Abstract
Seabirds are regarded as a group of species with relatively low levels or even complete lack of blood parasites. We used PCR to amplify a DNA fragment from the cytochrome b gene of the parasites to search for infections of the genera Plasmodium, Haemoproteus and Leucocytozoon in individuals of two sympatrically breeding gull species, the Herring Gull Larus argentatus, the Caspian Gull Larus cachinnans and their hybrids. Out of 56 analysed individuals, 53 (95 %) were identified as infected with Leucocytozoon, whereas three individuals carried double and triple infections with at least one Leucocytozoon and one Plasmodium lineages. No Haemoproteus lineage was detected. The most common lineage (LARCAC02), for the first time reported here, was found in 51 (96 %) of all infected birds, and 14 gulls carried two Leucocytozoon lineages. We analysed the evolutionary relationship of Leucocytozoon lineages from the Herring and Caspian Gull and other bird species. Our results show that (1) the two identified Leucocytozoon lineages are not closely related as they belong to two distinctly different clusters. Moreover, (2) seabirds breeding inland could be highly infected with blood parasites and (3) this high prevalence is probably associated with areas where parasite vectors are abundant. Further studies should explore the importance of environmental factors affecting parasite prevalence, in particular within species comparisons under different environment conditions, including vector monitoring and sampling.
Zusammenfassung
Hohe prävalenz von Leucozytozoon bei möwen in süsswasser-brutgebieten
Seevögel werden als eine Artengruppe angesehen, bei der relativ wenige oder gar keine Blutparasiten auftreten. Wir verwenden PCR, um ein DNA Fragment aus dem Cytochrom b Gen von Parasiten zu amplifizieren, um nach Infektionen mit den Genera Plasmodium, Haemoproteus und Leucocytozoon in Individuen zweier sympatrisch brütender Möwenarten zu suchen, nämlich der Silbermöwe (Larus argentatus), und der Steppenmöwe (L. cachinnans) und deren Hybride. Von 56 untersuchten Individuen waren 53 (95 %) mit Leucocytozoon infiziert, drei waren doppelt oder dreifach infiziert mit mindestens einer Leucocytozoon Linie und einer Plasmodium Linie. Es wurde keine Haemoproteus Linie gefunden. Die häufigste Linie (LARCA02), hier zum ersten mal beschrieben, wurde in 51 (96 %) aller infizierter Vögel gefunden, und 14 Tiere trugen zwei verschiedene Leucocytozoon Linien. Wir untersuchten die evolutionären Beziehungen von Leucocytozoon Linien der Silber- und Steppenmöwe und anderer Vogelarten. Unsere Ergebnisse zeigen (1), daß die zwei gefundenen Leucocytozoon Linien nicht eng miteinander verwandt sind, da sie zu zwei deutlich unterschiedlichen Clustern gehören. Darüberhinaus (2) können Seevögel im Binnenland schwer infiziert sein mit Blutparasiten und (3) steht diese hohe Prävalenz wahrscheinlich im Zusammenhang mit Gegenden, in denen Vektoren für die Parasiten ausreichend vorhanden sind. In weiteren Studien sollte die Rolle von Umwelteinflüssen auf die Prävalenz der Parasiten untersucht werden, insbesondere durch Vergleiche zwischen Arten und unter verschiedenen Umweltbedingungen, sowie Monitoring und Probennahme von Vektoren.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genera Plasmodium, Haemoproteus and Leucocytozoon represent a group of vector-borne blood parasites causing a malaria-like disease in birds (e.g. Bensch et al. 2000; Waldenstöm et al. 2004; Bensch and Åkesson 2003; Scheuerlein and Ricklefs 2004; Wood et al. 2007; Jenkins and Owens 2011; Swanson et al. 2014; Zhao et al. 2014). One of the features of such infections is that host species vary substantially in parasite prevalence. Seabirds represent a diverse group; there are many species with an apparent absence or scarcity of these parasites (Piersma 1997; Figuerola 1999; Jovani et al. 2001; Quillfeldt et al. 2010, 2011; Krams et al. 2012), but also some species with relatively high blood parasite prevalence (Ruiz et al. 1995; Martínez-Abraín et al. 2002; Quillfeldt et al. 2010). Several hypotheses have been proposed to explain why blood parasites are rare in some birds species. These include absence or low abundance of vectors in saline or arid habitats (Engström et al. 2000; Sol et al. 2000; Jovani et al. 2001; Wojczulanis-Jakubas et al. 2010), competitive exclusion by ectoparasites (Martínez-Abraín et al. 2004), specific host–parasite assemblage (Earle and Underhill 1993; González-Solís and Abella 1997; Jones and Shellam 1999; Martínez-Abraín and Urios 2002; Fokidis et al. 2008) or host immune capabilities (Merino and Minguez 1998; Tella et al. 1999).
