Abstract
Research on the occurrence and community composition of vector-transmitted protozoan haemoparasites in birds is heavily skewed toward passerines with many other orders underrepresented. In caprimulgids, a family of primarily ground-nesting, crepuscular/nocturnal birds occupying a wide range of dry habitats, research on protozoan haemoparasites is limited and in most cases based on only a few individuals. Here, using the molecular approach, the occurrence and diversity of parasites from four genera (Haemosporida: Haemoproteus, Plasmodium, Leucocytozoon; Trypanosomatida: Trypanosoma) were investigated in a representative of the family—the Eurasian Nightjar (Caprimulgus europaeus). Birds were sampled at a breeding location in south-eastern Poland at the beginning of the breeding season. Overall, 20 individuals, including 17 males and 3 females, were screened. Only 10% of birds were infected and in total, two parasite lineages—both representing Plasmodium genus—were identified. Detected parasite lineages were previously registered in a wide range of avian hosts. Known transmission areas of these lineages indicate that breeding populations of Eurasian Nightjars from south-eastern Poland contract infections on non-breeding grounds. This study reinforces earlier observations of the low prevalence of haemosporidians and trypanosomes in caprimulgids.
Zusammenfassung
Geringe Prävalenz von Hämosporidien- und Trypanosomeninfektionen bei der Nachtschwalbe Caprimulgus europaeus
Forschung zum Auftreten und zur Zusammensetzung vektorübertragener protozoischer Blutparasiten bei Vögeln findet vor allem an Singvögeln statt; viele andere Ordnungen sind dagegen deutlich unterrepräsentiert. Bei den Caprimulgiden, einer Familie vorwiegend bodenbrütender, dämmerungs-/nachtaktiver Vögel, die ein breites Spektrum trockener Habitate besiedeln, sind Forschungsergebnisse zu protozoischen Blutparasiten begrenzt und basieren in den meisten Fällen nur auf wenigen Individuen. Wir wählten einen molekularen Ansatz, um Auftreten und Diversität von Parasiten aus vier Gattungen (Haemosporida: Haemoproteus, Plasmodium, Leucocytozoon; Trypanosomatida: Trypanosoma) bei einem Vertreter dieser Familie, der Nachtschwalbe Caprimulgus europaeus, zu untersuchen. Die Vögel wurden zu Beginn der Brutsaison in einem Brutgebiet in Südostpolen beprobt. Insgesamt untersuchten wir 20 Individuen, darunter 17 Männchen und drei Weibchen. Nur 10% der Vögel waren infiziert und es wurden insgesamt zwei Parasitenlinien—beide zur Gattung Plasmodium gehörig—festgestellt. Die beobachteten Parasitenlinien wurden zuvor bereits bei den verschiedensten Wirtsvögeln nachgewiesen. Die bekannten Übertragungsgebiete dieser Abstammungslinien deuten darauf hin, dass sich die südostpolnischen Brutpopulationen der Nachtschwalbe nicht in den Brutgebieten infizieren. Diese Studie bestätigt frühere Beobachtungen der geringen Befallshäufigkeit mit Hämosporidien und Trypanosomen bei Caprimulgiden.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Infections with vector-transmitted protozoan haemoparasites, such as haemosporidians and trypanosomes, are widespread among birds (Valkiūnas 2005). Despite the great interest in the interaction between these parasites and their avian hosts, and the steep increase in the number of studies focusing on this phenomenon, promoted by the introduction of molecular diagnostics (reviewed in Bensch and Hellgren 2020), our knowledge on the frequency of occurrence and diversity of haemoparasites is heavily skewed toward passerines with some other bird orders largely underexplored (MalAvi database, Bensch et al. 2009).
