Abstract
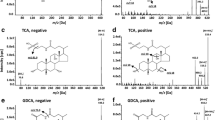
Bile acids are essential materials in the fatty acid cycle. Thus, understanding the metabolism of bile acids is significant. In this study, LC–MS was employed to analyze the composition and content of bile acids in rat cecal contents. Six individual bile acids were determined simultaneously. The mobile phase consisted of acetonitrile and 10 mmol L−1 of ammonium acetate solution. The flow rate was 0.2 mL min−1, and the total analysis time was 34 min. Under the developed conditions, all calibration curves for the six individual bile acids were achieved, and all correlation coefficients exceeded 0.99. Meanwhile, satisfactory results were obtained in the range of 50–500 µg g−1 for the six individual bile acids in the rat cecal contents. The LOD and LOQ of the bile acids were as low as 2.5 and 10 µg g−1 in the rat cecal contents, respectively, and the relative standard deviations of the peak area were within 1.25–2.01%. The dried residues after extraction were selected as the blank matrix. The overall recovery efficiencies of the bile acids in biological samples were increased to 97.2–103.2%. Therefore, the developed method can be used to quantify individual bile acids at extremely low levels in rat cecal contents.
Graphical Abstract



Similar content being viewed by others
References
Bhowmik SK, An JH, Lee SH, Jung BH (2012) vMAlteration of bile acid metabolism in pseudo germ-free rats. Arch Pharmacal Res 35(11):1969–1977
Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, Schoonjans K (2006) Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 439(7075):484–489
Jahnel J, Zöhrer E, Alisi A, Ferrari F, Ceccarelli S, De Vito R, Scharnagl H, Stojakovic T, Fauler G, Trauner M, Nobili V (2015) Serum bile acid levels in children with nonalcoholic fatty liver disease. J Pediatr Gastroenterol Nutr 61(1):85–90
Thomas LA, Veysey MJ, French G, Hylemon PB, Murphy GM, Dowling RH (2001) Bile acid metabolism by fresh human colonic contents: a comparison of caecal versus faecal samples. Gut 49(6):835–842
Bortolini O, Fantin G, Fogagnolo M, Mari L (2004) Control of the enantioselectivity by keto bile acid derivatives in the epoxidation of alkenes with Oxone. Tetrahedron Asymmetry 15(24):3831–3833
Kollerov VV, Monti D, Deshcherevskaya NO, Lobastova TG, Ferrandi EE, Larovere A, Gulevskaya SA, Riva S, Donova MV (2013) Hydroxylation of lithocholic acid by selected actinobacteria and filamentous fungi. Steroids 78(3):370–378
Hyun JJ, Lee HS, Kim CD, Dong SH, Lee S, Ryu JK, Lee DH, Jeong S, Kim TN, Lee J, Dong HK, Park ET, Lee I, Yoo BM, Kim JH (2015) Efficacy of magnesium trihydrate of ursodeoxycholic acid and chenodeoxycholic acid for gallstone dissolution: a prospective multicenter trial. Gut Liver 9(4):547–555
Ridlon JM, Kang DJ, Hylemon PB (2006) Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47(2):241–259
Park JY, Park BK, Ko JS, Bang S, Song SY, Chung JB (2006) Bile acid analysis in biliary tract cancer. Yonsei Med J 47(6):817–825
Jurek D, Fleckl E, Marian B (2005) Bile acid induced gene expression in LT97 colonic adenoma cells. Food Chem Toxicol 43(1):87–93
Ghaffarzadegan T, Nyman M, Jönsson JÅ, Sandahl M (2014) Determination of bile acids by hollow fibre liquid-phase microextraction coupled with gas chromatography. J Chromatogr B 944:69–74
Shi Y, Xiong J, Sun D, Liu W, Wei F, Ma S, Lin R (2015) Simultaneous quantification of the major bile acids in Artificial Calculus bovis by high-performance liquid chromatography with precolumn derivatization and its application in quality control. J Sep Sci 38(16):2753–2762
Mi S, Lim DW, Turner JM, Wales PW, Curtis JM (2016) Determination of bile acids in piglet bile by solid phase extraction and liquid chromatography–electrospray tandem mass spectrometry. Lipids 51(3):359–372
Stojakovic T, Putz-Bankuti C, Fauler G, Scharnagl H, Wagner M, Stadlbauer V, Gurakuqi G, Stauber RE, Marz W, Trauner M (2007) Atorvastatin in patients with primary biliary cirrhosis and incomplete biochemical response to ursodeoxycholic acid. Hepatology 46(3):776–784
Griffiths WJ, Sjovall J (2010) Bile acids: analysis in biological fluids and tissues. J Lipid Res 51(1):23–41
Bobeldijk I, Hekman M, de Vries van der Weij J, Coulier L, Ramaker R, Kleemann R, Kooistra T, Rubingh C, Freidig A, Verheij E (2008) Quantitative profiling of bile acids in biofluids and tissues based on accurate mass high resolution LC–FTMS: compound class targeting in a metabolomics workflow. J Chromatogr B 871:306–313
Sarafian MH, Lewis MR, Pechlivanis A, Ralphs S, McPhail MJ, Patel VC, Dumas ME, Holmes E, Nicholson JK (2015) Bile acid profiling and quantification in biofluids using ultra-performance liquid chromatography tandem mass spectrometry. Anal Chem 87(19):9662–9670
Lee G, Lee H, Hong J, Lee SH, Jung BH (2016) Quantitative profiling of bile acids in rat bile using ultrahigh-performance liquid chromatography–orbitrap mass spectrometry: alteration of the bile acid composition with aging. J Chromatogr B 1031:37–49
Hagio M, Matsumoto M, Fukushima M, Hara H, Ishizuka S (2008) Improved analysis of bile acids in tissues and intestinal contents of rats using LC/ESI-MS. J Lipid Res 50(1):173–180
Amplatz B, Zöhrer E, Haas C, Schäffer M, Stojakovic T, Jahnel J, Fauler G (2017) Bile acid preparation and comprehensive analysis by high performance liquid chromatography–high-resolution mass spectrometry. Clin Chim Acta 464:85–92
Zhang C, Huang L, Wu Z, Chang C, Yang Z (2016) Determination of sulfonate ester genotoxic impurities in imatinib mesylate by gas chromatography with mass spectrometry. J Sep Sci 39(18):3558–3563
Acknowledgements
The author gratefully acknowledges the National Natural Science Foundation of China (No. 31400092) for their financial support of this study.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Funding
This study was funded by the National Natural Science Foundation of China (No. 31400092).
Conflict of interest
All authors declare that they no conflict of interest.
Human and animal rights statement
This study does not involve any experiment with animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Zhang, C., Zheng, Y., Ma, S. et al. Determination of Bile Acids in Rat Cecal Contents by LC–MS. Chromatographia 80, 1733–1739 (2017). https://doi.org/10.1007/s10337-017-3395-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10337-017-3395-y