Abstract
Linear scaling of generic shoulder models leads to substantial errors in model predictions. Customisation of shoulder modelling through magnetic resonance imaging (MRI) improves modelling outcomes, but model development is time and technology intensive. This study aims to validate 10 MRI-based shoulder models, identify the best combinations of anthropometric parameters for model scaling, and quantify the improvement in model predictions of glenohumeral loading through anthropometric scaling from this anatomical atlas. The shoulder anatomy was modelled using a validated musculoskeletal model (UKNSM). Ten subject-specific models were developed through manual digitisation of model parameters from high-resolution MRI. Kinematic data of 16 functional daily activities were collected using a 10-camera optical motion capture system. Subject-specific model predictions were validated with measured muscle activations. The MRI-based shoulder models show good agreement with measured muscle activations. A tenfold cross-validation using the validated personalised shoulder models demonstrates that linear scaling of anthropometric datasets with the most similar ratio of body height to shoulder width and from the same gender (p < 0.04) yields best modelling outcomes in glenohumeral loading. The improvement in model reliability is significant (p < 0.02) when compared to the linearly scaled-generic UKNSM. This study may facilitate the clinical application of musculoskeletal shoulder modelling to aid surgical decision-making.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Validated computational models of the musculoskeletal (MSK) system can be used to understand normal and pathological human movement by predicting articular and tissue loading, parameters that cannot currently be measured directly. The precise knowledge of musculoskeletal loading is essential for clinical applications in order to improve surgical and rehabilitative treatment planning, assistive device design and analysis of joint arthroplasty design.
The use of musculoskeletal models in clinical practice has been hampered by the dependency of modelling results on model input, in particular the anatomical geometry.4,6 Linearly scaled-generic models, derived from the dissection of cadaveric specimens, are widely used to represent a subject’s anatomical geometry.5,8,14,17,26,32 These models accommodate geometric variation across subjects through linear scaling, based on three-dimensional positions of anatomical landmarks.25 As linearly scaled-generic models do not account for individual variations in anthropometry such as muscle attachment sites and muscle volumes, they lead to errors in muscle path estimations that will result in substantial inaccuracies in calculated muscle and joint forces.18,38
Driven by the need for more accurate model predictions in clinical settings and facilitated by advances in medical imaging technology, subject-specific modelling techniques are becoming more widely developed.4,46 Muscle moment arm predictions from a personalised musculoskeletal model constructed from magnetic resonance imaging (MRI) have been compared to those from a linearly scaled-generic model of the lower limb,14 demonstrating significantly improved model estimations when compared to cadaveric measurements.38,39 The use of an EMG-driven neuromusculoskeletal lower limb model has shown improvements in joint contact force predictions through subject-specific digitisation of joint centres and muscle attachment sites from medical imaging.19 Similarly, a customised musculoskeletal modelling framework for the evaluation of shoulder muscle and joint function during activities of daily living by Wu et al.46 demonstrated that linearly scaled-generic models do not reproduce muscle loading obtained from linearly-scaled subject-specific models. The uncertainty of musculoskeletal model predictions by linearly scaled-generic models may be as high as 20% when compared to highly-detailed, linearly scaled subject-specific musculoskeletal models,29 with modelling errors being dependent on anthropometric differences between scaled-generic and subject-specific models.4
Customisation of musculoskeletal modelling through medical imaging significantly improves model reliability, when compared to an individual linearly scaled-generic model.19,39 However, the development of subject-specific computational models is time, labour and technology intensive. In order to overcome the drawbacks of generating a personalised dataset for each subject, linear scaling of musculoskeletal shoulder models with high anthropometric similarity from a model database, or atlas, has the potential to yield modelling results that are close to predictions of subject-specific models and more accurate than a single, scaled-generic model. As the number of openly available anatomical datasets with quantification of all upper limb muscle attachment sites and muscle volumes for shoulder modelling in the literature is small,42 this study aims to develop and validate 10 MRI-based shoulder models against measured muscle activations and muscle moment arms, demonstrate the dependency of modelling results (glenohumeral joint contact and muscle forces) on anatomical geometry, identify the best combinations of anthropometric parameters that yield the smallest error in model estimations of glenohumeral joint contact force and muscle forces through scaling of personalised musculoskeletal shoulder models, and quantify the improvement in model reliability through anthropometric scaling of anatomical datasets when compared to a single, scaled-generic model. The MRI-based musculoskeletal models as well as anatomical datasets developed in this study are available at www.msksoftware.org.uk.
Materials and Methods
Experimental Data
Ten healthy volunteers (five male, five female) with a wide variation in height participated in this study (Table 1). Subjects were classified as tall (height > 90th percentile), medium (height 50th–90th percentile) and short (height < 50th percentile) according to anthropometric estimates for British adults.35 The study was granted ethical approval by the NHS Health Research Authority and the Imperial College Research Ethics Committee.