Caprimulgids, represented by 97 species classified in 19 genera, is a family of primarily ground-nesting, crepuscular/nocturnal, and insectivorous birds (Winkler et al. 2020). They have a worldwide distribution (except for Antarctica), occupying a wide range of habitats: deserts, semi-deserts, open country, forests as well as suburban areas. They have been shown to host various vector-transmitted haemoparasites, including representatives of the commonly studied genera: Haemoproteus, Plasmodium, Leucocytozoon and Trypanosoma (Williams et al. 1975; Greiner et al. 1975; White et al. 1978; Peirce 1981; Earle et al. 1991; Bennett et al. 1992; Valkiūnas 2005; Savage et al. 2009; Pori 2018). However, in only a few studies the number of examined birds was large enough (over 15 individuals) to allow avoiding the high statistical uncertainty of prevalence estimates (Jovani and Tella 2006). In all these cases prevalence was low or very low, ranging from 0 in the Red-necked Nightjar (Caprimulgus ruficollis) (Forero et al. 1997) and Swamp Nightjar (Caprimulgus natalensis) (Bennett et al. 1992), through 2.9–6.2% in the Square-tailed Nightjar (Caprimulgus fossii) (Earle et al. 1991; Bennett et al. 1992) and 5.6 in the Eurasian Nightjar (Caprimulgus europaeus) (Valkiūnas 2005), up to 7.1–10.0% in the Common Nighthawk (Chordeiles minor) (Williams et al. 1975; Greiner et al. 1975). However, given that to date, most studies of haemosporidian and trypanosome parasites in caprimulgids were based on blood smear screening, which is a less sensitive method than molecular screening, prevalence and/or diversity of parasites may be underestimated, especially in the case of low-intensity infections (Jarvi et al. 2002; Durrant et al. 2006; Garamszegi 2010, but see Valkiūnas et al. 2008; Argilla et al. 2013).
Here, we present data on the occurrence and community composition of four genera of vector-transmitted protozoans in a representative of the caprimulgid family—the Eurasian Nightjar (hereafter referred to as Nightjar). This crepuscular and nocturnal aerial insectivore is a long-distance migrant with breeding grounds ranging across Europe (except for northern regions beyond ca 64° N), parts of Asia and north-western Africa, and wintering grounds–across sub-Saharan Africa (Cramp 1985; Cleere et al. 2021). It breeds in diverse open-ground dry habitats including heathlands, clear-cuts, and burned areas in pine forests, producing one or two broods per breeding season (Cleere et al. 2021). The eggs, most commonly two, are laid in a shallow depression on the sparsely vegetated ground. The nesting cycle lasts for approximately 36 days.
We screened Nightjars for the presence in their bloodstream of parasites representing three genera of the order Haemosporida: Haemoproteus–vectored by hippoboscid flies (Hippoboscidae) and biting midges (Ceratopogonidae), Plasmodium–vectored by mosquitoes (Culicidae), Leucocytozoon–vectored by black flies (Simuliidae) and biting midges and parasites from a genus representing order Trypanosomatida: Trypanosoma–vectored by black flies, hippoboscid flies, biting midges, Culex spp. mosquitoes and dermanyssid mites (Dermanyssidae) (Molyneux 1977; Votýpka et al. 2012; Santiago-Alarcon et al. 2012). To date, parasites from all four genera have been detected in the Eurasian Nightjar (Peirce 1981; Shurulinkov and Golemansky 2002; Valkiūnas 2005). However, all studies except for one were based on the small number of examined birds (less than eight individuals) and used primarily blood smear screening to detect infections (Table 1). Here, we employed the molecular approach to screen for the presence of haemosporidians and trypanosomes in Nightjars sampled at a breeding location in south-eastern Poland.
Methods
Sampling site and sample collection
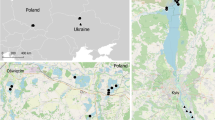
The sampling site—the Sobibór Landscape Park (hereafter called “Sobibór”) was selected based on the breeding habitat preferences of the Eurasian Nightjar (Cramp 1985; Cleere et al. 2021). It is located in the southeast of Poland, near the city of Włodawa, on the border with Ukraine and Belarus (51.42 N, 23.6 E). It is located in the UNESCO Cross-Border Biosphere Reserve “West Polesie”. The Sobibór Landscape Park, with an area of 10,000 ha (plus 9500 ha of a buffer zone), covers the most valuable parts of the Sobiborskie Forests with notable large wet areas, including well-preserved peat bogs and mid-forest lakes. Coniferous habitats, including dry, fresh, wet, and marshy coniferous forests are the most common type of habitat. Nightjars were caught mostly on dry clearings adjacent to wetlands.
Birds were sampled within a framework of a project focusing on the link between sexual characters (the white spots) and morphological and behavioural traits in Eurasian Nightjar males. To that end, chosen method of capturing nightjars targeted males during territory establishment. In Poland, Eurasian Nightjars arrive in April–May and depart to Africa in August–September. Birds were caught between 1 and 6 May (in 2018), a timing which corresponds to the beginning of species breeding season at the sampling area. Catching sessions were conducted from sunset to sunrise in the likely breeding territories. The Nightjars’ activity was usually the highest during dusk and dawn and moonlit nights. Birds were caught using an ultra-thin net (dimensions: 12 × 3 m, Ecotone, Gdańsk, Poland) fixed on two 6-m-long poles and playback of the song of a male unknown to the territorial individuals. Because mostly males react to playbacks, females were caught only occasionally. Birds were sexed and aged (2-year-old, 2-year-old or older, 3-year-old or older) based on plumage characteristics (Ottenby Bird Observatory 2015). All birds received a metal ring with a unique alphanumerical code. Finally, blood samples (up to 20 μl) were taken from the wing vein and stored in 96% ethanol. After transporting to the laboratory, the samples were kept at 4 °C until molecular analysis.
