Abstract
In the present study, we investigated the genetic structure and diversity of P. nigra populations in Bulgaria, using simple sequence nuclear repeats. Among-population structure was studied with distance and Bayesian frequency methods, assuming geometric distance and a “non-admixture” model. The “NJ” and “non-admixture” clusters confirm the “mountain effect” hypothesis of the black pine genetic structure in the study region. The analyses showed moderate among-population divergence (13.31 %; AMOVA) and evidence of genetic bottlenecks. The coalescent analyses suggest that P. nigra has survived for a long period (thousands of generations) under strong selection pressure and that its populations continued to be exposed to stochastic factors like climate fluctuation, forest fire and disease. The combination of recent and historic changes is responsible for the present population size and genetic diversity. Our results suggest that conservation and management practices should strive to maintain this genetic differentiation, specifically by emphasising reforestation efforts with stocks from local provenances to avoid non-local introductions.



Similar content being viewed by others
References
Alexandrov A, Rafailov G, Nedelin G, Canov K, Bogdanov B, Spasov C (1988) Coniferous forest in Bulgaria. Zemizdat, Sofia [in Bulgarian]
Auckland LD, Bui T, Zhou Y, Shepherd M, Williams C (2002) Conifer microsatellite handbook. Corporate Press, Raleigh
Beaumont MA (1999) Detecting population expansion and decline using microsattelites. Genetics 153:2013–2029
Belkhir K (2002) GENETIX V. 4.01 A software for population genetics data analysis. Laboratoire Génome et populations, Université de Montpellier II, France
Bondev I (1973) Atlas of Bulgaria. Bulgaria Academy of Sciences, Sofia [in Bulgarian]
Cengel B, Tayanc Y, Kandemir G, Velioglu E, Alan M, Kaya Z (2012) Magnitude and efficiency of genetic diversity captured from seed stands of Pinus nigra (Arnold) subsp. pallasiana in established seed orchards and plantations. New For 43(3):303–317
Derory J, Mariette S, Gonzalez-Martınez SC, Chagne D, Madura D, Gerber S, Brach J, Persyn F, Ribeiro MM, Plomion C (2002) What can nuclear microsatellites tell us about maritime pine genetic resources conservation and provenance certification strategies? Ann For Sci 59:699–708
Dobrinov I (1983) Genetic and selection of forest trees. Zemizdat, Sofia [in Bulgarian]
Dobrinov I, Doykov G, Gagov V (1982) Forest genetic pool in Bulgaria. Zemizdat, Sofia [in Bulgarian]
Evanno G, Madec L, Arnaud JF (2005) Multiple paternity and post-copulatory sexual selection in a hermaphrodite what influences sperm precedence in the garden snail Helix aspersa? Mol Ecol 14:805–812
Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Excoffier L, Schneider S, Roessli D (2002) ARLEQUIN V.2.001 A software for population genetics data analysis. Department of Anthropology and Ecology, University of Geneva, Geneva
Falush DM, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data linked loci and correlated allele frequencies. Genetics 164:1567–1587
Fave MJ, Turgeon J (2008) Patterns of genetic diversity in Great Lakes bloaters (Coregonus hoyi) with a view to future reintroduction in Lake Ontario. Cons Gen. doi:10.1007/s10592-007-9339-6
Felsenstein J (1985) Confidence limits on phylogenies an approach using the bootstrap. Evolution 39(4):783–791
Fukarek P (1958a) Prilog poznavanju cgnog bora. Radovi, Poljoprivredno-sumarskog fakulteta Univerziteta u Sarajevu 3:3–146 [in Serbian]
Fukarek P (1958b) Die Standortsrassen der Schwarzfohre (Pinus nigra Arn.). Centralbl F d ges Forstwesen 75:203–207 [in German]
