Abstract
Having a system capable of obtaining high resolution, quantitative, subcellular information of individual live cells through imaging, while minimizing cell damage, is of great importance in many fields of research. In recent years, optofluidic light-sheet fluorescent microscopy (LSFM) has emerged as a powerful tool providing a low photo-toxic imaging method utilising the fluidic environment offered by microfluidics. Here, the benefits of LSFM were integrated with an acoustic single cell prison for precise single cell handling and 3D multi-imaging in a semi-automated manner. The compact, monolithic, acousto-optofluidic platform employed standing surface acoustic waves (SSAWs) to sequentially trap an individual cell on either side of an imaging region, which gathered planar, cross-sectional images of the cell. A cytoplasmic stain was utilised to not only visualize the cell trajectory throughout the imaging process, but also to verify the cell viability post-acoustic exposure. A mitochondrial stain was also used to better demonstrate the resolution capabilities of the device. Through post-image processing, 3D volumetric images of the cell were reconstructed and the results between the first and second rounds of imaging were directly comparable. Furthermore, the acoustic prison advantageously positions the cells in the upper region of the channel, enabling the ability to accurately compare temporal changes in cell morphology; a capability that can lead to advancements in therapeutics and drug delivery to access responses of cells to stimuli over time.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Obtaining high-resolution, three-dimensional, information pertaining to intracellular architecture is a key task required for understanding complex biological mechanisms in a variety of research disciplines in cell biology and developmental biology (Jensen 2013). In order to study and understand the diverse anatomical and functional complexities of living organisms it is important to be able to visualise how living cells dynamically organise their intracellular organelles while they undergo certain processes (Gao et al. 2020). As such, single cell imaging of live cells, with high spatial and temporal resolution, is required. Although current available technologies offer advanced imaging of fixed cells (dead but morphologically preserved), there is still a gap in microfluidic chip-compatible methods for the three- dimensional imaging of living cells to access temporal responses to stimuli. On-chip imaging is desirable for a wide number of applications due to potential for the integration of other sample preparation methods.
Imaging techniques, such as fluorescent microscopy, have been optimised for imaging fixed cells, meaning that long recording times and exposure times are often used without fear of adverse effects on cell health. In contrast, for successful imaging of live cells, a balance between image quality and illumination intensity must be found while not compromising cell viability. Recent developments in live cell imaging have predominantly focused on high-throughput systems for accurate counting and sorting (Cho et al. 2010; Nitta et al. 2018; LaBelle et al. 2021) or imaging of large cell populations (Sesen and Whyte 2020). One of the most common methods is imaging flow cytometry, which utilises optofluidics for fast, two-dimensional imaging of live cells, down to a resolution of 500 nm (Doan et al. 2018; Holzner et al. 2021). However, obtaining high-resolution volumetric images, capable of showing internal cellular organisation, of individual living cells can still be difficult. To tackle these limitations, the advantages of microfluidics and light-sheet fluorescence microscopy (LSFM) (Deschout et al. 2014; Stelzer 2015; Lin et al. 2018; Yordanov et al. 2021) have been combined, offering 3D single cell imaging capabilities.
LSFM employs the use of a thin (in the order of a few hundred of nanometers to a few micrometers), planar monochromatic light sheet to illuminate a sample perpendicular to the observation direction (Weber and Huisken 2011; Strobl et al. 2017). In a microfluidics setting, the sample is carried by fluid flow through the light-sheet and fluorescent images of the cell are collected from each plane. Volumetric information can then be obtained by image reconstruction. Although this illumination method provides low photo-toxicity and deeper penetration, current LSFM techniques are also limited by inadequate resolving power along the optical axis, leading to difficulties in obtaining high quality volumetric information. These devices also employ optical components that are external to the imaging chip and hence need careful, expert alignment.
To further improve on-chip LSFM, Ordaz et al. developed a 3D optofluidic LSFM device capable of generating a sub-micron thick light-sheet allowing for volumetric images of living cells to be acquired (Vargas-Ordaz et al. 2021). The micro-optical lenses were incorporated within the microfluidic chip by means of soft lithography processes, allowing for the simplification of the imaging platform and offering the potential of integration of cell handling approaches. They were able to achieve a lateral resolution of 0.65 µm and an axial resolution limited by the light-sheet thickness, allowing for the quantification of changes in endosome numbers after the cells had undergone a drug treatment (Vargas-Ordaz et al. 2021). This system has the potential to offer repeated 3D imaging of live adherent cells at low exposure, hence accessing temporal data. However, to unlock this potential, it is necessary to add single cell handling capabilities.
Within the realm of microfluidic-based technologies, surface acoustic waves (SAWs) have been successfully employed for a wide range of manipulation applications for things such as fluid mixing (Rezk et al. 2012; Hsu and Chang 2022; Lv and Chen 2022), concentration (Shilton et al. 2008; Habibi and Neild 2019; Akther et al. 2020), sorting (Mutafopulos et al. 2019; Richard et al. 2019; Ng and Neild 2021), patterning (Nguyen et al. 2018; Maramizonouz et al. 2022), and droplet production (Castro et al. 2018; Rambach et al. 2018; Jin et al. 2019). The ability to precisely manipulate target cells using acoustic forces has been utilised in therapeutics and diagnostics (Salari et al. 2021; Agostini et al. 2022; Hao et al. 2022) where the ability to control individual cells to study cell-cell interactions (Li et al. 2014; Guo et al. 2015), or to perform single cell analysis (Link and Franke 2020; Mejía Morales et al. 2022), is of particular interest. The capacity of SAW devices in terms of single cell control was further demonstrated in the work published by Richard et al. where a single cell micro-dispenser was developed to trap, release and direct individual cells (Richard et al. 2022).
Building and expanding upon the acoustic trapping mechanism discovered and exploited in (Richard et al. 2022), an acoustic single cell prison was developed and integrated into a single chip with a 3D optofluidic LSFM device (Vargas-Ordaz et al. 2021). The resulting hybrid acousto-optofluidic chip, the first of its kind, allowed for the sequential trapping of an individual cell on either side of the imaging region to gather 3D images of the cell over two separate rounds of imaging in a controlled and reproducible manner. The focus of this work was to demonstrate the additional single cell handling capabilities offered by the acoustic components and not on the resolution proficiency of the optical components, which was demonstrated in an earlier publication. Furthermore, given the requirement of a piezoelectric substrate for the excitation of SAWs, this novel platform demonstrates the compatibility of these two technologies (acoustic and optical). Not only do the acoustic components allow for targeted single cell control, but they also lead to the realisation of a semi-automated, imaging platform with the ability to obtain temporal data in real-time. The acoustic control was demonstrated using two different cell lines, and stains, and 3D reconstruction of individual live cells were obtained. A Calcein stain was first used to help visualise the trajectory of the cell along the horizontal direction throughout the imaging process and to validate the size of the cell being imaged. This stain also allowed the cell viability post-acoustic exposure to be verified. An intracellular stain, MitoTracker, was then used to visualise the mitochondrial activity within the cell, demonstrating the resolution capabilities of the device as well as the ability to use different stains requiring different wavelength. The means to image an individual, living cell back-to-back and obtain comparable data is an advancement in on-chip LSFM technology.