In total, 20 birds were sampled for blood. The proportion of males was 0.85, while the proportion of 2-year-old, 2-year-old or older and 3-year-old or older was 0.15, 0.7 and 0.15, respectively.
Molecular analyses
DNA was isolated using the ammonium acetate method (Bruford et al. 1998). The following procedures were used to screen for haemosporidian and trypanosome infections:
-
1)
In the case of haemosporidians the samples were first screened following the Ciloglu et al. (2019) protocol. This protocol is based on a multiplex polymerase chain reaction (PCR), which allows for simultaneous amplification of the DNA sequences of Haemoproteus, Plasmodium and Leucocytozoon and their unambiguous identification based on the length of the amplified sequences: 525–533 bp in the case of Haemoproteus, 377–379 bp in the case of Plasmodium and 218 bp in the case of Leucocytozoon. The multiplex PCR protocol shows slightly higher detection rates of haemosporidian parasites and, importantly, is superior at detecting multiple infections in comparison with the commonly used nested PCR protocol (Ciloglu et al. 2019). The multiplex PCR reactions were set up using 2 × Qiagen Multiplex PCR Master Mix (Qiagen, Hilden, Germany). In the next step, all samples were screened with a nested PCR using primers targeting a 478 bp long fragment of the cytochrome b of haemosporidians (Hellgren et al. 2004). Firstly, this step allowed us to verify whether both screening protocols yield the same results, and secondly, PCR products amplified with this protocol were used for sequencing to identify parasite lineages as listed in the MalAvi database (Bensch et al. 2009). The concentration of PCR reagents followed Kubacka et al. (2019) except for 0.625 units of Taq DNA polymerase (GoTaq G2 Hot Start Polymerase, Promega, Madison, USA) as recommended by the manufacturer, while PCR thermal profiles followed Hellgren et al. (2004).
-
2)
The presence of Trypanosoma parasites was confirmed with a nested PCR targeting a 326 bp long fragment of the 18 S rRNA gene (Sehgal et al. 2001). PCR reaction mix was prepared according to Kubacka et al. (2019) and thermal profiles followed Sehgal et al. (2001).
All samples were run twice with each protocol. In all cases, PCR reactions contained approximately 50 ng of total genomic DNA. Each run contained a negative control (ddH20 instead of DNA isolate) to check for contamination and a positive control (either DNA isolate of the Great Tit (Parus major) with confirmed infection with all three haemosporidian genera or with Trypanosoma) for a possible PCR failure.
The amplicons (6 μl) were run on 2% agarose gel stained with SimplySafe (Eurx, Gdańsk, Poland) and visualised under UV light. To check the quality of DNA isolates all samples scored as negative were tested with primers P2 and P8, which amplify the fragments of the sex-linked CHD1 gene (Griffiths et al. 1998). Although this primer set amplified in males and females (as identified based on external characteristics) only one fragment (or two fragments of a very similar size, which were not resolved on an agarose gel), the amplification of the product was used as an indication that DNA isolate was of good quality. PCR reactions and thermal conditions for amplification of fragments of the CHD1 gene followed Cichoń et al. (2003).
PCR products amplified with the protocol of Hellgren et al. (2004) were cleaned enzymatically (Exo-sap) and sequenced bidirectionally by Genomed (Warsaw, Poland). Chromatograms were inspected visually for the presence of multiple peaks which indicate mixed infections and annotated and aligned with BioEdit software ver. 7.2.3 (Hall 1999). Consensus sequences were next searched against the MalAvi database (Bensch et al. 2009) and GenBank. If the sequence did not match with 100% accuracy any of the sequences deposited in the MalAvi database/GenBank, PCR and sequencing were repeated to exclude the possibility that a novel sequence was a product of PCR or sequencing error(s).
95% confidence intervals (95% CIs) for prevalence were calculated with Quantitative Parasitology (QPweb) ver. 1.0.15 software using the Sterne’s method (Reiczigel et al. 2019; Klaschka and Reiczigel 2021). The difference in the proportion of infected individuals between males and females was tested with Fisher’s exact test. The analysis was performed with R v. 4.1.1 using fisher. Test command implemented in stats package (R Core Team 2021).