All subjects were instructed to perform sixteen functional activities of daily living with three sets per activity (Table 2).28 Kinematic data collection was performed using a 10-camera optical motion tracking system (Vicon Motion Tracking System, Oxford, UK) with three load cells (Kistler Type 9286B, Kistler Instrument Corp., Winterthur, Switzerland).
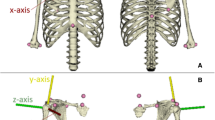
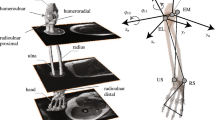
The marker set comprised anatomical landmarks of the upper limb including markers on the radial/ulna styloid (RS/US), medial/lateral epicondyles (ME/LE), right/left Acromioclavicular joint (RAC/LAC), right/left Sternoclavicular joint (RSC/LSC), Xiphoid Process (PX), Manubrium (MA), Jugular Notch (IJ) and 7th cervical vertebra (C7) as well as clusters of three markers on the forearm and upper arm.41 Scapula kinematics were measured using a scapula tracker.37
Surface electromyography (EMG; Myon 320, Myon AG, Switzerland) was recorded at 1000 Hz from five muscles during the activities as well as for maximum voluntary contraction tests: deltoid (DELT), pectoralis (PEC), trapezius (TRAP), biceps (BIC) and triceps (TRI). The electrodes were placed with an orientation parallel to the muscle fibres; full details for electrode placement and maximum voluntary contraction tests are provided in De Groot et al.11
MRI data of the upper limb were collected from study participants within 2 months of motion analysis. A 3D T1-weighted VIBE (volumetric interpolated breath-hold examination) sequence was used to acquire axial images of the subjects in the supine position using a 3T Siemens Verio MRI scanner (Siemens AG, Erlangen, Germany). The signal was received through a spine coil and two body matrix coils, with MRI protocol settings as follows: imaging field of view 450 × 450 mm,2 matrix = 384 × 384, axial plane resolution 1.17 × 1.17 mm, slice thickness 1 mm. The field of view (FOV) included the right half of the upper limb, scanning from skull to finger tips in axial blocks of length 22.5 cm, with 2.5 cm overlap between adjacent sections. The MRI scan consisted of five sections for the tallest participant and 3 sections for the shortest subject, with 5 min, 43 s acquisition time per section.
Musculoskeletal Geometry
For each volunteer, the anatomical geometry of the upper limb was described through a linearly scaled-generic model. The anatomical geometry of the scaled-generic model consists of 6 rigid bone elements and 87 muscle elements, crossing five functional joints (sternoclavicular, acromioclavicular, scapulathoracic, glenohumeral and elbow). The skeletal geometry of the linearly scaled-generic model as fully described in Charlton and Johnson8 was obtained from the male visible human dataset,42 while muscle properties, including muscle attachment sites, physiological cross-sectional area (PCSA), optimal fiber lengths and pennation angles, were taken from three different cadaveric studies.24,44,45 The segments (thorax, scapula, clavicle, humerus, radius, ulna) of each linearly scaled-generic model were scaled to ensure that intersegmental distances of the model match the corresponding distances of the participants, measured based on anatomical landmarks during a static trial in a motion analysis lab. The segment scaling utilises linear scaling for all segments except the thorax, which involves scaling of thorax height as the distance between PX and the midpoint of IJ and C7, thorax width as the distance between IJ and RAC, and thorax depth as the distance between C7 and the midpoint of PX and IJ. The optimal fiber length and tendon slack length are scaled with segment length and this is accounted for in the calculation of the PCSA as described in Charlton and Johnson.8
In addition to the linearly scaled-generic model to describe the musculoskeletal geometry of each study participant, a subject-specific model from MRI was developed for each study volunteer (Fig. 1). The MRI-based models were developed following exactly the same topology of the generic dataset as presented in the previous paragraph and fully described by Charlton and Johnson.8 All upper limb muscles for each study participant were manually segmented and the segmentations served to compute subject-specific PCSA for each muscle. All segmentations were performed by one individual. The calculation of PCSA included scaling of each muscle by cosine of the pennation angle, where values for pennation angles and optimal fiber length were obtained from the literature,34 thus this study did not include personalisation of pennation angles and optimal fiber lengths as these were not measured directly in this study. Similarly to the generic model, the scaling of optimal fiber length and tendon slack length with segment length is accounted for in the calculation of subject-specific PCSA for each muscle as described in Charlton and Johnson.8 In addition to subject-specific PCSA calculation from MRI, muscle origins, via points and insertion points were obtained through manual digitisation from MRI. The muscles were simulated to wrap around bony segments of the upper limb that were described with geometric wrapping objects between muscle origins and insertions. The thorax and the scapulathoracic gliding plane were modelled with two wrapping ellipsoids, while humerus, radius and ulna were described with wrapping cylinders, with full details being provided in Charlton and Johnson.8 The glenohumeral joint center of rotation was defined as the centre of the humeral head which was determined with a sphere fit.8 The wrapping object parameters were determined in two steps: an initial estimation from the scaled-generic model and then a manual adjustment based on the segmented MRI to ensure that the muscles wrapped smoothly over the subject-specific bony contour. All image segmentations and processing steps were performed using Mimics (Mimics Research 17.0, Materialise NV, Leuven, Belgium). Subject-specific segmental parameters including centre of mass and moments of inertia were determined through regression equations as described by De Leva,12 rather than through the use of subject-specific MRI.