Leucocytozoon is a genus that infects numerous species of avian hosts, including the domestic chicken, pigeons, owls and many passerine species (Valkiūnas 2005; Bunbury et al. 2007; Ishak et al. 2008; Dezfoulian et al. 2013; Zhao et al. 2014). The parasites have a complex life cycle by having merogony in fixed tissues of vertebrate hosts, sexually differentiated gametocytes in blood cells, and sporogony in simuliid flies or Culicoides midges (Valkiūnas 2005). In a global review of blood parasites in seabirds (453 species in 13 families), Leucocytozoon parasites were found to be reported in three species within the families Phalacrocoracidae and Spheniscidae (Quillfeldt et al. 2011). In Laridae there were no previous records of infections with these blood parasites. In this group, within 36 examined taxa, the only detected parasites were Plasmodium sp. (in a single species; Coatney 1938), Haemoproteus sp. (in four species; Lowery 1971; Padilla et al. 2006; Ishtiaq et al. 2007; Quillfeldt et al. 2010), Haemoproteus larae (in seven species; Peirce 1981; Valkiūnas 2005) and H. passeris (in one species; Berdyev 1979).
The Herring Gull Larus argentatus and the Caspian Gull L. cachinnans are closely related species that in the past decades have shown a rapid expansion and widespread colonization of new breeding habitats. Originally, the Herring Gull bred on the coasts of western Europe, whereas the Caspian Gull inhabited inland habitats in south-eastern Europe (Snow and Perrins 1998). In central Europe, including Poland, these species became established as breeders at inland freshwater habitats about three decades ago (Snow and Perrins 1998; Neubauer et al. 2006; Lenda et al. 2010). It is therefore likely that the change in breeding location exposed them to a new set of vector species of the families Culicidae and Simuliidae. These dipteran, blood-feeding insects are regarded as basic vectors transmitting blood parasites and differ by their ecological requirements for larvae development including water level, temperature and vegetation. Culicidae transmit mainly Plasmodium and Haemoproteus, whereas Leucocytozoon is carried by Simuliidae (Valkiūnas 2005). We analysed samples of the two gull species from one inland colony where massive outbreaks of Simuliidae black flies were observed and we expected to find blood parasites connected with those typically transmitted by river valley vectors.
Methods
We monitored a mixed species gull colony in central Poland, at the Włocławek reservoir (52°39′12″N, 19°08′18″E), where ca. 130 pairs of Herring Gulls, Caspian Gulls and their hybrids breed annually (Neubauer et al. 2009; Zagalska-Neubauer and Neubauer 2012). In the present study, we report analysis of blood samples collected between 25 April and 10 May in both 2007 and 2008. All samples were collected from adult breeding gulls captured during incubation with walk-in nest-traps. This analysis includes a random sample of 56 gulls, of which 29 were trapped in 2007 and 27 in 2008. The sample includes 22 Herring Gulls (13 females, 9 males), 17 Caspian Gulls (11 females, 6 males) and 17 hybrids (6 females, 11 males) (for details of species assignment see, Gay et al. 2007; Neubauer et al. 2009; Zagalska-Neubauer and Neubauer 2012).
Blood smear analyses are crucial to assign parasites to morphospecies and to quantify the degree of parasitemia in the host (Valkiūnas et al. 2009). This allows confirmation of the presence of mature gametocytes in order to establish whether the gulls are competent hosts for the encountered lineages. We prepared blood smears for all birds and assessed them microscopically for blood parasites. We detected Leucocytozoon in eight individuals and Plasmodium in one individual. However, for reasons related to smear preparation the quality of the smears was rather poor and not sufficient for detailed morphological analyses.