Results
Nested and multiplex PCR protocols employed to screen for haemosporidian infections produced the same results in terms of the identity of infected individuals and parasite genera.
Only 2 out of 20 Nightjars one male and one female—were positive for the presence of haemosporidian/trypanosome parasites (prevalence and 95% CIs 0.100, 0.018–0.320). The proportion of infected individuals was over five times lower in males (prevalence and 95% CIs 0.059, 0.003–0.287, n = 17) than in females (prevalence and 95% CIs 0.333, 0.017–0.865, n = 3), however, this difference was not statistically significant (Fisher’s exact test, p = 0.284). The male carried a single infection and the female—at least a single infection. It was not possible to unequivocally assess the number and identity of all lineages carried by the female, because of the inconsistency in the presence and location of double peaks in chromatograms of amplicons produced in three PCR replicates. The infections were caused by Plasmodium. Parasites representing three other genera—Haemoproteus, Leucocytozoon and Trypanosoma—were not detected.
In total, 2 lineages—ACCTAC01 and SW5—were identified, both previously recovered in other bird species. Lineage SW5 represents Plasmodium circumflexum morphospecies, while lineage ACCTAC01 has not been assigned to any morphospecies yet. Identified lineages were recovered to date from a wide range of families and orders: ACCTAC01—from 15 families in 7 orders and SW5—from 10 families in 9 orders (Table 2).
Discussion
We show using molecular screening that Eurasian Nightjars caught at the breeding site in south-eastern Poland rarely carry haemosporidian and trypanosome infections. Specifically, only 10% of birds at this location were infected and all infections belong to Plasmodium. Haemoproteus, Leucocytozoon and Trypanosoma were not detected.
Low prevalence of haemosporidian infections in the breeding population of Eurasian Nightjars is in accordance with the findings of Valkiūnas (2005) who reported two infected among 36 examined individuals of this species sampled during migration in the Curonian Spit in the Baltic Sea. Such low prevalence is also in accordance with infection rates found in other caprimulgids (Williams et al. 1975; Greiner et al. 1975; Earle et al. 1991; Bennett et al. 1992; Forero et al. 1997). Although the Eurasian Nightjar is known to host parasites from all four genera considered in this study–including Haemoproteus caprimulgi and Leucocytozoon caprimulgi, which are regarded as specific for caprimulgids (Williams et al. 1975; Shurulinkov and Golemansky 2002; Valkiūnas 2005)–we confirmed the presence of parasites belonging only to Plasmodium. Given the lack of blood smears in this study and limited knowledge about the taxonomic affiliation of specific lineages to morphospecies (MalAvi database, Bensch et al. 2009), only one morphospecies could be identified based on DNA sequence, namely Plasmodium circumflexum (Valkiūnas et al. 2014).
Males and females did not differ in infection rates with haemosporidians and trypanosomes. This finding falls in line with the results of a recent meta-analysis showing that in birds there is no difference between sexes in the prevalence of infection with any of the following categories of haemoparasites: microfilaria, Trypanosoma, Plasmodium, Leucocytozoon and Haemoproteus (Valdebenito et al. 2020). However, given the very small number of females tested for the presence of haemosporidian and trypanosome parasites in this study, no strong conclusions on sex-specific patterns of infection in the Eurasian Nightjar may be drawn.
Currently, there is scarce information about the molecular identity of haemosporidan parasites in caprimulgids. Apart from two lineages detected in the current study, only six other lineages (Haemoproteus: CHOMIN01, PAPOL01, TROAED20; Plasmodium: COPMAL02; Leucocytozoon: CAPLON01, CAPLON03) have been identified to date, all in species sampled in South America, North America and Asia (Ishtiaq et al. 2007; Lacorte et al. 2013; McNew et al. 2021). It has to be noted that in the case of lineages ACCTAC01 and SW5 detected in Nightjars in Sobibór, the lack of blood smears precludes the possibility to unequivocally show that the Eurasian Nightjar is a competent host for these parasites, e.g. an organism, in which the parasite completes its development and therefore the host may transmit the parasite to vectors (Valkiūnas 2005). Only the presence of haemosporidian gametocytes or erythrocytic meronts, which is verified with blood smears, is used as a confirmation that a species is a competent host (Valkiūnas et al. 2009). Consequently, it may not be excluded that for some of the parasites detected in this study, Eurasian Nightjars are dead-end hosts. Additionally, while molecular methods are highly sensitive at detection of parasites in the host’s blood (Sehgal et al. 2001; Hellgren et al. 2004), by using only molecular screening some parasites may be missed as has been shown in studies in which both the molecular approach and blood smear screening have been employed (Durrant et al. 2006). This may arise either because of limitations imposed by used primers or because of the preferential amplification of one of the lineages in the case of multiple infections (Pérez-Tris and Bensch 2005; Valkiūnas et al. 2006).