Musculoskeletal Simulation
The motion data and external forces for each functional activity served as inputs into the UK National Shoulder Model (UK NSM) as described by Charlton and Johnson,8 which represents a 3D musculoskeletal modelling software written in Matlab (The Mathworks Inc., Cambridge, UK) that models forces at the glenohumeral joint. The version of the UK NSM used in this study additionally incorporates the effect of glenohumeral concavity compression (through simulation of the labral contribution to joint stability)27 as well as subject-specific scapular tracking (personalised scapula motion as model input).37 Each functional activity of daily living was simulated using the linearly scaled-generic model and the MRI-based model.
The UK NSM is an inverse dynamics musculoskeletal model, with intersegmental moments being calculated with measured kinematics and a partially closed chain method in order to optimise scapula and clavicle kinematics.36 The load-sharing optimisation minimises the sum of squared muscle stresses to solve the natural muscle load-sharing redundancy, with maximum muscle forces being unbounded. The model predictions of the original UKNSM have been validated through comparison to instrumented implant measurements and electromyography for functional activities with a similar range of motion compared to the functional tasks of this study.8
Evaluation of Generic and MRI-Based Models
Surface EMG signals of the upper limb were processed to evaluate muscle activations predicted by the MRI-based and scaled-generic model. The raw EMG signals were high-pass filtered at 30 Hz, fourth order Butterworth filtered and rectified. The rectified signals were low-pass filtered at 10 Hz.2 For each subject, data were normalised based on maximum EMG signals during maximum voluntary contraction tests.2
The muscle force predictions of the MRI-based and linearly scaled-generic models were analysed as described in Klemt et al.28 In brief, once the start and end point of each functional activity was established, the muscle forces and joint forces were quantified at each time point. Interpolation using a cubic spline function was used to allow comparison between measurements for different activities and trials by representing each of them by 100 time points. The muscle force predictions were normalised by the maximum muscle force of each muscle.
Quantitative evaluation between EMG data and predicted muscle activations by MRI-based and generic models was conducted using Sprague and Geers metrics of magnitude (M), phase (P) and combined error (C).40 The Sprague and Geers metrics quantify the magnitude and phase error independently, while C combines the two errors and is computed as the root of the sum of squares of M and P. The interpretation of the Geers metric is as follows:
-
0 < Geers Metric < 0.15 excellent similarity;
-
0.15 < Geers Metric < 0.30 very good similarity;
-
0.30 < Geers Metric < 0.45 good similarity;
-
0.45 < Geers Metric < 0.60 moderate similarity; and
-
Geers Metric > 0.6 no similarity.
The similarity of waveforms between EMG data and predicted muscle activations by MRI-based and generic models was assessed using the coefficient of multiple correlation (CMC). The CMC quantifies waveform similarity between 0 and 1,16 with the interpretation of results as follows:
-
CMC < 0.65 no similarity;
-
0.65 < CMC < 0.75 moderate similarity;
-
0.75 < CMC < 0.85 good similarity;
-
0.85 < CMC < 0.95 very good similarity; and
-
0.95 < CMC < 1 excellent similarity.
The similarity metrics of EMG data and predicted muscle activations were calculated for each muscle and averaged over all trials, functional activities and study participants.
The muscle moment arm predictions of the MRI-based model were normalised to allow comparison to literature data.1,18,20,21,22,23 The normalisation involved the multiplication of each moment arm with the ratio of average humeral head radius of all participants to the humeral head radius of a given participant. This method has been used in the literature1 in order to eliminate inter-subject moment arm variation. The humeral head radius for each study participant was obtained through a manual sphere fit.
Anthropometric Model Scaling
The dependency of model predictions on the anatomical geometry was assessed in a tenfold cross-validation (Fig. 2). In the course of this cross-validation, each anatomical dataset from the 10 MRI-based model in the database served as input for each MRI-based shoulder model. This means that 9 anatomical datasets (skeletal geometry and muscle properties) as obtained from MRI served as input into each MRI-based model. For example, in the first run, the anatomical datasets (skeletal geometry and muscle properties) of MRI-based models 2 to 10 served as input into the MRI-based Model 1. Thereby, the predicted glenohumeral loading of MRI-based Model 1 during 16 functional daily tasks is serving as ground truth. Based on inputting 9 different anatomical datasets into MRI-based Model 1, the difference in predicted glenohumeral loading with different anatomical datasets based on the comparison of ground truth predictions from MRI-based Model 1 with those of the 9 different MRI-based models 2 to 10 was assessed. This procedure was performed 10 times, taking each of the 10 MRI-based models as ground truth in one of the 10 runs.