Garza JC, Williamson EG (2001) Detection of reduction in population size using data from microsatellite DNA. Mol Ecol 10:305–318
Goncharenko GG, Silin AE (1997) Populyatsionnaya i evolyutsionnaya genetika sosen Vostochnoi Evropy i Sibiri. [Population and Evolutionary Genetics of Pine in Eastern Europe and Siberia.] Tekhnalogiya, Minsk, Belarus [in Russian]
Hartl DL, Clark AG (1997) Principles of population genetics, 3rd edn. Sinauer Associates Inc., Sunderland
Hedrick P (1996) Bottleneck (s) or metapopulation in cheetahs. Cons Biol 10:897–899
Hedrick PW (2000) Genetics of populations, 2nd edn. Jones and Bartlett, Boston
Holmes S (2003) Bootstrapping phylogenetic trees theory and methods. Stat Sci 18:241–255
Ihaka R, Gentleman R (1996) R A language for data analysis and graphics. J Comput Graph Stat 5:299–314
Jones ME, Paetkau D, Geffen E, Moritz C (2004) Genetic diversity and population structure of Tasmanian devils, the largest marsupial carnivore. Mol Ecol 13:2197–2209
Kalinowski ST (2009) How well do evolutionary trees describe genetic relationships between populations? Heredity 102:506–513
Keller LF, Waller DM (2002) Inbreeding effects in wild populations. Trends Ecol Evol 17:230–241
Kimura M, Crow JF (1964) The number of alleles that can be maintained in a finite population. Genetics 49:725–738
Kostov K (1974) A very insect-resistant form of Pinus nigra (Arn.) in Bulgaria. For Manag (Bulgaria) 3:6–16 [in Bulgarian]
Kostov G, Paligorov I, Petrov S, Bogdanov Z (2005) Illegal logging in Bulgaria. In: Hirschberger P (ed) WWF European Forest programme and the Danube Carpathian Programme (DCP)
Krutovskii KV, Erofeeva SY, Aagaard JE, Strauss SH (1999) Simulation of effects of dominance on estimates of population genetic diversity and differentiation. J Hered 60:499–502
Leberg PL, Firmin BD (2008) Role of inbreeding depression and purging in captive breeding and restoration programmes. Mol Ecol 17:334–343
Lehman N (1998) Conservation biology genes are not enough. Curr Biol 8:R722–R724
Lian C, Miwa M, Hogetsu T (2000) Isolation and characterization of microsatellite loci from the Japanese red pine, Pinus densiflora. Mol Ecol 9:1171–1193
Lippe C, Dumont P, Bernatchez L (2006) High genetic diversity and no inbreeding in the endangered copper redhorse, Moxostoma hubbsi (Catostomidae, Pisces) the positive sides of a long generation time. Mol Ecol 15:1769–1780
Mariette S, Chagnea D, Lezier C, Pastuszka P, Raffin A, Plomion C, Kremer A (2001) Genetic diversity within and among Pinus pinaster populations: comparison between AFLP and microsatellite markers. Heredity 86:469–479
Mihailov V (1993) Biological and morphological study of European Black pine (Pinus nigra Arn.) seeds from different provenances and selection structures in Pirin and Slavianka mountains in Bulgaria. Ph.D. dissertation, Forest Research Institute, Bulgarian Academy of Sciences, Sofia, Bulgaria [in Bulgarian]
Mihailov V (1998) Variability of European Black pine (Pinus nigra Arn.) according to the size, weight and shape of cone apophyses in Pirin and Slavianka mountains. For Sci (Bulgaria) 1–2:24–37 [in Bulgarian]
Mirov NT (1967) The genus Pinus. Ronald Press, New York
Naydenov KD, Velkov D, Alexandrov A, Genov K, Asparuchova E, Iliev I (1993/1996) Research on chemophenotypic variation of representatives in the genus PINUS with regards to their preservation—Rapport N CC-318/93 NFNI-MONT, Bulgaria [in Bulgarian]
Naydenov KD, Tremblay MF, Ganchev P (2003) Karyotype diversity in of European Black pine (Pinus nigra Arn.) from Bulgarian provenances. Phyton 43(1):9–28