2 Results and discussion
2.1 Light-sheet generation and device characterisation
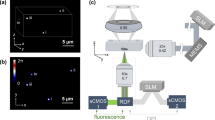
The acousto-optofluidic chip was designed to handle and image cells flowing within a microfluidic channel (see design schematic in Fig. 1). The two-component system consists of a PDMS layer containing all the self-aligned micro-optical and microfluidic components in a monolithic assembly, and a piezoelectric substrate (lithium niobate (LN)) patterned with electrodes in order to create the required acoustic waves. The key features of the optical components have been described previously (Vargas-Ordaz et al. 2021), where the optofluidic LSFM chip was entirely fabricated of PDMS. Here, we ensure that the addition of the LN substrate doesn’t cause detrimental effects to the light-sheet generation and image capture. Hence, it is the acoustic trapping part of this work, and the demonstration of the benefits it affords in gaining temporal data which is the primary focus. As such, the compatibility of the substrate was first verified by determining the lateral and axial resolutions of the hybrid device, followed by the characterisation of the trapping mechanism responsible for the double imaging process (within the acoustic prison).
Device schematic and working principle. i Device inlet depicting the sheath flow providing focusing of the cells from the sample inlet from both sides, as well as from beneath. ii Loading region depicting interdigital transducers (IDTs) patterned on a lithium niobate (LN) substrate and actuated to excite surface acoustic waves (SAWs). The resulting acoustic radiation force (ARF), denoted Frad, overcoming the drag force (Fdrag), is utilised for cell trapping and vertical positioning. iii Imaging region where the single cell flows through a light-sheet created by an on-chip optical fibre, and accompanying holder (1), micro-lens (2) and mirror (3). The side view inset depicts the obtained cross-sectional planar images of the cell. iv Collection region where the imaged cell is trapped for a second time, the flow is reversed via an external control system and the cell is then pulled through the extraction channel and undergoes a second round of imaging
A detailed analysis of the trapping mechanism can be found in (Richard et al. 2022), but briefly, interdigital transducers (IDTs) patterned on a LN substrate are actuated via an externally applied electrical signal leading to the excitation of surface vibrations. These SAWs will couple into the fluid leading to regions of local pressure minima and maxima. When an object, such as a cell, is present within the acoustic field it will interact with the field, giving rise to acoustic radiation forces (i.e., Frad in Fig. 1ii). By applying a sufficient amount of power to the IDTs resulting in an acoustic radiation force (ARF) with a magnitude larger than or equal to the drag force (Fdrag), the cell will cease to move. This mechanism can be utilised for a variety of cell types, sizes, and offers a non-contact, biocompatible, and precise manipulation method.
The potential effects of the presence of this optically nonlinear crystal on the light-sheet generation and imaging resolution were investigated. Foremost, the lateral resolution of the device was verified using fluorescent sub-diffraction polystyrene (PS) beads (210 nm diameter). The beads were passed through the device without acoustic excitation and planar images were acquired using a 40x/0.6 NA objective external to the chip. Although this objective is generally used in combination with a glass coverslip (n = 1.51), here, an optically flat PDMS channel sidewall (n = 1.41) was used in lieu. The quantification of the experimental lateral resolution of the device was provided by capturing the point spread function (PSF) of the bead. The PSF also determines any aberrations potentially brought about by the detection path and the substrate. The side view of the produced light-sheet can be seen in Fig. 2b i, where two locations of interest are high- lighted; a region near the upper-left area (yellow) of the channel and a region in the lower-middle (green) of the channel nearer to the LN substrate. At the region closer to the LN substrate (green), it is observed that the quality of the PSF (Fig. 2b ii) renders a lateral resolution of 1.17 µm, which is diminished when compared to our prior work (Vargas-Ordaz et al. 2021). However, with the employment of the acoustic prison, when the acoustic actuation is utilised, the cells being imaged will be pushed further away from the plane of the substrate by the vertical components of that acoustic radiation force (ARF), further aided by the presence of the sheath flow beneath the sample (Fig. 1 i). The lateral component of the ARF is responsible for the trapping (Richard et al. 2019). This effect can be observed from experimental data at the location of the cell undergoing sequential imaging (Fig. 4c). Hence, the PSF (Fig. 2b iii) was also examined at a location further away from the substrate (yellow box), this shows a focused central plane of the bead with visible Airy rings, similar to those obtained in (Vargas-Ordaz et al. 2021). The measured lateral resolution at this location was found to be 1.08 µm. It is this PSF that was applied as a kernel for imaging deconvolution of the cells. However, to more precisely compare the resolution near to the LN substrate with the resolution more towards the middle, or upper regions of the channel, PSFs at different locations were sampled (Nlocations = 6). It was found that nearer to the LN substrate, the mean resolution was 1.24 ± 0.05 µm whereas further away from the substrate the mean was 1.21 ± 0.08 µm. The results show no significant difference between these values (unpaired t-test, P = 0.49). We can thus conclude that the LN substrate does not cause any significant scattering effects.
Light-Sheet generation in an acousto-optofluidic chip. a Top view image of the fabricated device depicting all optical components (1: optical fibre, 2: micro-lens, 3: mirror) and the generated light-sheet. b Side view of the imaging region showing the light-sheet formation by flowing fluorescein through the sample inlet (i). The point spread functions (PSFs) were obtained using fluorescent sub-diffraction polystyrene (PS) beads, at regions of interest near the substrate (green) and at a distance away from the substrate (yellow). (ii) The lateral resolution at this location was determined to be 1.17 µm whereas in (iii) it was 1.08 µm. The visible Airy rings seen in (iii) demonstrate the lack of scattering due to interactions of the light-sheet with the lithium niobate (LN) substrate at this distance from the substrate (color figure online)
The axial resolution of the device dictates the minimum distance required between cross-sectional images to obtain high-quality data and is dependent on the numerical aperture (NA) of the detection objective. The axial resolution was determined to be 1.31 µm (detection objective NA = 0.6). Although individual cells are loaded within the system through the means of a standing surface acoustic wave (SSAW) gate (see Fig. 1ii), their movement is governed by the laminar flow regime provided by the microfluidic environment meaning that the spacing between each planar image is controlled by the applied flow rate, the frame rate of the off-chip imaging camera, and the light-sheet thickness (Fig. 1iii). The micro-lens used here is designed to produce a light-sheet with a thickness of 1.8 ± 0.2 µm. The flow rate of the sample should then be chosen based on the diameter of the cell of interest to maximise the number of planar images obtained, where the distance between consecutive images should be less than, or equal to, the axial resolution of the device. As a result, the device was first characterised based on the loading flow rate and the corresponding number of planar images (slices) acquired. The device was characterised with COS-7 cells (African green monkey kidney fibroblast-like cell line), rather than with particles, to obtain appropriate device operating parameters for high-resolution imaging. Foremost, the cell concentration was fixed at approximately 200,000 cells ml−1 and flow rate experiments were conducted with varying sample and sheath flow rates (Fig. 3a). For all cases the data collection time was fixed at 85 s.