Demonstration of one run of the tenfold cross-validation, whereby anatomical datasets from MRI-based models 2 to 9 served as input into MRI-based model 1. The comparison of predicted glenohumeral loading between ground truth predictions from MRI-based Model 1 and the 9 anatomical datasets enabled the identification of the closest MRI-based model, defined as the MRI-based model that yields the smallest RMSE in predicted glenohumeral loading, when compared to ground truth predictions.
In order to assess the dependency of model predictions on the anatomical geometry in a tenfold cross-validation, anthropometric parameters were selected based on studies in the literature, demonstrating the effect of musculoskeletal geometry on modelling results.6,9 These parameters were measurable from MRI and include shoulder width (distance between LAC and RAC; only parameter obtained from optical motion marker data), humeral length (distance between midpoint of ME and LE and the humeral head center), humeral width (distance between the ME and LE), forearm length (distance between midpoint of ME and LE and midpoint of RS and US), forearm width (distance between RS and US), scapula length (distance between inferior scapula angle and superior scapula angle), scapula width (distance of superior scapula angle and acromion angle) as well as gender. For the analysis of the results of the cross-validation, Pearson correlations and multiple linear regressions were used to test for relationships between anthropometric measurements and changes in a compound measure that took into account both the accuracy of the glenohumeral contact force as well as all shoulder muscle forces (normalised root mean square error, RMSE, normalised by body mass [N/kg]). This was calculated by taking the average of all shoulder muscle force errors, summing this with the errors of the glenohumeral joint contact force and dividing those by two, so that both measures contributed an equal amount to the target value. Partial correlation coefficients were used to identify which variables contribute to significant changes in RMSE in the presence of all variables. Multiple regression models were identified when taking the significant predictors into account, with these models serving to determine anthropometric parameters or combinations of these that yield best modelling outcomes through scaling of personalised shoulder models. The final multiple regression model was identified using stepwise forward regression, with a significance level of p < 0.05 being used throughout. The appropriateness of the stepwise forward regression models was evaluated by examining histograms of the residuals as well as plots of the residuals against the independent and predicted values.
The final multiple regression model was utilised to identify the closest dataset for each MRI-based model. The closest dataset for each MRI-based model is defined as the linearly linearly-scaled MRI-based model from the template database that yields the smallest RMSE in glenohumeral loading during 16 functional daily tasks. This is termed the closest MRI-based model. The model predictions of the linearly scaled-closest MRI-based model were compared to the linearly scaled-generic model in order to assess improvements in modelling outcomes through the use of linearly scaled-closest MRI-based model. These improvements in modelling outcomes were assessed for the glenohumeral joint contact force as well the rotator cuff muscles. A non-parametric Mann–Whitney test with a level of significance of 0.05 was used to test for significant differences in predicted glenohumeral loading between the MRI-based model predictions and those from the scaled-closest MRI-based model and the scaled-generic model. A Bonferroni correction was used to account for multiple comparisons.
Results
The quantitative evaluation of EMG data and predicted muscle activations using Sprague and Geers metric is shown in Table 3. The MRI-based model predictions demonstrate good agreement with EMG measurements for deltoid, biceps, triceps and pectoralis, while the trapezius shows moderate agreement with EMG measurements. The subject-specific MRI-based model predictions demonstrate better agreement with EMG measurements, when compared to scaled-generic model as well as scaled-closest MRI-based model. The predictions of the scaled-closest MRI-based model compare better to the subject-specific MRI-based model predictions, when compared to the scaled-generic model.
The analysis of waveform similarity between EMG data and predicted muscle activations using the coefficient of multiple correlation is shown in Table 4. The subject-specific MRI-based model predictions demonstrate good to very good similarity with EMG measurements for deltoid, biceps and triceps, while trapezius and pectoralis show moderate similarity with EMG measurements. The MRI-based model predictions demonstrate better agreement with EMG measurements, when compared to scaled-generic model as well as scaled-closest MRI-based model. The predictions of the scaled-closest MRI-based model compare better to the subject-specific MRI-based model predictions, when compared to the scaled-generic model.
The comparison of muscle moment arms between MRI-based models and cadaveric measurements for functional daily activities is shown in Figs. 3 and 4. The MRI-based muscle moment arms compare well with in vitro measurements for shoulder muscle prime movers, with cadaveric measurements being largely within one standard deviation of the MRI-based model predictions.