The low infection rate observed in the Eurasian Nightjar may be driven by a few not mutually exclusive mechanisms: susceptibility to infection, exposure to competent vectors and exposure to parasites for which Nightjars are competent hosts. Based on the currently available data it is not possible to assess to what extent susceptibility of Nightjars shapes low infection rates by vector-transmitted haemoparasites. The factor which may play an important role is exposure to vectors. On breeding grounds, Nightjars occupy dry soil habitats such as dry pine forests or heathlands (Cleere et al. 2021), and on wintering grounds in Africa–tropical grassland, savannah, shrubland and steppe vegetation with scattered trees and dense tree stands (Evens et al. 2017; Norevik et al. 2017). The common feature of many of these habitats is low water retention in the soil and low humidity. Because the majority of blood-feeding arthropods which vector Haemoproteus, Plasmodium, Leucocytozoon and Trypanosoma require access to water, either still or running, or humid substrate for oviposition and larvae development (Valkiūnas 2005; Santiago-Alarcon et al. 2012), habitats occupied by Nightjars, may not provide appropriate conditions for these arthropods. Therefore, such habitats may be expected to hold only low numbers of vectors if any.
Because the study species is migratory, it may contract infections on breeding grounds, wintering grounds and stopover sites during migration. Based on the requirements necessary to identify a geographic region as a transmission area (at least one of the three conditions has to be met: (1) the presence of infections in juvenile birds of migratory species, (2) the presence of infections in juvenile and/or adult birds of resident species, (3) the presence of infective parasite stages in salivary glands of vectors), Plasmodium lineage ACCTAC01 is currently considered to be transmitted only in Africa, while SW5 is in Africa, southern Europe and Asia (MalAvi database, Njabo et al. 2009; Ventim et al. 2012). Although currently there is no data on migration routes of Nightjars breeding in central Europe, birds from western and northern Europe (Belgium, Denmark, France, Sweden, UK) are known to stop in southern Europe and on several stopover sites in Africa before reaching wintering sites in the central sub-Saharan region (Evens et al. 2017; Norevik et al. 2017; Jacobsen et al. 2017). Given transmission areas of the parasites detected in this study, it is highly probable, that Eurasian Nightjars breeding in central Europe contract infections only outside of breeding grounds: on stopover and wintering sites in Africa and potentially also on stopover sites in southern Europe. Such a pattern of infection contraction is in accordance with the pattern suggested for long-distance Palearctic migrants wintering in Africa, which spend most of the year (ca 9 months) on non-breeding grounds (Valkiūnas 2005).
Summing up, we found that Eurasian Nightjars sampled at the breeding site in south-eastern Poland are in the vast majority free of haemospordians and trypanosomes. To what extent this pattern is mediated by high resistance to these parasites and to what extent by the choice of habitats occupied by the species, remains to be explored. More studies, based on molecular screening and large sample size, in other Eurasian Nightjar populations as well as in other caprimulgids are needed, to confirm the generality of the low frequency of haemosporidian and trypanosome infections in this family.
Data availability
Data associated with this study are available at the Open Science Framework repository: osf.io/87gyj.