Naydenov KD, Tremblay F, Fenton N, Alexandrov A (2006) Structure of Pinus nigra Arn. populations in Bulgaria revealed by chloroplast microsatellites and terpenes analysis Provenance tests. Biochem Syst Ecol 34:562–574
Naydenov KD, Naydenov MK, Tremblay F, Alexandrov A, Aubin-Fournier LD (2011) Patterns of genetic diversity that result from bottlenecks in Scots Pine and the implications for local genetic conservation and management practices in Bulgaria. New For 42:179–193
Naydenov KD, Alexandrov A, Matevski V, Vasilevski K, Naydenov MK, Gyuleva V, Carcaillet C, Wahid N, Kamary S (2014) Range-wide genetic structure of maritime pine predates the last glacial maximum: evidence from nuclear DNA. Hereditas 151:1–13
Nei M (1975) Molecular population genetics and evolution. North-Holland, Amsterdam
Nikolic D, Tucic N (1983) Isoenzyme variation within and among populations of European Black pine (Pinus nigra Arn.). Silvae Genet 32:80–89
Peakall R, Smouse PE (2006) GENALEX V. 6 genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pertoldi C, Bijlsma R, Loeschcke V (2007) Conservation genetics in a globally changing environment present problems, paradoxes and future challenges. Biodivers Conserv 16:4147–4163
Pimm SL, Gittleman JL, McCracken GF, Gilpin M (1989) Plausible alternatives to bottlenecks to explain reduced genetic diversity. Trends Ecol Evol 4:176–178
Pritchard JK, Wen W (2003) Documentation for structure software version 2 [online]. http://pritch.bsd.uchicago.edu/software/readme_2_1/readme.html. Accessed 17 June 2005
Raymond M, Rousset F (1995a) An exact test for population differentiation. Evolution 49:1280–1283
Raymond M, Rousset F (1995b) GENEPOP: a population genetics software for exact test and ecumenicism. J Hered 86:248–249
Ribeiro MM, Plomion C, Petit RJ, Vendramin GG, Szmidt AE (2001) Variation in chloroplast single-sequence repeats in Portuguese maritime pine (Pinus pinaster Ait.). Theor Appl Genet 102:97–103
Rogers JS (1972) Measures of genetic similarity and genetic distance. Univ Tex Publ 7213:145–153
Rubio-Moraga A, Candel-Perez D, Lucas-Borja ME, Tiscar PA, Viñegla B, Linares JC, Gómez-Gómez L, Ahrazem O (2012) Genetic diversity of Pinus nigra Arn. Populations in Southern Spain and Northern Morocco revealed by inter-simple sequence repeat profiles. Int J Mol Sci 13:5645–5658
Salim K, Naydenov KD, Benyounes H, Tremblay F, Latifa EH, Wahid N, Valeria O (2010) Genetic signals of ancient decline in Aleppo pine populations at the species’ southwestern margins in the Mediterranean Basin. Hereditas 147:165–175
Scaltsoyiannes A, Rohr R, Panetsos K, Tsaktsira M (1994a) Allozyme frequency distributions in five European populations of Black pine (Pinus nigra Arnold). I. Estimation of genetic variation within and among populations. Silvae Genet 43:20–25
Scaltsoyiannes A, Rohr R, Panetsos K, Tsaktsira M (1994b) Allozyme frequency distributions in five European populations of Black pine (Pinus nigra Arnold). II. Contribution of isozyme analysis to the taxonomic status of the species. Silvae Genet 43:25–30
Shannon CE (1948a) A mathematical theory of communication. Bell Syst Tech J 27:379–423
Shannon CE (1948b) A mathematical theory of communication. Bell Syst Tech J 27:623–656
Slatkin M (1987) Gene flow and geographic structure of natural populations. Science (Washington, D.C.) 236:787–792
Slatkin M, Barton N (1989) A comparison of three indirect methods for estimating average levels of gene flow. Evolution 43:1349–1368
Stefanov B (1941/1942) Geographical distribution of coniferous species and their form in nature. Godichnik na Sofiiskia Darjaven Universitet, Sofia, (Bulgaria), XIX and XX, 1–88 [in Bulgarian]