The dependency of model predictions of joint contact force and shoulder muscle forces on anatomical geometry is illustrated in Table 5, through correlation of anthropometric differences between subjects with the RMSE in the compound measure that took equally into account both the accuracy of the glenohumeral contact force as well as all shoulder muscle forces. The best predictor for changes in RMSE is the ratio of body height to shoulder width (Pearson correlation coefficient, R = 0.46, p = 9.67E−5), followed by gender (R = 0.42, p = 1.44E−5). The next best predictors include the ratios of segment length to width (R = 0.35, p = 6.21E−3).
The final regression model was developed to take into account the significant predictors (Table 6). As identified by the forward multiple linear regression model, the ratio of body height to shoulder width as well as gender were in the final set of predictors for the RMSE in glenohumeral loading, with both variables combined explaining 45% of the variance (coefficient of determination, R2 = 0.45, adjusted R2 = 0.43). The inclusion of additional anthropometric variables such as the ratios of segment length to width only explained an additional 4% of the variation in RMSE. The closest MRI-based model, with the minimum error in joint contact force is derived from the following multiple regression model:
The regression equation was utilised to identify the linearly scaled-closest MRI-based model, with the comparison of the linearly scaled-closest MRI-based model and the linearly scaled-generic model demonstrating a significant reduction in RMSE of the compound measure that took equally into account both the accuracy of the glenohumeral contact force as well as all shoulder muscle forces (p < 0.02) for the linearly scaled-closest MRI-based model (Tables 7, 8).
Residual analysis of the forward stepwise regression models indicated this linear model was an appropriate choice with the residuals being normally distributed and the residual plots indicating there was constant variance and no evidence of nonlinearity.
Discussion
Customisation of musculoskeletal modelling through medical imaging has demonstrated significant improvements in model reliability when compared to linearly scaled-generic models,19,38,39 but the model development is time, labour and technology intensive. Linear scaling of musculoskeletal shoulder models with high anthropometric similarity from an anatomical atlas has the potential to yield modelling results that are close to predictions of subject-specific models and more accurate than a single, scaled-generic model. This study has developed and validated 10 subject-specific shoulder models to demonstrate that linear scaling of MRI-based shoulder models with the closest ratio of body height to shoulder width and from the same gender yield best modelling outcomes in glenohumeral loading. These model predictions are significantly improved when compared to a linearly scaled-generic dataset.
The validation of musculoskeletal shoulder models is essential for utility and clinical applicability, but the difficulty in measuring in vivo muscle forces makes model validation a challenging task. The subject-specific models presented in this study were validated against measurements of muscle activations using EMG as well as measurements of muscle moment arms as reported in the literature. Muscle forces are the main determinant for joint contact forces, making EMG measurements a justified comparator for model validation that has been widely used in the literature.10,31 The predicted muscle activations of the MRI-based models yield coefficients of multiple correlation that show very good to moderate agreement with EMG measurements, with comparable coefficients for waveform similarity being reported in the literature.30 Similarly, the subject-specific models show combined errors of the Geers metric that are comparable to other validation results in the literature (ranging from 0.13 to 0.41),10 with a time lag being observed relative to measured EMG data, representing the neuromechanical delay between muscular activation and force production.7 The predicted muscle activations of the MRI-based models are also superior to those of the scaled-generic model.
The comparison of MRI-based muscle moment arms with in vitro measurements as reported in the literature demonstrates that experimental data are largely within one standard deviation of model estimations (Figs. 3 and 4). The differences between computational simulation and experimental testing may be explained with the definition of muscle subregions that were chosen during cadaveric testing by Ackland et al.,1 Hughes et al.23 and Herrmann et al.20 While musculoskeletal shoulder models separate muscles into individual muscle lines of action, cadaveric studies divided shoulder muscles into functionally distinct muscle fiber bundles, making a direct comparison challenging. In addition, the small number of human cadavera used during in vitro testing may contribute to the differences observed between MRI-based model predictions and experimental measurements.1,20,23 Furthermore, the age difference between the elderly cadaveric specimens and the young study participants mat also contribute to this. Nevertheless, MRI-based model estimations for shoulder muscle moment arms compare well to in vitro measurements from different experimental studies1,20,23), providing further evidence for physiological model estimations of the developed MRI-based shoulder models.