References
Argilla LS, Howe L, Gartrell BD, Alley MR (2013) High prevalence of Leucocytozoon spp. in the endangered Yellow-eyed Penguin (Megadyptes antipodes) in the sub-Antarctic regions of New Zealand. Parasitology 140:672–682. https://doi.org/10.1017/S0031182012002089
Bennett GF, Earlé RA, Du Toit H, Huchzermeyer FW (1992) A host-parasite catalogue of the haematozoa of the sub-Saharan birds. Onderstepoort J Vet Res 59:1–73
Bensch S, Hellgren O (2020) The use of molecular methods in studies of avian haemosporidians. In: Santiago-Alarcon D, Marzal A (eds) Avian malaria and related parasites in the tropics: ecology, evolution and systematics. Springer International Publishing, Cham, pp 113–135
Bensch S, Hellgren O, Pérez-Tris J (2009) MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol Ecol Resour 9:1353–1358. https://doi.org/10.1111/j.1755-0998.2009.02692.x
Billerman SM, Keeney BK, Rodewald PG, Schulenberg TS (eds) (2022) Birds of the world. Cornell Lab of Ornithology, Ithaca, NY, USA
Bruford MW, Hanotte O, Brookfield JFY, Burke T (1998) Multilocus and single-locus DNA fingerprinting. In: Hoelzel AR (ed) Molecular genetic analysis of populations: a practical approach. IRL Press, Oxford, pp 287–336
Carlson ML, Proudfoot GA, Gentile K et al (2018) Haemosporidian prevalence in Northern Saw-whet Owls Aegolius acadicus is predicted by host age and average annual temperature at breeding grounds. J Avian Biol 49:e01817. https://doi.org/10.1111/jav.01817
Cichoń M, Dubiec A, Stoczko M (2003) Laying order and offspring sex in Blue Tits Parus caeruleus. J Avian Biol 34:355–359. https://doi.org/10.1111/j.0908-8857.2003.03201.x
Ciloglu A, Ellis VA, Bernotienė R et al (2019) A new one-step multiplex PCR assay for simultaneous detection and identification of avian haemosporidian parasites. Parasitol Res 118:191–201. https://doi.org/10.1007/s00436-018-6153-7
Cleere N, Christie DA, Rasmussen PC (2021) Eurasian Nightjar (Caprimulgus europaeus) version 1.1. In: Rasmussen PC (ed) Birds of the world. Cornell Lab of Ornithology, Ithaca, NY, USA
Cramp S (ed) (1985) Birds of the Western Palearctic, vol 4. Terns to Woodpeckers, (1st Edn). Oxford University Press, Oxford
Durrant KL, Beadell JS, Ishtiaq F et al (2006) Avian hematozoa in South America: a comparison of temperate and tropical zones. Ornithol Monogr 60:98–111. https://doi.org/10.2307/40166831
Earle RA, Bennett GF, Hester DT et al (1991) Regional and seasonal distribution of avian blood parasites from northern South Africa. South Afr J Wildl Res 21:47–53. https://doi.org/10.10520/EJC116868
Evens R, Conway GJ, Henderson IG et al (2017) Migratory pathways, stopover zones and wintering destinations of Western European Nightjars Caprimulgus europaeus. Ibis 159:680–686. https://doi.org/10.1111/ibi.12469
Forero MG, Tella JL, Gajon A (1997) Absence of blood parasites in the Red-necked Nightjar. J Field Ornithol 68:575–579
Gangoso L, Gutiérrez-López R, Martínez-de la Puente J, Figuerola J (2016) Genetic colour polymorphism is associated with avian malarial infections. Biol Lett 12:20160839. https://doi.org/10.1098/rsbl.2016.0839
Ganser C, Monadjem A, McCleery RA et al (2020) Is it best on the nest? Effects of avian life-history on haemosporidian parasitism. Int J Parasitol Parasites Wildl 13:62–71. https://doi.org/10.1016/j.ijppaw.2020.07.014
Garamszegi LZ (2010) The sensitivity of microscopy and PCR-based detection methods affecting estimates of prevalence of blood parasites in birds. J Parasitol 96:1197–1203. https://doi.org/10.1645/GE-2531.1
Greiner EC, Bennett GF, White EM, Coombs RF (1975) Distribution of the avian hematozoa of North America. Can J Zool 53:1762–1787. https://doi.org/10.1139/z75-211
Griffiths R, Double MC, Orr K, Dawson RJ (1998) A DNA test to sex most birds. Mol Ecol 7:1071–1075. https://doi.org/10.1046/j.1365-294x.1998.00389.x
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Harvey JA (2018) Avian haemosporidians: detection, host, and climate association across contrasting regions of Africa. PhD dissertation, Texas A&M University
Hellgren O, Waldenström J, Bensch S (2004) A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium, and Haemoproteus from avian blood. J Parasitol 90:797–802. https://doi.org/10.1645/GE-184R1
Ishtiaq F, Gering E, Rappole JH et al (2007) Prevalence and diversity of avian hematozoan parasites in Asia: a regional survey. J Wildl Dis 43:382–398. https://doi.org/10.7589/0090-3558-43.3.382