Characterisation of the device based on sample flow rate and the resulting number of planar images (a). Each data point represents a single cell imaged at the corresponding flow rate. The experiments were conducted over a fixed period of 85 s. Inset depicting the relation between the number of planar images (i.e. slices) is inversely related to the applied flow rate (i.e. velocity). b Characterisation of the acoustic loading capabilities based on the applied actuation time of the loading IDTs for the accurate and reproducible loading of a single cell into the imaging region. The flow rates were held constant at 0.20 µl min−1 for both the cells and the sheath flow. A control system was used to trigger the IDTs in a cyclical manner with a 1 s release time. Data was collected for N = 18 cycles. The trapping outcomes are defined as follows: − 1 is a no trap occurrence (either a trapping failure or cells flow through during the release time), 0 is when no cell enters the region, and 1 is when only a single cell is trapped
The optimal flow rate was chosen based on the consistent flow of cells into the imaging region balanced with the number of planar images obtained of the cell from the side view (Fig. 3a inset). To obtain efficient optical sectioning, the cell must pass through the light-sheet with a velocity allowing for a maximum of the cell volume to be exposed. As expected, at lower flow rates fewer cells are able to enter the channel within the same time frame as compared to higher flow rates, not only due to the lower velocity but also due to the fact that the cells will have more time to sediment and interact with the channel walls at the entry point (channel inlet) (observed at 0.10 µl min−1 cells/0.20 µl min−1 sheath). After flowing at relatively low flow rates over a period of time, the cells will again almost cease to flow entirely, potentially due to a build-up of adhered cells at the inlet (observed at 0.30 µl min−1 for both cell and sheath flows). As the flow rates are increased (i.e. equal to and larger than 0.25 µl min−1 and 0.5 µl min−1, for the sample and sheath flow respectively), more cells begin to flow into the channel and in a more homogeneous manner. However, as the flow rates increase, the number of planar images obtained decreases. As such, the optimum flow rates were chosen based on a balance between the two, cell flow consistency and sufficient planar images. This was found to occur at flow rates of 0.20 µl min−1 for both the cells and the sheath flow resulting in a mean of approximately 14 planar images for a singular cell. At these flow rates, the corresponding cell velocity (136 µl s−1) along with a capture frame rate of 100 frames per second (fps) (detection objective), allows for an image to be captured every 1.36 µm. Although this value is slightly above the device’s axial resolution of 1.31 µm, the chosen parameters still avoid under-sampling for the 3D imaging of the cell. Note that more freedom can be afforded in the process of identifying the desired flow rate by use of a camera with a wider range of available frame rates.
Once the flow rate was determined, the acoustic loading mechanism was tested to determine the trapping time (the SSAW excitation time) that allows for the most consistent loading of an individual cell into the imaging region. For accurate and reproducible operation, a control system was used to trigger the actuation of the loading IDTs (Fig. 1ii) in a cyclical manner; the chosen actuation time (4 s, 6 s, or 8 s) with a fixed release time of 1 s. Data was collected for N = 18 cycles for all three cycle times and the number of occurrences for each of the trapping outcomes was plotted (Fig. 3b). The trapping outcomes are defined as follows: 1 represents the desired scenario when only a single cell is trapped and released, 0 is when no cell enters the trapping region, and −1 represents the case where a cell flows through the loading region but is not trapped (either a trapping failure or during the release time). The actuation time resulting in the most desired trapping outcomes at the flow rates for imaging, in this case, 8 s, was chosen for the subsequent automated double imaging experiments.
2.2 Acoustic single cell double-pass imaging
The acoustofluidic loading capabilities of the device were then utilised to image a single live cell multiple times in a semi- automated manner. The COS-7 cells were stained with the most common live-cell viability dye, Calcein AM, to demonstrate the biocompatibility of the acoustics. A control system consisting of an Arduino and accompanying circuitry was used to control the cell loading, reimaging, and extraction process. A pump was used to inject the cell sample and the sheath flow media at the fixed flow rate previously determined. The loading IDTs were actuated for 8 s, followed by a 1 s release to load the individual cell of interest to the imaging region (Fig. 4a) to undergo the first imaging process (Fig. 4a ii).
Overview of the double-imaging of an individual COS-7 cell. a Top view experimental images of the first imaging process; (i) the single cell is loaded via the actuation of the first pair of IDTs (8 s on), followed by the triggered release (1 s off) into the imaging region (ii) showing the cell trajectory. Scale in inset is 10 µm. b Top view experimental images of the second imaging process; (i) collection of the same cell via the second pair of actuated IDTs, a triggered flow reversal (5 µl min−1 pull for 3 s) leading to the cell being re-loaded (ii) to the imaging region (iii). c Side view fluorescent images obtained of the stained cell showing the vertical location of the cell during the first imaging (i) and second imaging (ii), with their overlap shown in (iii). The white lines in the images represent the PDMS channel sidewalls. An external control system was used to trigger the first pair of IDTs and the syringe pump responsible for the re-imaging while the second IDTs were triggered manually. The flow rate was held constant at 0.20 µl min−1 for both the cell sample and sheath flow. See ESI Movie S1 for the corresponding video of this process (color figure online)
A delay of 15 s was programmed to allow for the first imaging to take place, whereby the same cell was collected by a second pair of IDTs (Fig. 4bi) which was manually triggered. Concurrently, the loading IDTs were actuated during the imaging time to withhold any unwanted cells from entering the system using the acoustic trapping mechanism as before (Fig. 5aii). After the first imaging, a second syringe pump connected to the extraction channel was triggered to reverse the flow with a pull flow rate of 5 µl min−1 for 3 s, simultaneously re-loading the same cell to the imaging region for a second imaging (Fig. 4bii and iii) and extracting the unwanted cells withheld during the imaging process (Fig. 5b). See ESI Movie S1 and Movie S2 for the corresponding videos of these processes.
