4C) was the only strain showing a high tolerance and even a significant increase in bacterial growth efficiency after UV exposure. This strain probably has effective repair mechanisms that allows for maintaining cell production and thus to increase bacterial growth efficiency after UV exposure. The second strain showing signs of UV tolerance and a relatively low decrease in bacterial growth efficiency after UV exposure was GKS10 (Sphingomonas sp. B14, Fig. 4c).
Overall, strain-specific effects on bacterial growth efficiency were mainly driven by the changes in bacterial production. These results agree with those of other studies (Toolan 2001; Reinthaler and Herndl 2005) that showed growth efficiencies of natural bacterial assemblages mainly reflect changes in bacterial production rather than in respiration. Nevertheless, as shown in our study, the effect of UVR on bacterial respiration was strain-specific and rates decreased or increased after exposure. Though in the experiment with the natural bacterial community of Schwarzsee ob Sölden, we did not measure bacterial production concomitantly; previous results from a transparent alpine lake such as this showed that this process is strongly inhibited by UVR (Sommaruga et al. 1997). Thus, the most probable outcome of the effect of UVR on this natural bacterial community was a reduction in bacterial growth efficiency.
In summary, our findings indicate that changes in metabolic rates caused by UVR were highly strain-specific and that variations in bacterial growth efficiency mainly reflected the response in production of new bacterial biomass to this environmental stressor. Our results underline the role UVR has in potentially altering the efficiency by which the dissolved organic matter pool is transformed into bacterial biomass in aquatic ecosystems. Furthermore, the alterations caused by UVR may have consequences for the efficiency of carbon cycling, especially considering that climate change will affect the UV transparency of freshwaters (Adrian et al. 2009).
References
Adrian R, O’Reilly CM, Zagarese H, Baines SB, Hessen DO, Keller W, Livingstone DM, Sommaruga R, Straile D, Van Donk E, Weyhenmeyer GA, Winder M (2009) Lakes as sentinels of climate change. Limnol Oceanogr 54:2283–2297
Aertsen A, Michiels C (2004) Stress and how Bacteria cope with death and survival. Crit Rev Microbiol 30:263–273
Agogué H, Joux F, Obernosterer I, Lebaron P (2005) Resistance of marine bacterioneuston to solar radiation. Appl Environ Microbiol 71:5282–5289
Alonso-Sáez L, Gasol JM, Lefort T, Hofer J, Sommaruga R (2006) Effect of natural sunlight on bacterial activity and differential sensitivity of natural bacterioplankton groups in NW Mediterranean coastal waters. Appl Environ Microbiol 72:5806–5813
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res 25:3389–3402
Amato P, Parazols M, Sancelme M, Laj P, Mailhot G, Delort AM (2007) Microorganisms isolated from the water phase of tropospheric clouds at the Puy de Dome: major groups and growth abilities at low temperatures. FEMS Microbiol Ecol 59:242–254
Arrieta JM, Weinbauer MG, Herndl GJ (2000) Interspecific variability in sensitivity to UV radiation and subsequent recovery in selected isolates of marine bacteria. Appl Environ Microbiol 66:1468–1473
Biddanda B, Opsahl S, Benner R (1994) Plankton respiration and carbon flux through bacterioplankton on the Louisiana shelf. Limnol Oceanogr 39:1259–1275
Boelen P, Veldhuis MJW, Buma AGJ (2001) Accumulation and removal of UVBR-induced DNA damage in marine tropical plankton subjected to mixed and simulated non-mixed conditions. Aquat Microb Ecol 24:265–274
Briand E, Pringault O, Jacquet S, Torreton JP (2004) The use of oxygen microprobes to measure bacterial respiration for determining bacterioplankton growth efficiency. Limnol Oceanogr Meth 2:406–416
Buesing N, Marxsen J (2005) Theoretical and empirical conversion factors for determining bacterial production in freshwater sediments via leucine incorporation. Limnol Oceanogr Meth 3:101–107
Buma AGJ, Boelen P, Jeffrey WH (2003) UVR-induced DNA damage in aquatic organsims. In: Helbling EW, Zagarese HE (eds) UV effects in aquatic organisms and ecosystems. Royal Society of Chemistry, Cambridge, UK, pp 293–327
Chatila K, Demers S, Mostajir B, Gosselin M, Chanut JP, Monfort P, Bird D (2001) The responses of a natural bacterioplankton community to different levels of ultraviolet-B radiation: a food web perspective. Microb Ecol 41:56–68