Jacobsen LB, Jensen NO, Willemoes M et al (2017) Annual spatiotemporal migration schedules in three larger insectivorous birds: European Nightjar, Common Swift and Common Cuckoo. Anim Biotelemetry 5:4. https://doi.org/10.1186/s40317-017-0119-x
Jarvi SI, Schultz JJ, Atkinson CT (2002) PCR diagnostics underestimate the prevalence of avian malaria (Plasmodium relictum) in experimentally-infected passerines. J Parasitol 88:153–158. https://doi.org/10.1645/0022-3395(2002)088[0153:PDUTPO]2.0.CO;2
Jovani R, Tella JL (2006) Parasite prevalence and sample size: misconceptions and solutions. Trends Parasitol 22:214–218. https://doi.org/10.1016/j.pt.2006.02.011
Klaschka J, Reiczigel J (2021) On matching confidence intervals and tests for some discrete distributions: methodological and computational aspects. Comput Stat 36:1775–1790. https://doi.org/10.1007/s00180-020-00986-0
Krausová S. (2015) Ptačí malárie vlaštovky obecné [Avian malaria in the Swallow]. MSc dissertation, Univerzita Karlova
Kubacka J, Gerlée A, Foucher J et al (2019) Correlates of blood parasitism in a threatened marshland passerine: infection by kinetoplastids of the genus Trypanosoma is related to landscape metrics of habitat edge. Parasitology 146:1036–1046. https://doi.org/10.1017/S0031182019000350
Lacorte GA, Félix GMF, Pinheiro RRB et al (2013) Exploring the diversity and distribution of neotropical avian malaria parasites – a molecular survey from Southeast Brazil. PLoS One 8:e57770. https://doi.org/10.1371/journal.pone.0057770
McNew SM, Barrow LN, Williamson JL et al (2021) Contrasting drivers of diversity in hosts and parasites across the tropical Andes. Proc Natl Acad Sci 118:e2010714118. https://doi.org/10.1073/pnas.2010714118
Mohammed AHH, Al-Taqi MMS (1975) A general survey of blood parasites of birds from Kuwait. Kuwait J Sci 2:167–176
Molyneux DH (1977) Vector relationships in the Trypanosomatidae. Adv Parasitol 15:1–82. https://doi.org/10.1016/s0065-308x(08)60526-6
Njabo KY, Cornel AJ, Sehgal RN et al (2009) Coquillettidia (Culicidae, Diptera) mosquitoes are natural vectors of avian malaria in Africa. Malar J 8:193. https://doi.org/10.1186/1475-2875-8-193
Norevik G, Åkesson S, Hedenström A (2017) Migration strategies and annual space-use in an Afro-Palaearctic aerial insectivore – the European Nightjar Caprimulgus europaeus. J Avian Biol 48:738–747. https://doi.org/10.1111/jav.01071
Nourani L, Djadid ND, Rabiee K et al (2020) Detection of haemosporidian parasites in wild and domestic birds in northern and central provinces of Iran: Introduction of new lineages and hosts. Int J Parasitol Parasites Wildl 13:203–212. https://doi.org/10.1016/j.ijppaw.2020.10.001
Ottenby Bird Observatory (2015) Ringers’ DigiGuide–Caprimulgus europaeus. www.ringersdigiguide.ottenby.se
Peirce MA (1981) Distribution and host-parasite check-list of the haematozoa of birds in Western Europe. J Nat Hist 15:419–458. https://doi.org/10.1080/00222938100770321
Pellegrino I, Ilahiane L, Boano G et al (2021) Avian haemosporidian diversity on Sardinia: a first general assessment for the insular Mediterranean. Diversity 13:75. https://doi.org/10.3390/d13020075
Pérez-Tris J, Bensch S (2005) Diagnosing genetically diverse avian malarial infections using mixed-sequence analysis and TA-cloning. Parasitology 131:15–23. https://doi.org/10.1017/s003118200500733x
Pori T (2018) Avian haemoparasite prevalence in Kruger National Park and the surrounding human settlements. MSc dissertation, University of the Witwatersrand
R Core Team (2021) R: A language and environment for statistical computing. R Foundation for Statistical Computing
Reiczigel J, Marozzi M, Fábián I, Rózsa L (2019) Biostatistics for parasitologists–a primer to quantitative parasitology. Trends Parasitol 35:277–281. https://doi.org/10.1016/j.pt.2019.01.003
Santiago-Alarcon D, Palinauskas V, Schaefer HM (2012) Diptera vectors of avian Haemosporidian parasites: untangling parasite life cycles and their taxonomy. Biol Rev Camb Philos Soc 87:928–964. https://doi.org/10.1111/j.1469-185X.2012.00234.x
Savage AF, Robert V, Goodman SM et al (2009) Blood parasites in birds from Madagascar. J Wildl Dis 45:907–920. https://doi.org/10.7589/0090-3558-45.4.907