Top view experimental images of the semi-automated double-cell imaging process from the loading and extraction regions. a Demonstrates the single cell loading (i), the desired cell emphasised by the red arrow, and the withholding of the undesired cells during the first imaging (ii), encircled in red. b The flow reversal extracts the undesired cells that were trapped (i) and pulls the imaged cell back for the second round of imaging and can be seen being removed from the system through the extraction channel following the second imaging (ii). In this experiment, MG63 cells were used, which have slightly smaller diameters compared to COS-7 cells. See ESI Movie S2 for the corresponding video of this process (color figure online)
A side view of the double-imaging can be seen in Fig. 4c showing the location of the cell in the vertical direction (along the z-axis) during the first imaging (Fig. 4c i) and the second imaging (Fig. 4c ii). As observed by the overlapping of the two imaging processes (Fig. 4c iii) the cell remains localised within the same channel height in both instances. As previously discussed, the ARF acts to push the cell toward the channel roof, with the aid of the sheath flow, thus positioning the cell at an elevated vertical location. When the cell arrives at the collection region, the vertical force of the ARF from the second pair of IDTs works to maintain the cell’s vertical position in the channel.
This mechanism allows for some predictability of the cell position in the vertical direction, allowing for the cell to be imaged sequentially in the same area of the light-sheet, and therefore maintaining the same optical characteristics to facilitate image-to-image comparison. Being able to maintain a similar imaging trajectory for both rounds of imaging enables obtaining precise and comparable information. This is of particular importance for future applications such as in drug uptake studies where changes within the cell before and after drug uptake can be accurately observed with the same resolution.
2.3 Light-sheet fluorescence microscopy (LSFM) on chip
During the acoustic single cell double-imaging process, planar images were collected at 100 fps using an off-chip objective and a high-speed camera aligned with the sidewall of the imaging region (as shown in Fig. 1iii). A select few of the obtained, deconvoluted cross-sectional images from the imaging plane (xz) can be seen in Fig. 6a for the first imaging and in Fig. 6c for the second imaging. Although only 7 images are shown here, the full stack consisting of 16 and 15 images, are available in ESI Movie S3 and S4 for the first and second imaging respectively. The Calcein dye stains the cytoplasm of the cell (the cell content between the membrane and the nucleus), leaving the nucleus unaffected. This is in agreement with the results obtained and what is known in the literature (i.e. the nucleus of COS-7 cells is approximately 25 µm) (Son et al. 2020). Based on the spacing between the obtained images (1.36 µm), the total number of images obtained (16 slices), and the average range of COS-7 cell diameters (between 20 and 25 µm (Sagi et al. 2009)), the majority of the cell volume could be imaged with good resolution during the first round of imaging.
3D double imaging of a single cell. Deconvoluted cross-sectional micrographs of a Calcein AM-stained COS-7 cell in the zx plane for the first imaging (a) and second imaging (c). The 3D reconstruction of the cell from these images are shown in (b) and (d) for both instances. Red arrows point to regions of higher intensity. See ESI Movie S3 and S4 for the complete stack of planar cross-sectional images obtained for the first and second imaging, respectively (color figure online)
As a reference, a 2D image of the same cell obtained using a standard fluorescent microscope can be seen in contrast to the 3D image obtained here in ESI Fig. S1, demonstrating the advancements enabled by this device.
For the second imaging process, the applied flow rate required to reverse the flow to pull the cell results in an increase in the cell velocity, and thus, a slight increase in the distance between consecutively captured images (approximately 1.48 µm). Unfortunately, as the camera used for these experiments was limited to 100 fps, it can be observed in the acquired cross-sectional images (Fig. 6c) that there is a slight loss in resolution in the form of blurring predominantly around the edge of the cell in comparison to the first set of images. While this can be improved for future applications of the demonstrated device, the resolution obtained here is still sufficient to display similarities between both rounds of imaging and prove the capacity for accurate comparison for single cell studies. For instance, along the bottom of the cell there is a region of higher intensity (labelled as 1 in Fig. 6) that can be seen throughout all of the images in both cases, along with two smaller high-intensity regions in the upper left and right region (predominantly seen in both Fig. 6a and c, labelled 2). The deconvoluted stacks were used to obtain a 3D reconstruction of the cell, as shown in Fig. 6b and d, where the high-intensity regions (labelled 1 and 2), and the overall structure of the cell, are emphasised further. The reproducibility of the sequential imaging is critical for accurately observing temporal changes in the cell, which is a need for applications in therapeutics and drug delivery to observe morphological changes within the cell over time.
To further demonstrate the resolution and application capabilities of the device, a mitochondrial stain, MitoTracker, was used to gather volumetric images of the intracellular organelles. Since mitochondria are essential to many cellular processes, monitoring their activity within a cell is valuable. COS-7 cells were again used and were stained attached to the culture flask surface. As a reference, inverted fluorescent images of the attached cells were obtained to verify the activity of the mitochondria within the cells (Fig. 7a and c). As expected, the mitochondria are primarily located within the vicinity of the cell nucleus, and their activity (correlated to the intensity of the stain) varies from cell to cell based on which stage of the cell cycle (Mitosis, G1, S, or G2) they’re in (Antico Arciuch et al. 2012; Herrera et al. 2018). The double imaging experiments were performed as previously described, and planar images were obtained for both rounds of imaging. The mitochondrial activity within the cell was visualised via 3D reconstruction and the results can be seen Fig. 7b and c, where the insets show the cell undergoing a rotation about the z-axis. Similarly to the adhered cells in Fig. 7a, the volumetric data collected for the single cell in suspension depicts a high localisation of the mitochondria near to the nucleus (empty space depicted by the yellow arrow), and concentrated to one side. Between the first and second rounds of imaging there is no clear change in overall mitochondrial localisation, or area (approximately 9.8 µm, by 6.5 µm, by 12 µm (x, y, z)), and the same regions of higher intensity can still be seen. However, as seen with the Calcein stain, the second round of imaging has a slightly lower resolution due to the slight increase in cell velocity during the flow reversal. Based on the degree of rotation and the corresponding view of the mitochondria in the inset images, the cell has undergone an overall rotation between the first and second rounds of imaging but can still be directly compared. Given that mitochondria, on average, have diameters ranging from 0.5 to 1 µm (Alberts et al. 2002), the ability to image these intracellular organelles demonstrates the high resolution capabilities of our acousto-optofluidic LSFM device. See ESI Movie S5 for the top view of the double cell imaging process at the imaging region of the device, and ESI Movie S6 and S7 for the complete 3D reconstruction of the cell for the first and second imaging, respectively.
High-resolution 3D double imaging of mitochondria within a single cell. COS-7 cells were stained with MitoTracker deep red (MTDR). a Adhered cells in a culture flask stained for comparison, image taken with an inverted fluorescent microscope. b 3D images obtained of a single cell during the first round of imaging. c 3D images of the same cell during the second round of imaging. In both cases, subset images demonstrate the cell at different rotations along the z-axis. Yellow arrow points to the empty space where the nucleus should be located. See ESI Movie S5 for the top view of the double cell imaging process at the imaging region of the device, and ESI Movie S6 and S7 for the complete 3D reconstruction of the cell for the first and second imaging, respectively (color figure online)
3 Conclusions
A monolithic acousto-optofluidic chip was successfully developed to perform single cell double imaging of live cells utilising LSFM technology; also demonstrating the compatibility of the piezoelectric substrate with the optical components. Through the actuation of IDTs, surface acoustic waves were used to perform reliable, accurate and controllable single cell loading, and to re-collect the same cell for a second round of imaging. This process was semi-automated using an external control system capable of triggering the first pair of IDTs and the extraction syringe pump responsible for the flow reversal to pull the re-collected cell for a second imaging. Furthermore, during the imaging process, the loading IDTs behave as a gate to keep any unwanted cells from entering the imaging region and interrupting the imaging process of the cell of interest.