The 10 validated MRI-based shoulder models were utilised to identify the effect of anthropometrics on musculoskeletal modelling results and to provide an atlas for customisation of shoulder models in future applications. There are several studies assessing changes in model estimations with alterations in muscle physiology parameters,3,4,6 with no study reported in the literature having investigated the effect of anthropometrics on musculoskeletal modelling outcomes. The precise understanding of this effect would be essential in order to evaluate the errors associated with scaling of generic cadaveric datasets to subjects with different anthropometry, particularly as some generic datasets in the literature (including Charlton and Johnson,8 Klein Breteler et al.26 and Carbone et al.5) may not necessarily be representative of an average person. This study has found small correlation coefficients of body height, body mass as well as upper limb segment length with changes in root mean square error in glenohumeral loading. Best model estimations of those measures tested are yielded for anatomical geometries with the closest ratio of body height to shoulder width and from the same gender. The significance of these two anthropometric parameters is supported by studies in the literature,6 demonstrating significant alterations in model estimations with changes in moment arms through variations in muscle attachment sites. While the ratio of body height to shoulder width has a direct impact on moment arms due to the bony contours provided for muscle wrapping, there is evidence in the literature that muscle physiology parameters including muscle attachment sites are gender dependent.9,15
The regression equation was utilised to identify the closest anatomical dataset for each MRI-based model, with the comparison of linearly scaled-closest MRI-based model and the linearly scaled-generic model demonstrating a significant reduction in root mean square error in the compound measure that took equally into account both the accuracy of the glenohumeral contact force as well as all shoulder muscle forces for the scaled-closest MRI-based model. This suggests that establishing a database of anatomical models for anthropometric scaling is essential to improve model reliability, when compared to linearly scaling a single, generic model that has demonstrated a maximum RMSE of 11 N/kg for the glenohumeral joint contact force and 3 N/kg for the rotator cuff muscle forces. Future research should aim to improve the number of anatomical datasets for scaling to see if a reduction in the mean RMSE (3.8 N/kg, 1.4 N/kg) and maximum RMSE (7.2 N/kg, 2.2 N/kg) of the joint contact forces and muscle forces, respectively, could be achieved. The additional anatomical geometries should be male and female volunteers with different ratios of body height to shoulder width as expressed by the multiple linear regression model.
This study has some limitations. Firstly, the identification of the scaled-closest MRI-based model is only applicable for musculoskeletal shoulder models with the same scaling law and changes in scaling law are likely to alter the multiple linear regression model. Nevertheless, the scaling law as implemented in the UKNSM is widely used (including Garner and Pandy,17 Delp et al.14) and the data provided in this study provide the basis to adapt the regression model to musculoskeletal shoulder models with different scaling laws.33 Secondly, the number of the MRI-based shoulder models developed in this study is limited. While a larger number of subject-specific models would increase statistical power of the findings, we believe that the 10 datasets are representative for a wide variety of subjects across the population and future work should aim to add MRI-based models to the atlas that will be towards the more extreme end of anthropometrics. Finally, the musculoskeletal model did not define an upper bound for each muscle during functional tasks involving external loads to quantify the maximum force that each muscle can contribute to shoulder movement. This has led to muscle force predictions that exceed 100% of the muscle force capacity in 9 of the 16 functional daily tasks. These tasks involve lifting of a 3 kg object to shoulder and head height, as well as lifting this object from the floor and placing this object with an outstretched arm. In additional, these tasks involve opening a jam jar, sit-to-stand and activities with large ranges of motion such as reaching across the body, abduction and flexion. These force estimations have yielded peak contact forces of 3 to 5 times the body weight for the opening jam jar task, with the lifting tasks leading to peak glenohumeral contact forces of 1–2 times the body weight. Around 1–1.5 times the body weight glenohumeral loading was observed for daily activities involving large ranges of motion. While these predictions are non-physiological, this unbounded approach has been widely used in the literature13,17 to account for the inherent difficulty in measuring muscle stresses.43
In summary, this is the first study to develop and validate 10 subject-specific shoulder models from MRI in order to demonstrate the effect of anthropometrics on modelling outcomes. The study findings show that linear scaling of subject-specific models with the closest ratio of body height to shoulder width and from the same gender yield best modelling results for glenohumeral joint loading, with significant improvements in model estimations when compared to a linearly-scaled generic model. This suggests that establishing an atlas with MRI-based models for scaling significantly improves model predictions. This should facilitate the translation of musculoskeletal models in clinical practice in order to assist surgical and rehabilitative planning, assistive device design and analysis of shoulder arthroplasty design.
References
Ackland, D. C., P. Park, M. Richardson, and M. G. Pandy. Moment arms of the muscles crossing the anatomical shoulder. J. Anat. 213:383–390, 2008.
Arnold, E. M., S. R. Hamner, A. Seth, M. Millard, and S. L. Delp. How muscle fiber lengths and velocities affect muscle force generation as humans walk and run at different speeds. J. Exp. Biol. 216:2150–2160, 2013.
Bolsterlee, B., A. N. Vardy, F. C. T. van der Helm, and H. E. J. Veeger. The effect of scaling physiological cross-sectional area on musculoskeletal model predictions. J. Biomech. 48:1760–1768, 2015.
Bolsterlee, B., and A. A. Zadpoor. Transformation methods for estimation of subject-specific scapular muscle attachment sites. Comput. Meth. Biomech. Biomed. Eng. 17:1492–1501, 2014.
Carbone, V., R. Fluit, P. Pellikaan, M. M. van der Krogt, D. Janssen, M. Damsgaard, L. Vigneron, T. Feilkas, H. F. J. M. Koopman, and N. Verdonschot. TLEM 2.0-a comprehensive musculoskeletal geometry dataset for subject-specific modelling of lower extremity. J Biomech. 48:734–741, 2015.