Davidson AT, van der Heijden A (2000) Exposure of natural Antarctic marine microbial assemblages to ambient UV radiation: effects on bacterioplankton. Aquat Microb Ecol 21:257–264
del Giorgio PA (1992) The relationship between ETS (electron transport system) activity and oxygen consumption in lake plankton: a cross-system calibration. J Plankton Res 14:1723–1741
del Giorgio PA, Cole JJ (1998) Bacterial growth efficiency in natural aquatic systems. Annu Rev Ecol Syst 29:503–541
del Giorgio PA, Williams PJleB (2005) Respiration in Aquatic Ecosystems. Oxford University Press, Oxford 326 p
Fernández RO, Pizarro RA (1996) Lethal effect induced in Pseudomonas aeruginosa exposed to Ultraviolet-A radiation. Photochem Photobiol 64:334–339
Ferreyra GA, Demers S, del Giorgio PA, Chanut JP (1997) Physiological responses of natural plankton communities to ultraviolet-B radiation in Redberry Lake (Saskatchewan, Canada). Can J Fish Aquat Sci 54:705–714
Fuhrman JA, Azam F (1980) Bacterioplankton secondary production estimates for coastal waters of British Columbia, Antarctica, and California. Appl Environ Microbiol 39:1085–1095
Garde K, Gustavson K (1999) The impact of UV-B radiation on alkaline phosphatase activity in phosphorus-depleted marine ecosystems. J Exp Mar Biol Ecol 238:93–105
Gattuso JP, Peduzzi S, Pizay MD, Tonolla M (2002) Changes in freshwater bacterial community composition during measurements of microbial and community respiration. J Plankton Res 24:1197–1206
Gottschalk G (1986) Bacterial metabolism. Springer-Verlag, New York 359 p
Gustavson K, Garde K, Wangberg SA, Selmer JS (2000) Influence of UV-B radiation on bacterial activity in coastal waters. J Plankton Res 22:1501–1511
Hahn MW, Lünsdorf H, Wu Q, Schauer M, Höfle MG, Boenigk J, Stadler P (2003) Isolation of novel ultramicrobacteria classified as actinobacteria from five freshwater habitats in Europe and Asia. Appl Environ Microbiol 69:1442–1451
Hecker M, Pane-Farre J, Volker U (2007) SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu Rev Microbiol 61:215–236
Helbling EW, Marguet ER, Villafañe VE, Holm-Hansen O (1995) Bacterioplankton viability in Antarctic waters as affected by solar ultraviolet-radiation. Mar Ecol Prog Ser 126:293–298
Hernandez KL, Quiñones RA, Daneri G, Farias ME, Helbling EW (2007) Solar UV radiation modulates daily production and DNA damage of marine bacterioplankton from a productive upwelling zone (36 degrees S), Chile. J Exp Mar Biol Ecol 343:82–95
Herndl GJ, Obernosterer I (2002) UV radiation and pelagic bacteria. In: Hessen DO (ed) UV Radiation and Arctic Ecosystems. Ecological Studies, vol 153. Springer-Verlag, Berlin, pp 245–259
Herndl GJ, Muller-Niklas G, Frick J (1993) Major role of ultraviolet-B in controlling bacterioplankton growth in the surface-layer of the Ocean. Nature 361:717–719
Hervàs A, Camarero L, Reche I, Casamayor EO (2009) Viability and potential for immigration of airborne bacteria from Africa that reach high mountain lakes in Europe. Environ Microbiol 11:1612–1623
Jeffrey WH, Kase JP, Willhelm SW (2000) UV radiation effects on heterotrophic bacterioplankton and viruses in marine environment. In: de Mora S, Demers S, Vernet M (eds) The effects of UV radiation in the marine environment. Cambridge University Press, Cambridge, pp 206–236
Joux F, Jeffrey WH, Lebaron P, Mitchell DL (1999) Marine bacterial isolates display diverse responses to UV-B radiation. Appl Environ Microbiol 65:3820–3827
Kaiser E, Herndl GJ (1997) Rapid recovery of marine bacterioplankton activity after inhibition by UV radiation in coastal waters. Appl Environ Microbiol 63:4026–4031
Kane MD, Poulsen LK, Stahl DA (1993) Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl Environ Microbiol 59:682–686
Kidambi SP, Booth MG, Kokjohn TA, Miller RV (1996) RecA-dependence of the response of Pseudomonas aeruginosa to UVA and UVB irradiation. Microbiology 142:1033–1040
Kirchman D, K’Nees E, Hodson R (1985) Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl Environ Microbiol 49:599–607
La Duc MT, Nicholson W, Kern R, Venkateswaran K (2003) Microbial characterization of the Mars Odyssey spacecraft and its encapsulation facility. Environ Microbiol 5:977–985
Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR (1985) Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA 82:6955–6959