The acoustics allows for the targeted and tailored control of cells and is applicable to a range of cell types with varying dimensions and characteristics. Although hydrodynamic forces have been used for particle and cell trapping and manipulation in other applications (Tanyeri et al. 2010; Kumar et al. 2020a, b), the use of acoustic forces allows for a more flexible level of control. The on-chip trajectory control allows for the possibility of fully automating the device for cell loading and multi-pass imaging. In the future, the control system could also be used to trigger the camera for imaging.
Appropriate sampling of the cell volume was achieved to obtain high-resolution planar cross-sectional images of the cell cytoplasm and mitochondria, which were then used for 3D reconstruction. Thanks to the acoustic radiation force, utilised not only for loading and collecting the cell but also for positioning and maintaining the cell location in the vertical direction (away from the LN substrate), further aided by the sheath flow, the cell could be imaged within the same intensity region of the light-sheet in both sequences. This positioning mechanism is important not only for reducing potential scattering effects when the imaging takes place near the LN substrate, but more importantly for repeatability. The accurate and reproducible imaging capabilities of the device allow for sequential imaging of a single cell where temporal data can be obtained. This is of critical importance for comparing morphological changes within the cell over a period of time, for instance when monitoring cell drug intake or efficacy. Furthermore, observing mitochondrial levels within cells is important in applications like cancer research, where mitochondrial populations, or activity, are increased in cancerous cells. Being able to better probe and understand the role of mitochondria would provide researchers with more therapeutic opportunities.
4 Methods
4.1 Device fabrication
To ensure that the optical fibre holder and the micro-lens used to form the light-sheet are aligned with the channel, the device was fabricated from a single PDMS mould. The cylindrical micro-lens is formed by an air pocket (with a refractive index of n = 1), within the PDMS (n = 1.41). The optical fibre used to couple the light in the system is held within an on-chip fibre holder (open-ended channel) such that it is centred on the optical axis of the micro-lens (see Fig. 2a for top view images of the fabricated device depicting all optical components (i) and the generated light-sheet (ii), and Fig. 2bi for a side view image of the light-sheet). The fabrication process of this optofluidic system overcomes the effects of drift experienced by traditional imaging platforms (Louvergneaux et al. 2004; Gustavsson et al. 2018) since the alignment accuracy is only dependent on the microfabrication tolerances. Since the micro-lens and fluidic channels are fixed, the only spatial variable is the distance of the fibre tip to the micro-lens.
The acoustic components, microfluidic channels and all optofluidic components were fabricated utilising microfabrication techniques as described by Richard et al. (2022) and Vargas-Ordaz et al. (2021), respectively. Briefly, the focused IDTs were patterned on a 128° YX lithium niobate (LiNbO3) piezoelectric substrate via a standard photolithography and lift-off process. The metal deposition consisted of a 10 nm thick chrome adhesion layer and a 290 nm aluminium layer. To prevent corrosion of the metal layers, as well as to improve bonding of the PDMS microfluidic channel, a 300 nm thick layer of SiO2 was deposited using Electron beam evaporation. The SAW wavelength of the IDTs was designed to be 30 µm. For actuation of the IDTs, the contact pads of the IDTs were electrically connected to a signal generator via copper wire and conductive silver paint.
The microfluidic channels were fabricated from PDMS (5:1) (Sylgard 184; Dow Corning, Midland, MI, USA) via soft lithography using an SU-8 (MicroChemicals GmbH, Ulm, Germany) master mould. The channel design incorporated all inlets, outlets, trapping microchannels, as well as all of the required imaging components. The microchannels had varying channel heights, the imaging components (imaging chamber, optical fibre holder, and the micro-lens) were 120 µm in height whereas the remainder of the channels, where cell loading and collection takes place, were 40 µm in height. As such, the SU-8 mould was fabricated in a two-step lithography process where a mask aligner (EVG620, EV Group, Austria) was utilised to align the features. Prior to casting, the mould is silanised using trichloro (1H,1H,2H,2H perfluorooctyl) silane (Sigma-Aldrich, Missouri, USA) for improved detachment.
The PDMS was then cast over the SU-8 master, degassed to remove all air-bubbles and cured overnight at 70°C on a hotplate. The microfluidic channel was then bonded directly to the lithium niobate chip using plasma activation (PDC 32G, Harrick Plasma, USA). To ensure high-resolution imaging, optical aberrations and scattering must be eliminated by ensuring that the sidewall facing the detection lens is as smooth as possible. Thus, uncured, degassed PDMS was cast onto the sidewall and moulded using a silanised glass slide pressed up to the side of the device with a 90° angle. Curing was done as before.
4.2 Experimental setup and light-sheet characterisation
For experiments, the acoustofluidic imaging chip was mounted on a stage of a custom optical setup on an optical table (Thorlabs, New Jersey, USA. ITEM #T1220D). The optical stage was a 3-axis linear translation stage (Thorlabs, New Jersey, USA. ITEM #XR50C/M, XR25C/M, XR25C/M, XR25-YZ/M). The loading IDTs were actuated at a frequency of 127.4 MHz using A/C pulses by a signal generator (APSIN3000HC, Anapico, Switzerland). The output signal was amplified by a high-power amplifier (ZHL-5W- 1+, Mini-Circuits, USA). The output power applied to each loading IDT was 63 mW (126 mW total). The actuation was triggered via a custom control system composed of an Arduino (Arduino Mega 2560) and accompanying circuitry. The second IDT pair used for collecting the cell for re-imaging was actuated at 132.3 MHz via a separate signal generator (PowerSAW, BelektroniG GmbH) with a total applied power of 185 mW (which was then split between the two IDTs). The cell sample and sheath flow media were continuously introduced into the microfluidic channel using a syringe pump with two modules (Cetoni GmbH, Korbussen, Germany. ITEM #NEM-B101-03 A)) via 1 ml syringes (Tuberculin, Becton Dickinson). The extraction channel was connected to a separate syringe pump (KDS100, KD Scientific, Holliston, MA, USA) that could be triggered using the control system.