Carbone, V., M. M. Van der Krogt, H. Koopman, and N. Verdonschot. Sensitivity of subject-specific models to errors in musculo-skeletal geometry. J. Biomech. 45:2476–2480, 2012.
Cavanagh, P. R., and P. V. Komi. Electromechanical delay in human skeletal muscle under concentric and eccentric contractions. Eur. J. Appl. Physiol. 42:159–163, 1979.
Charlton, I. W., and G. R. Johnson. A model for the prediction of the forces at the glenohumeral joint. Proc. Inst. Mech. Eng. 220:801–812, 2006.
Clark, E. M., A. R. Ness, and J. H. Tobias. Gender differences in the ratio between humerus width and length are established prior to puberty. Osteoporos. Int. 18:463–470, 2007.
Cleather, D. J., and A. M. J. Bull. The development of a musculoskeletal model of the lower limb: introducing FREE BODY. R. Soc. Open Sci. 2:140449, 2015.
De Groot, J. H., L. A. Rozendal, C. G. Meskers, and H. J. Arwert. Isometric shoulder muscle activation patterns for 3-D planar forces: a methodology for musculoskeletal model validation. Clin. Biomech. 19:790–800, 2004.
De Leva, P. Adjustments to zatsiorsky-seluyanov’s segment inertia parameters. J. Biomech. 29:1223–1230, 1996.
Delp, S. L., F. C. Anderson, A. S. Arnold, P. Loan, A. Habib, C. T. John, E. Guendelman, and D. G. Thelen. OpenSim: open-source software to create and analyse dynamic simulations of movement. IEEE Trans. Biomed. Eng. 54:1940–1950, 2007.
Delp, S. L., P. Loan, M. G. Hoy, F. E. Zajac, E. L. Topp, and J. M. Rosen. An interactive graphics-based model of the lower extremity to study orthopaedic surgical procedures. IEEE Trans. Biomed. Eng. 37:757–767, 1990.
Edama, M., H. Onishi, M. Kubo, T. Takabayashi, E. Yokoyama, T. Inai, H. Watanabe, S. Nashimoto, Y. Koga, and I. Kageyama. Gender differences of muscle and crural fascia origins in relation to the occurrence of medial tibial stress syndrome. Scand. J. Med. Sci. Sports 27:203–208, 2017.
Ferrari, A., A. G. Cutti, and A. Capello. A new formulation of the coefficient of multiple correlation to assess the similarity of waveforms measured synchronously by different motion analysis protocols. Gait Posture. 31:540–542, 2010.
Garner, B. A., and M. G. Pandy. Musculoskeletal model of the upper limb based on the visible human male dataset. Comput. Methods Biomech. Biomed. Eng. 4:93–126, 2001.
Gatti, C. J., C. R. Dickerson, E. K. Chadwick, A. G. Mell, and R. E. Hughes. Comparison of model-predicted and measured moment arms for the rotator cuff muscles. Clin. Biomech. 22:639–644, 2007.
Gerus, P., M. Sartori, T. F. Besier, B. J. Fregly, S. L. Delp, S. A. Banks, M. G. Pandy, and D. G. Lloyd. Subject-specific knee joint geometry improves predictions of medial tibiofemoral contact forces. J. Biomech. 46:2778–2786, 2013.
Herrmann, S., C. Konig, M. Heller, C. Perka, and S. Greiner. Reverse shoulder arthroplasty leads to significant biomechanical changes in the remaining rotator cuff. J. Orthop. Surg. Res. 6:1–7, 2011.
Hogfors, C., D. Karlsson, and B. Peterson. Structure and internal consistency of a shoulder model. J. Biomech. 28:767–777, 1995.
Holzbaur, K. R. S., W. M. Murray, and S. L. Delp. A model of the upper extremity for simulating musculoskeletal surgery and analysing neuromuscular control. Ann. Biomed. Eng. 33:829–840, 2005.
Hughes, R. E., G. Niebur, J. Liu, and K. N. An. Comparison of two methods for computing abduction moment arms of the rotator cuff. J. Biomech. 31:157–160, 1998.
Johnson, G. R., D. Spalding, A. Nowitzke, and N. Bogduk. Modelling the muscles of the scapula, morphometric and coordinate data and functional implications. J. Biomech. 29:1039–1051, 1996.
Kaptein, B. L., and F. C. T. van der Helm. Estimating muscle attachment contours by transforming geometrical bone models. J. Biomech. 37:263–273, 2004.
Klein Breteler, M. D., C. W. Spoor, and F. C. van der Helm. Measuring muscle and joint geometry parameters of a shoulder for modelling purposes. J Biomech. 32:1191–1197, 1999.