Stefanov B (1943) The phyto-geographical elements of Bulgaria. Thesis of Bulgarian Academy of Sciences, Faculty of Nature and Mathematics, Sofia, (Bulgaria), Vol. XXXIX, 19:1–121 [in Bulgarian]
Tallmon DA, Beaumont MA, Luikart GH (2004) Effective population size estimation using approximate Bayesian computation. Genetics 167:977–988
Tekezaki N, Nei M (1996) Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics 144:389–399
Van Oosterhout C, Hutchinson WF, Wills DP, Shipley P (2004) Program note: MICRO—CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Velkov D, Mihailov V, Dobrev R (1983) Taxonomical and biological study of seed production practices for Pinus nigra (Arn.). In Nauchno-technicheska konferencia s mejdunarodno uchastie na tema “Nasoki I problemi na izgrajdaneto na gorskata semeproizvoditelna baza”, Borovec, Bulgaria. Vol 1, pp 12–13 [in Bulgarian]
Vidakovic M (1974) Genetics of European Black pine (Pinus nigra Arn.). Ann For (Zagreb) 6:57–86
Vidakovic M (1991) Conifers—morphology and variation. Graficki Zavod Hrvatske, Croitia
Watts BD, Byrd MA, Watts MU (2004) Status and distribution of breeding Ospreys in the Chesapeake Bay 1995–1996. J Raptor Res 38:47–54
Whitehouse AM, Harley EH (2001) Post-bottleneck genetic diversity of elephant populations in South Africa, revealed using microsatellite analysis. Mol Ecol 10:2139–2149
Willi Y, Van Buskirk J, Hoffmann AA (2006) Limits to the adaptive potential of small populations. Annu Rev Ecol Evol Syst 37:433–458
Williams CG, Elsik CG, Barnes RD (2000) Microsatellite analysis of Pinus taeda L. in Zimbabwe. Heredity 84:261–268
Wrigth S (1965) The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 19:395–420
Zhelev P (1992) Ecolo-biological and selection—genetic Scots Pine investigations from the Rhodope Mountains. Ph.D. Thesis of forest genetic, selection and tree breeding. University of Forestry, Sofia, Bulgaria [in Bulgarian]
Zhelev P, Longauer R, Ladislav P, Gomory D (1994) Genetic variation of the indigenous Scots Pine (Pinus sylvestris L.) populations from the Rhodope Mountains. For Sci (Bulgaria) 3:68–76 [in Bulgarian]
Acknowledgments
We thank Irena M. Naydenova and T&T for their technical assistance. We are grateful to the staff of the Bulgarian forest service for their assistance with the plant material; the Ministry of Education and Science of Bulgaria (EU) for their financial support (CC-318/1993–1996); and the Fond KAZAROVI-Geneva (Switzerland) for a student grant to K. D. Naydenov during an internship at the University of Geneva, Switzerland (Dept. of Botany). We also wish to thank Dan MacKay (CFLS-St-Jean, Qc., Canada) and Translation-Group (UK) for his careful revision of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Jarmo Holopainen.
This article is dedicated to the memory of Prof. Dr. Kosta Kostov from Forest Research Institute, Bulgarian Academy of Science (1924–2009).
Appendices
Appendix 1
See Table 3.
Appendix 2
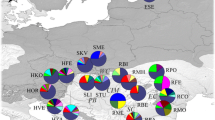
The neighbour-joining (NJ) dendrogram of Rogers’s (1972) genetic distance assumes the “non-mutation” model (i.e. geometric distance). The ring on the nodes indicates bootstrap values of ≥50 % at 1000 replications (R 2av = 0.94 for F st; Kalinowski 2009).

Rights and permissions
About this article
Cite this article
Naydenov, K.D., Mladenov, I., Alexandrov, A. et al. Patterns of genetic diversity resulting from bottlenecks in European black pine, with implications on local genetic conservation and management practices in Bulgaria. Eur J Forest Res 134, 669–681 (2015). https://doi.org/10.1007/s10342-015-0881-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10342-015-0881-3