The light-sheet was generated using a 488 nm (green) benchtop laser source (Thorlabs, New Jersey, USA. ITEM #S4FC488) was used for imaging the Calcein, and a 637 nm (red) benchtop laser source (Thorlabs, New Jersey, USA. ITEM #S4FC637) was used for imaging the MitoTracker. Both were connected to a single-mode optical fibre (Thorlabs, New Jersey, USA. ITEM #P1-460B-FC). The side view microscope system consisted of a 40 × objective lens with NA = 0.6 (Olympus, Tokyo, Japan. ITEM #LUCPLFLN40X), a GFP emission filter (Thorlabs, New Jersey, USA. ITEM #MF525-39) for the green laser, which was swapped for a longpass filter (Thorlabs, New Jersey, USA. ITEM #FELH0650) for the red laser, a fixed tube lens of 200 mm (EHD imaging GmbH, Damme, Germany. ITEM #FT160) and a CMOS camera with a pixel size of 4.8 µm (Basler AG, Ahrensburg, Ger- many. ITEM#acA1920-150um– Basler ace). The top view microscope system utilised a 5 × objective with NA = 0.14 (Mitutoyo, Japan. Y86038815), with a fixed tube lens of 160 mm (EHD imaging GmbH, Damme, Germany. ITEM #FT160), a LED light source (EHD imaging GmbH, Damme, Germany. ITEM #IL100), and a second CMOS camera, with a pixel size of 1.465 µm (The Imaging Source Asia Co., Ltd. DMK 33UX174). Both imaging components were assembled on a rail system (Qioptiq, Rhyl, UK. X 95 Profile System).
The light-sheet was observed from the top view by flowing a solution of fluorescein (a contrast agent) and for the imaging resolution was characterised from the side view using a solution of 210 nm fluorescent beads (Bangs Laboratories, Inc., Indiana, USA. Catalog number CFDG001, streptavidin coated microspheres. (480, 520)) diluted in a 2% PEG solution (PEG-PPG-PEG, Aldrich Product Number 542342).
4.3 Image processing
All of the image post-processing and analysis were performed using Fiji. The collected planar images of the Calcein-stained cell were deconvoluted to obtain Fig. 6a and c using the DeconvolutionLab2 plugin with the PSF shown in Fig. 2b iii. The Richardson-Lucy algorithm was implemented four times, with an iteration of 10 for each. To obtain the volumetric 3D reconstruction of the cell (Fig. 6b and d), the CleanVolume Fiji plugin was used. The ClearVolume plugin requires the voxel depth to accurately recreate the 3D cell. The voxel depth is defined as the velocity of the cell times the time between planar images. Here, since the frame rate was 100 fps that means an image was obtained every 10 ms. The top view microscopic system was used to determine the velocity of the cell by relating the number of frames and time it took the cell to travel a 100 µm, and to measure the cell diameter.
For the MitoTracker-stained cell, the collected planar images were stacked using the 3D Project option in Fiji to obtain the 3D volumetric reconstructions of the cell mitochondria seen in Fig. 7. No deconvolution was performed. The same voxel depth was used.
4.4 Cell culturing and sample preparation
COS-7 cells (African green monkey kidney fibroblast-like cell line) were maintained in Dulbecco’s modified Eagle medium (DMEM) containing high levels of glucose (1 g L−1) and 110 mg L−1 of sodium pyruvate, supplemented with 100 U mL−1 of penicillin- streptomycin (P/S) (Life technologies) and 10% (v/v) fetal bovine serum (FBS) (Scientifix). The cells were conserved in humidified conditions at 37℃ with 5% CO2. The MG63 cells were cultured in the same manner.
For experimentation the cells were trypsinized, counted using a hemocytometer, re-suspended in a 1 ml solution of Calcein (2.5 µl of Calcein AM (Life Technologies, USA) in 5 ml of PBS), and incubated for 10 minutes at room temperature. The cells were then re-suspend in a fresh solution of PBS with 10 % of FBS, warmed to 37℃. The same media was used for the sheath flow. All solutions were filtered using a syringe filter (0.45 µm pore size, Acrodisc, Pall Corporation) prior to use to eliminate any unwanted debris (i.e. dust, leftover waste after a cell dies, precipitates of proteins, peptide aggregates, or phosphates) that would be large enough to clog the channels. For the Mitotracker staining, 2 ml of the staining solution (2.5 µl of MitoTracker deep red (MTDR) FM (Invitrogen, ThermoFisher Scientific, USA) in 12.5 ml of non-complete DMEM, high-glucose media) was added directly to the culture flask and the cells were incubated, adhered to the flask, in the incubator at 37℃ for 45 minutes. Non-complete media consists of only the DMEM, no added P/S or FBS so that oxidisation of the MTDR stain does not occur. The staining procedure was done for adhered cells due to the metabolic nature of the stain. After incubation the cells were trypsinized, counted and solutions were prepared as before.
Availability of data and material
Requests for additional material should be made to the corresponding authors.
References
Agostini Matteo, Lunardelli Francesco, Gagliardi Mariacristina, Miranda Alessio, Lamanna Leonardo, Luminare Antonella Giuliana, Gambineri Francesca, Lai Michele, Pistello Mauro, Cecchini Marco (2022) Surface-acoustic-wave (saw) induced mixing enhances the detection of viruses: Application to measles sensing in whole human saliva with a saw lab-on-a-chip. Adv Funct Mater. https://doi.org/10.1002/adfm.202201958
Akther A, Marqus S, Rezk AR, Yeo LY (2020) Submicron particle and cell concentration in a closed chamber surface acoustic wave microcentrifuge. Anal Chem 92(14):10024–10032. https://doi.org/10.1021/acs.analchem.0c01757
Alberts B, Johnson A, Lewis J et al (2002) The mitochondrion. In: Molecular biology of the cell, 4th edn. Garland Science, New York. https://www.ncbi.nlm.nih.gov/books/NBK26894/
Antico Arciuch VG, Elguero ME, Poderoso JJ, Carreras MC (2012) Mitochondrial regulation of cell cycle and proliferation. Antioxid Redox Signal 16(10):1150–80. https://doi.org/10.1089/ars.2011.4085
Castro JO, Ramesan S, Rezk AR, Yeo LY (2018) Continuous tuneable droplet ejection via pulsed surface acoustic wave jetting. Soft Matter 14(28):5721–5727. https://doi.org/10.1039/C7SM02534C