Klemt, C., D. Nolte, G. Grigoriadis, E. Di Federico, P. Reilly, and A. M. J. Bull. The contribution of the glenoid labrum to glenohumeral stability under physiological joint loading using finite element analysis. Comput. Methods Biomech. Biomed. Eng. 20:1613–1622, 2017.
Klemt, C., J. A. Prinold, S. Morgans, S. H. L. Smith, D. Nolte, P. Reilly, and A. M. J. Bull. Analysis of shoulder contact forces during functional activities of daily life. Clin. Biomech. 54:34–41, 2018.
Lerner, Z. F., M. S. Demers, and S. L. Delp. How tibiofemoral alignment and contact locations affect predictions of medial and lateral tibiofemoral contact forces. J. Biomech. 48:1–7, 2015.
McGinley, J. L., R. Baker, R. Wolfe, and M. E. Morris. The reliability of three dimensional kinematic measurements: a systematic review. Gait Posture. 7:360–369, 2009.
Modenese, L., A. T. Phillips, and A. M. J. Bull. An open source lower limb model: hip joint validation. J. Biomech. 44:2185–2193, 2011.
Nikooyan, A. A., H. E. J. Veeger, E. J. K. Chadwick, M. Praagman, and F. C. T. van der Helm. Development of a comprehensive musculoskeletal model of the shoulder and elbow. Med. Biol. Eng. Comput. 49:1425–1435, 2011.
Nikooyan, A. A., H. E. J. Veeger, P. Westerhoff, F. Graichen, G. Bergmann, and F. C. T. van der Helm. Validation of the Delft Shoulder and Elbow Model using in vivo glenohumeral joint contact forces. J. Biomech. 43:3007–3014, 2010.
Peterson, S. L., and G. M. Rayan. Shoulder and upper arm muscle architecture. J. Hand Surg. Am. 36:881–889, 2011.
Pheasant, S., and C. Haslegrave. Bodyspace: Anthropometry, Ergonomics, and the Design of Work. London: Taylor and Francis, p. 244, 2006.
Prinold, J. A. I., and A. M. J. Bull. Scaling and kinematics optimisation of the scapula and thorax in upper limb musculoskeletal models. J. Biomech. 47:2813–2819, 2014.
Prinold, J. A. I., A. F. Shaheen, and A. M. J. Bull. Skin-fixed scapula trackers: a comparison of two dynamic methods across a range of calibration positions. J. Biomech. 44:2004–2007, 2011.
Scheys, L., A. Spaepen, P. Suetens, and I. Jonkers. Calculated moment-arm and muscle-tendon lengths during gait differ substantially using MR based versus rescaled generic lower limb musculoskeletal models. Gait Posture 28:640–648, 2008.
Scheys, L., A. van Campenhout, A. Spaepen, P. Suetens, and I. Jonkers. Personalised MR-based musculoskeletal models compared to rescaled generic models in the presence of increased femoral anteversion. Effect on hip moment arm lengths. Gait Posture. 28:358–365, 2008.
Schwer, L. E. Validation metrics for response histories: perspectives and case studies. Eng. Comput. 23:295–309, 2007.
Shaheen, A. F., C. M. Alexander, and A. M. J. Bull. Effects of attachment position and shoulder orientation during calibration on the accuracy of the acromial tracker. J. Biomech. 44:1401–1418, 2011.
Spitzer, V. M., and D. G. Whitlock. The visible human dataset: the anatomical platform for human simulation. Anat. Rec. 253:49–57, 1998.
Van der Helm, F. C. T. A finite element musculoskeletal model of the shoulder mechanism. J. Biomech. 27:551–569, 1994.
van der Helm, F. C. T., H. E. J. Veeger, G. M. Pronk, L. H. V. van der Woude, and R. H. Rozendaal. Geometry parameters for musculoskeletal modelling of the shoulder mechanism. J. Biomech. 25:129–144, 1992.
Veeger, H. E. J., B. Yu, K. N. An, and R. H. Rozendal. Parameters for modelling the upper extremity. J. Biomech. 30:647–652, 1997.
Wu, W., P. V. Lee, A. L. Bryant, M. Galea, and D. C. Ackland. Subject-specific musculoskeletal modelling in the evaluation of shoulder muscle and joint function. J. Biomech. 49:3626, 2016.
Acknowledgments
This study was funded by the Engineering and Physical Sciences Research Council through an industrial case PhD studentship with JRI Orthopaedics [Grant Number EP/M507878/1]. Dr. Ziyun Ding is funded through the Royal British Legion Centre for Blast Injury Studies. The musculoskeletal model and anatomical datasets used in this study are available at www.msksoftware.org.uk.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Michael R. Torry oversaw the review of this article.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Klemt, C., Nolte, D., Ding, Z. et al. Anthropometric Scaling of Anatomical Datasets for Subject-Specific Musculoskeletal Modelling of the Shoulder. Ann Biomed Eng 47, 924–936 (2019). https://doi.org/10.1007/s10439-019-02207-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-019-02207-2