Sehgal RN, Jones HI, Smith TB (2001) Host specificity and incidence of Trypanosoma in some African rainforest birds: a molecular approach. Mol Ecol 10:2319–2327. https://doi.org/10.1046/j.1365-294x.2001.01339.x
Shurulinkov P, Golemansky V (2002) Haemoproteids (Haemosporida: Haemoproteidae) of wild birds in Bulgaria. Acta Protozool 41:359–374
Shurulinkov P, Golemansky V (2003) Plasmodium and Leucocytozoon (Sporozoa: Haemosporida) of wild birds in Bulgaria. Acta Protozool 42:205–214
Šíma M (2011) Hostitelská specifita, diverzita a distribuce malarických parazitů v kontaktní zóně dvou druhů slavíků [Host specificity, diversity and distribution of avian malaria parasites in a contact zone of two nightingale species]. MSc dissertation, Univerzita Karlova
Valdebenito JO, Liker A, Halimubieke N et al (2020) Mortality cost of sex-specific parasitism in wild bird populations. Sci Rep 10:20983. https://doi.org/10.1038/s41598-020-77410-6
Valkiūnas G (2005) Avian malaria parasites and other haemosporidia. CRC Press, Boca Raton
Valkiūnas G, Bensch S, Iezhova TA et al (2006) Nested cytochrome b polymerase chain reaction diagnostics underestimate mixed infections of avian blood haemosporidian parasites: microscopy is still essential. J Parasitol 92:418–422. https://doi.org/10.1645/GE-3547RN.1
Valkiūnas G, Iezhova TA, Krizanauskiene A et al (2008) A comparative analysis of microscopy and PCR-based detection methods for blood parasites. J Parasitol 94:1395–1401. https://doi.org/10.1645/GE-1570.1
Valkiūnas G, Iezhova TA, Loiseau C, Sehgal RNM (2009) Nested cytochrome b polymerase chain reaction diagnostics detect sporozoites of hemosporidian parasites in peripheral blood of naturally infected birds. J Parasitol 95:1512–1515. https://doi.org/10.1645/GE-2105.1
Valkiūnas G, Palinauskas V, Ilgūnas M et al (2014) Molecular characterization of five widespread avian haemosporidian parasites (Haemosporida), with perspectives on the PCR-based detection of haemosporidians in wildlife. Parasitol Res 113:2251–2263. https://doi.org/10.1007/s00436-014-3880-2
Ventim R, Morais J, Pardal S et al (2012) Host-parasite associations and host-specificity in haemoparasites of reed bed passerines. Parasitology 139:310–316. https://doi.org/10.1017/S0031182011002083
Votýpka J, Szabová J, Rádrová J et al (2012) Trypanosoma culicavium sp. nov., an avian trypanosome transmitted by Culex mosquitoes. Int J Syst Evol Microbiol 62:745–754. https://doi.org/10.1099/ijs.0.032110-0
White EM, Greiner EC, Bennett GF, Herman CM (1978) Distribution of the hematozoa of Neotropical birds. Rev Biol Trop 26(Suppl 1):43–102
Williams NA, Bennett GF, Mahrt JL (1975) Avian haemoproteidae. 6. Description of Haemoproteus caprimulgi sp. nov., and a review of the haemoproteids of the family Caprimulgidae. Can J Zool 53:916–919. https://doi.org/10.1139/z75-106
Winkler DW, Billerman SM, Lovette IJ (2020) Nightjars and Allies (Caprimulgidae), version 1.0. In: Billerman SM, Keeney BK, Rodewald PG, Schulenberg TS (eds) Birds of the world. Cornell Lab of Ornithology, Ithaca, NY, USA
Yang G, He H, Zhang G et al (2021) Neglected parasite reservoirs in wetlands: prevalence and diversity of avian haemosporidians in waterbird communities in Northeast China. Int J Parasitol Parasites Wildl 15:177–183. https://doi.org/10.1016/j.ijppaw.2021.04.013
Acknowledgements
We thank Dr. Bartłomiej Woźniak and the Sobibór Research Group from the Warsaw University of Life Sciences for the support before and during this study and Edyta Podmokła for comments that improved the manuscript.
Funding
Molecular analyses were funded by the Museum and Institute of Zoology, Polish Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare that they have no conflict of interest.
Ethical approval
All applicable national and institutional guidelines for the care and use of animals were followed. Birds were sampled under permits from the 1st Local Ethical Committee in Warsaw (permit no 637/2018) and the Regional Directorate for Environmental Protection (permit no WPN.6401.77.2018.MPR) in Lublin, Poland.
Additional information
Communicated by I. Moore.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dubiec, A., Da Silva, A. & Celej, M. Low prevalence of haemosporidian and trypanosome infections in the Eurasian Nightjar (Caprimulgus europaeus). J Ornithol 164, 445–453 (2023). https://doi.org/10.1007/s10336-022-02031-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10336-022-02031-x