Cho Sung Hwan, Chen Chun H, Tsai Frank S, Godin Jessica M, Lo Yu-Hwa (2010) Human mammalian cell sorting using a highly integrated micro-fabricated fluorescence-activated cell sorter (facs). Lab Chip 10(12):1567–1573. https://doi.org/10.1039/C000136H
Deschout H, Raemdonck K, Stremersch S, Maoddi P, Mernier G, Renaud P, Jiguet S, Hendrix A, Bracke M, Van den Broecke R, Röding M, Rudemo M, Demeester J, De Smedt SC, Strubbe F, Neyts K, Braeckmans K (2014) On-chip light sheet illumination enables diagnostic size and concentration measurements of membrane vesicles in biofluids. Nanoscale 6(3):1741–1747. https://doi.org/10.1039/C3NR04432G
Doan M, Vorobjev I, Rees P, Filby A, Wolkenhauer O, Goldfeld AE, Lieberman J, Barteneva N, Carpenter AE, Hennig H (2018) Diagnostic potential of imaging flow cytometry. Trends Biotechnol 36(7):649–652. https://doi.org/10.1016/j.tibtech.2017.12.008
Gao W, Zhao L, Jiang Z, Sun D (2020) Advanced biological imaging for intracellular micromanipulation: Methods and applications. Appl Sci 10(20):7308
Guo Feng, Li Peng, French Jarrod B, Mao Zhangming, Zhao Hong, Li Sixing, Nama Nitesh, Fick James R, Benkovic Stephen J, Huang Tony Jun (2015) Controlling cell-cell interactions using surface acoustic waves. Proc Natl Acad Sci 112(1):43–48. https://doi.org/10.1073/pnas.1422068112
Gustavsson Anna-Karin, Petrov Petar N, Lee Maurice Y, Shechtman Yoav, Moerner WE (2018) 3d single-molecule super resolution microscopy with a tilted light sheet. Nat Commun 9(1):123. https://doi.org/10.1038/s41467-017-02563-4
Habibi R, Neild A (2019) Sound wave activated nano-sieve (swans) for enrichment of nanoparticles. Lab Chip 19(18):3032–3044. https://doi.org/10.1039/c9lc00369j
Hao Nanjing, Wang Zeyu, Liu Pengzhan, Becker Ryan, Yang Shujie, Yang Kaichun, Pei Zhichao, Zhang Peiran, Xia Jianping, Shen Liang, Wang Lin, Welsh-Bohmer Kathleen A, Sanders Laurie H, Lee Luke P, Huang Tony Jun (2022) Acoustofluidic multimodal diagnostic system for Alzheimer’s disease. Biosens Bioelectron 196:113730. https://doi.org/10.1016/j.bios.2021.113730
Herrera EL, Azzam SZ, Berger MC, Diaz-Martinez LA (2018) Genomic heterogeneity meets cellular energetics: crosstalk between the mitochondria and the cell cycle. J Cancer Metastasis Treat 4:42. https://doi.org/10.20517/2394-4722.2018.35
Holzner Gregor, Mateescu Bogdan, van Leeuwen Daniel, Cereghetti Gea, Dechant Reinhard, Stavrakis Stavros, de Mello Andrew (2021) High-throughput multiparametric imaging flow cytometry: toward diffraction-limited sub- cellular detection and monitoring of sub-cellular processes. Cell Rep 34(10):108824. https://doi.org/10.1016/j.celrep.2021.108824
Hsu Jin-Chen, Chang Chih-Yu (2022) Enhanced acoustofluidic mixing in a semicircular microchannel using plate mode coupling in a surface acoustic wave device. Sens Actuators A Phys 336:113401. https://doi.org/10.1016/j.sna.2022.113401
Jensen EC (2013) Overview of live-cell imaging: requirements and methods used. Anat Rec 296(1):1–8. https://doi.org/10.1002/ar.22554
Jin S, Wei X, Liu Z, Ren J, Jiang Z, Abell C, Ziyi Yu (2019) Focused surface acoustic waves induced microdroplets generation and its application for microgels. Sens Actuators B Chem 291:1–8. https://doi.org/10.1016/j.snb.2019.04.055
Kumar D, Richter CM, Schroeder CM (2020a) Conformational dynamics and phase behavior of lipid vesicles in a precisely controlled extensional flow. Soft Matter 16(2):337–347. https://doi.org/10.1039/C9SM02048A
Kumar D, Shenoy A, Deutsch J, Schroeder CM (2020b) Automation and flow control for particle manipulation. Curr Opin Chem Eng 29:1–8. https://doi.org/10.1016/j.coche.2020.02.006
LaBelle CA, Massaro A, Cortés-Llanos B, Sims CE, Allbritton NL (2021) Image-based live cell sorting. Trends Biotechnol 39(6):613–623. https://doi.org/10.1016/j.tibtech.2020.10.006
Li Sixing, Guo Feng, Chen Yuchao, Ding Xiaoyun, Li Peng, Wang Lin, Cameron Craig E, Huang Tony Jun (2014) Standing surface acoustic wave based cell coculture. Anal Chem 86(19):9853–9859. https://doi.org/10.1021/ac502453z
Lin M, Liu Q, Chao Liu Xu, Qiao CS, Xuantao Su (2018) Label-free light-sheet microfluidic cytometry for the automatic identification of senescent cells. Biomed Opt Express 9(4):1692–1703. https://doi.org/10.1364/BOE.9.001692
Link A, Franke T (2020) Acoustic erythrocytometer for mechanically probing cell viscoelasticity. Lab Chip 20(11):1991–1998. https://doi.org/10.1039/C9LC00999J
Louvergneaux Eric, Szwaj Christophe, Agez Gonzague, Glorieux Pierre, Taki Majid (2004) Experimental evidence of absolute and convective instabilities in optics. Phys Rev Lett 92(4):043901. https://doi.org/10.1103/PhysRevLett.92.043901
Lv H, Chen X (2022) Novel study on the mixing mechanism of active micromixers based on surface acoustic waves. Ind Eng Chem Res 61(28):10264–10274. https://doi.org/10.1021/acs.iecr.2c01539
Maramizonouz Sadaf, Jia Changfeng, Rahmati Mohammad, Zheng Tengfei, Liu Qiang, Torun Hamdi, Qiang Wu, YongQing Fu (2022) Acoustofluidic patterning inside capillary tubes using standing surface acoustic waves. Int J Mech Sci 214:106893. https://doi.org/10.1016/j.ijmecsci.2021.106893
Mejía Morales J, Glynne-Jones P, Vassalli M, Lippi GL (2022) Acoustofluidic interferometric device for rapid single-cell physical phenotyping. Eur Biophys J 51(2):185–191. https://doi.org/10.1007/s00249-021-01585-7
Mutafopulos K, Spink P, Lofstrom CD, Lu PJ, Lu H, Sharpe JC, Franke T, Weitz DA (2019) Traveling surface acoustic wave (tsaw) microfluidic fluorescence activated cell sorter (facs). Lab Chip 19:2435–2443. https://doi.org/10.1039/C9LC00163H
Ng Jia Wei, Neild Adrian (2021) Multiple outcome particle manipulation using cascaded surface acoustic waves (csaw). Microfluid Nanofluid 25(2):16. https://doi.org/10.1007/s10404-020-02417-4
Nguyen TD, Tran VT, Fu YQ, Du H (2018) Patterning and manipulating microparticles into a three-dimensional matrix using standing surface acoustic waves. Appl Phys Lett 112(21):213507. https://doi.org/10.1063/1.5024888