Laurion I, Ventura M, Catalan J, Psenner R, Sommaruga R (2000) Attenuation of ultraviolet radiation in mountain lakes: Factors controlling the among- and within-lake variability. Limnol Oceanogr 45:1274–1288
Maranger R, del Giorgio PA, Bird DF (2002) Accumulation of damaged bacteria and viruses in lake water exposed to solar radiation. Aquat Microb Ecol 28:213–227
Matallana-Surget S, Douki T, Cavicchioli R, Joux F (2009) Remarkable resistance to UVB of the marine bacterium Photobacterium angustum explained by an unexpected role of photolyase. Photochem. Photobiol. Sci. 8:1313–1320
Müller DR, Warwick VF, Bonilla S, Laurion I (2005) Extremotrophs, extremophiles and broadband pigmentation strategies in a high arctic ice shelf ecosystem. FEMS Microbiol Ecol 53:73–87
Pakulski JD, Aas P, Jeffrey W, Lyons M, Von Waasenbergen L, Mitchell D, Coffin R (1998) Influence of light on bacterioplankton production and respiration in a subtropical coral reef. Aquat Microb Ecol 14:137–148
Pérez MT, Hörtnagl P, Sommaruga R (2010) Contrasting ability to take up leucine and thymidine among freshwater bacterial groups: implications for bacterial production measurements. Environ Microbiol 12:74–82
Porter KG, Feig YS (1980) The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr 25:943–948
Pringault O, Tesson S, Rochelle-Newall E (2009) Respiration in the light and bacterio-phytoplankton coupling in a coastal environment. Microb Ecol 57:321–334
Rae R, Vincent WF (1998) Effects of temperature and ultraviolet radiation on microbial foodweb structure: potential responses to global change. Freshwater Biol 40:747–758
Reche I, Pace ML, Cole JJ (1998) Interactions of photobleaching and inorganic nutrients in determining bacterial growth on colored dissolved organic carbon. Microb Ecol 36:270–280
Reinthaler T, Herndl GJ (2005) Seasonal dynamics of bacterial growth efficiencies in relation to phytoplankton in the southern North Sea. Aquat Microb Ecol 39:7–16
Reinthaler T, Winter C, Herndl GJ (2005) Relationship between bacterioplankton richness, respiration, and production in the southern North Sea. Appl Environ Microbiol 71:2260–2266
Revsbech NP (1989) An oxygen microsensor with a guard cathode. Limnol Oceanogr 34:474–478
Russell JB, Cook GM (1995) Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol Rev 59:48–62
Sieracki ME, Sieburth JM (1986) Sunlight induced growth delay of planktonic marine bacteria in filtered seawater. Mar Ecol Prog Ser 33:19–27
Simonson CS, Kokjohn TA, Miller RV (1990) Inducible UV repair potential of Pseudomonas aeruginosa PAO. J Gen Microbiol 136:1241–1249
Sommaruga R, Oberleiter A, Psenner R (1996) Effect of UV radiation on the bacterivory of a heterotrophic nanoflagellate. Appl Environ Microbiol 62:4395–4400
Sommaruga R, Obernosterer I, Herndl GJ, Psenner R (1997) Inhibitory effect of solar radiation on thymidine and leucine incorporation by freshwater and marine bacterioplankton. Appl Environ Microbiol 63:4178–4184
Toolan T (2001) Coulometric carbon-based respiration rates and estimates of bacterioplankton growth efficiencies in Massachusetts Bay. Limnol Oceanogr 46:1298–1308
Warkentin M, Freese HM, Karsten U, Schumann R (2007) New and fast method to quantify respiration rates of bacterial and plankton communities in freshwater ecosystems by using optical oxygen sensor spots. Appl Environ Microbiol 73:6722–6729
Zenoff V, Heredia J, Ferrero M, Sineriz F, Farias M (2006a) Diverse UV-B resistance of culturable bacterial community from high-altitude wetland water. Curr Microbiol 52:359–362
Zenoff VF, Sineriz F, Farias ME (2006b) Diverse responses to UV-B radiation and repair mechanisms of bacteria isolated from high-altitude aquatic environments. Appl Environ Microbiol 72:7857–7863
Acknowledgments
We thank J. Hofer for helping with the isolation and cultivation of bacterial strains, S. Morales-Gómez for helping to determine bacterial numbers, M. Hahn for providing isolate MHWTa3, and two anonymous reviewers for their comments. This work was supported by the Austrian Science Fund (FWF) through a research project (P19245-BO3) to R. Sommaruga.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Piet Spaak.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Hörtnagl, P., Pérez, M.T. & Sommaruga, R. Contrasting effects of ultraviolet radiation on the growth efficiency of freshwater bacteria. Aquat Ecol 45, 125–136 (2011). https://doi.org/10.1007/s10452-010-9341-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10452-010-9341-9