Nitta Nao, Sugimura Takeaki, Isozaki Akihiro, Mikami Hideharu, Hiraki Kei, Sakuma Shinya, Iino Takanori, Arai Fumihito, Endo Taichiro, Fujiwaki Yasuhiro, Fukuzawa Hideya, Hase Misa, Hayakawa Takeshi, Kotaro Hiramatsu Yu, Hoshino Mary Inaba, Ito Takuro, Karakawa Hiroshi, Kasai Yusuke, Koizumi Kenichi, Lee SangWook, Lei Cheng, Li Ming, Maeno Takanori, Matsusaka Satoshi, Murakami Daichi, Nakagawa Atsuhiro, Oguchi Yusuke, Oikawa Minoru, Ota Tadataka, Shiba Kiyotaka, Shintaku Hirofumi, Shirasaki Yoshitaka, Suga Kanako, Suzuki Yuta, Suzuki Nobutake, Tanaka Yo, Tezuka Hiroshi, Toyokawa Chi-hana, Yalikun Yaxiaer, Yamada Makoto, Yamagishi Mai, Yamano Takashi, Yasumoto Atsushi, Yatomi Yutaka, Yazawa Masayuki, Di Carlo Dino, Hosokawa Yoichiroh, Uemura Sotaro, Ozeki Yasuyuki, Goda Keisuke (2018) Intelligent image-activated cell sorting. Cell 175(1):266-276.e13. https://doi.org/10.1016/j.cell.2018.08.028
Rambach RW, Biswas P, Yadav A, Garstecki P, Franke T (2018) Fast selective trapping and release of picoliter droplets in a 3d microfluidic pdms multi-trap system with bubbles. Analyst 143(4):843–849. https://doi.org/10.1039/C7AN01100H
Rezk Amgad R, Qi Aisha, Friend James R, Li Wai Ho, Yeo Leslie Y (2012) Uniform mixing in paper-based microfluidic systems using surface acoustic waves. Lab Chip 12(4):773–779. https://doi.org/10.1039/C2LC21065G
Richard C, Fakhfouri A, Colditz M, Striggow F, Kronstein-Wiedemann R, Tonn T, Medina-Sánchez M, Schmidt OG, Gemming T, Winkler A (2019) Blood platelet enrichment in mass-producible surface acoustic wave (saw) driven microfluidic chips. Lab Chip 19(24):4043–4051. https://doi.org/10.1039/C9LC00804G
Richard C, Devendran C, Ashtiani D, Cadarso VJ, Neild A (2022) Acoustofluidic cell micro-dispenser for single cell trajectory control. Lab Chip 22(18):3533–3544. https://doi.org/10.1039/D2LC00216G
Sagi Y, Basser P, Assaf Y (2009) Estimation of cell size using the composite hindered and restricted model of diffusion. In: International society for magnetic resonance in medicine, proceedings 17th scientific meeting: 1390. https://www.nichd.nih.gov/sites/default/files/inline-files/Cell_Size_Estimation.pdf
Salari Alinaghi, Appak-Baskoy Sila, Coe Imogen R, Abousawan John, Antonescu Costin N, Tsai Scott S. H, Kolios Michael C (2021) Dosage-controlled intracellular delivery mediated by acoustofluidics for lab on a chip applications. Lab Chip 21(9):1788–1797. https://doi.org/10.1039/D0LC01303J
Sesen M, Whyte G (2020) Image-based single cell sorting automation in droplet microfluidics. Sci Rep 10(1):8736. https://doi.org/10.1038/s41598-020-65483-2
Shilton Richard, Tan Ming K, Yeo Leslie Y, Friend James R (2008) Particle concentration and mixing in microdrops driven by focused surface acoustic waves. J Appl Phys 104(1):014910. https://doi.org/10.1063/1.2951467
Son J, Mandracchia B, Jia S (2020) Miniaturized modular-array fluorescence microscopy. Biomed Opt Express 11:7221. https://doi.org/10.1364/BOE.410605
Stelzer EHK (2015) Light-sheet fluorescence microscopy for quantitative biology. Nat Methods 12(1):23–26. https://doi.org/10.1038/nmeth.3219
Strobl Frederic, Klees Selina, Stelzer Ernst H. K (2017) Light sheet-based fluorescence microscopy of living or fixed and stained tribolium castaneum embryos. J Vis Exp 122:55629. https://doi.org/10.3791/55629
Tanyeri Melikhan, Johnson-Chavarria Eric M, Schroeder Charles M (2010) Hydrodynamic trap for single particles and cells. Appl Phys Lett 96(22):224101. https://doi.org/10.1063/1.3431664
Vargas-Ordaz EJ, Gorelick S, York HM, Liu B, Halls ML, Arumugam S, Neild A, de Marco A, Cadarso VJ (2021) Three-dimensional imaging on a chip using optofluidics light-sheet fluorescence microscopy. Lab Chip 21(15):2945–2954. https://doi.org/10.1039/D1LC00098E
Weber M, Huisken J (2011) Light sheet microscopy for real-time developmental biology. Curr Opin Genet Dev 21(5):566–572. https://doi.org/10.1016/j.gde.2011.09.009
Yordanov S, Neuhaus K, Hartmann R, Díaz-Pascual F, Vidakovic L, Singh PK, Drescher K (2021) Single-objective high-resolution confocal light sheet fluorescence microscopy for standard biological sample geometries. Biomed Opt Express 12(6):3372–3391. https://doi.org/10.1364/BOE.420788
Acknowledgements
This work was partially supported by an Ideas Grant (no 2004126) from the National Health and Medical Research Council (NHMRC). This work was performed in part at the Melbourne Centre for Nanofabrication (MCN) in the Victorian Node of the Australian National Fabrication Facility (ANFF). The authors would like to thank Melati Abdul Halim for her help with freezing and thawing the cells and to Bonan Liu for providing the COS-7 cells.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions.
Author information
Authors and Affiliations
Contributions
C.R. designed the acoustic aspects of the system, conducted the experiments and wrote the manuscript. E. J. V-O designed with the optical aspects of the system and edited the manuscript. V.J.C. and A.N. conceived the research directions, guided the project and edited the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
E. J. V-O, V. J. C. and A. N. hold a patent on the optical aspects for this work for a specific application which is outside the scope of this work. There are no conflicts to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file2 (MP4 25868 KB)
Supplementary file3 (MP4 25868 KB)
Supplementary file4 (MP4 8332 KB)
Supplementary file5 (MP4 186 KB)
Supplementary file6 (MP4 168 KB)
Supplementary file7 (MP4 59484 KB)
Supplementary file8 (MP4 421 KB)
Supplementary file9 (MP4 348 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Richard, C., Vargas-Ordaz, E.J., Cadarso, V.J. et al. Acoustic prison for single live cell 3D multi-imaging enabled by light-sheet microscopy. Microfluid Nanofluid 27, 35 (2023). https://doi.org/10.1007/s10404-023-02649-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10404-023-02649-0