For molecular analysis blood samples were taken by tarsal venepuncture, stored in 96 % ethanol and DNA was extracted using a NucleoSpin® blood kit (Macherey–Nagel). An assessment of the presence and quality of the extracted DNA was made with the use of a Colibri microvolume spectrophotometer and by electrophoresing 4 µl of the extract in 2 % agarose gel. We used nested PCR for parallel detection of Leucocytozoon, Plasmodium and Haemoproteus (Hellgren et al. 2004). The method used primers located in conserved regions of the cytochrome b gene of malaria parasites to identify the sequence of interest, by amplification of a 478-bp fragment of the gene for three parasite genera. In the first PCR we used the primers HaemNF1 and HaemNR2 to amplify parasitic DNA from all three genera. The second step included a PCR assay with primers specific for particular genera, for Haemoproteus and Plasmodium primers: HaemF/HaemR2, and separately for Leucocytozoon: HaemFL/HaemR2L (Hellgren et al. 2004). Each plate contained a positive control, i.e. DNA from individuals with confirmed infection, and negative controls (ddH2O), to control for possible contamination or failures during PCRs. Both PCR rounds were performed in 25 µl volumes. The first round of PCR contained 25 ng genomic DNA, 0.125 mM of each dNTP, 0.6 µM of each primer, 25 mM MgCl2, 0.5 units of Taq DNA polymerase and 1× PCR buffer. The thermal profile consisted of 2 min denaturation at 94 °C, followed by 20 cycles of 94 °C for 30 s, 52 °C for 30 s, 72 °C for 45 s, and ended with final elongation at 72 °C for 10 min. The second PCR round was as above, except that as a template we used 2 µl of the product from the first PCR instead of genomic DNA. The thermal profile for the second round PCR was also similar, but the annealing temperature was 55 °C, and the number of cycles was increased to 35. PCR products from the second step were run on 2 % agarose gel, stained with GelRed and visualized under UV. Positive samples with ca. 600-bp products were prepared for sequencing by a differential precipitation step with ammonium acetate (NH4Ac). The purified PCR products were then sequenced by dye terminator cycle sequencing (v1.1, Applied Biosystems), prior to precipitation with an EDTA protocol. All positive samples were sequenced on ABI PRISM™ 3100 capillary sequencing robot (Applied Biosystems, USA).
Obtained fragments were edited and aligned in BioEdit and Geneious software (Hall 1999; Geneious ver6.1, Biomatters Ltd.). Obtained sequences were aligned and compared with sequences available in the MalAvi database (Bensch et al. 2009). Representatives of all lineages were also sequenced with the reverse primer. Multiple infections were resolved by examining if the most common lineage could be part of the multiple infection and from this comparison we subtracted the other sequence from the sites with double base calling (Pérez-Tris and Bensch 2005).
Phylogenetic analyses were made using MEGA5 (Tamura et al. 2011). First, to identify the clade of lineages including LARCAC02 and CIAE02, we constructed the phylogenetic relationship of all Leucocytozoon in MalAvi (N = 418) using the neighbour-joining method with maximum composite likelihood model (not shown). In the second step we used the lineages included in this clade (N = 21) together with ten morphologically described Leucocytozoon lineages to construct phylogenies by maximum-likelihood and a GTR+G substitution model of sequence evolution.
Results
The sampled gull population showed a high level of infection with haemosporidian parasites. Overall, 53 out of 56 individuals (95 %) were scored positive by PCR. We identified two different Leucocytozoon lineages and one Plasmodium lineage. Samples that tested positive for blood parasites were successfully sequenced. Of those, all were identified as Leucocytozoon sequences while 6 % were identified as co-infected. Two individuals were co-infected with one lineage of Leucocytozoon and with a Plasmodium, and one individual was co-infected with two Leucocytozoon lineages and with a Plasmodium (Tables 1, 2). The Plasmodium sequence was identical to the P. relictum lineage (SGS1) and detected in females, two Caspian Gulls and one Herring Gull. We did not detect any Haemoproteus lineages (Table 1). The Leucocytozoon sequences consisted of two distinct lineages. The most common lineage, LARCAC02 (GenBank KP271931), was detected in 96 % of infected birds and has not previously been found in other birds. The Leucocytozoon lineage CIAE02 (GenBank EF607287) occurred as a single infection in two birds and in multiple infections with LARCAC02 in 14 birds (Table 2). We detected that 100 % of the Herring Gulls were infected, and all but one Caspian Gull male and two hybrid females were infected (Table 2).
We used the MalAvi database to determine if the encountered lineages were restricted to the gull species or found in other bird hosts. The most prevalent Leucocytozoon lineage (LARCAC02) did not have any complete matches in the MalAvi database. The other Leucocytozoon lineage (CIAE02) had exact matches from a variety of European and Asian birds of prey Falconiformes: Marsh Harrier Circus aeruginosus, Griffon Vulture Gyps fulvus, Cinereous Vulture Aegypius monachus, Besra Accipiter virgatus, Black Kite Milvus migrans, Common Buzzard Buteo buteo and Long-legged Buzzard Buteo rufinus. CIAE02 was also recently detected in the Corn Crake Crex crex. The detected Plasmodium relictum lineage had exact matches from 97 host species, from different birds groups in Europe, Asia, Africa, South America and Oceania.
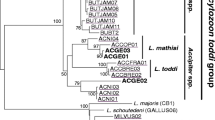
The bootstrapped phylogeny of the Herring and Caspian Gull Leucocytozoon lineages showed that they are distinctly different from the previously morphologically described lineages (Fig. 1). The Leucocytozoon lineages identified as similar to the two gull lineages in the present study formed two apparent clusters. The first, a more homogeneous cluster contained lineages specific for passerines, and the second, a more heterogeneous cluster with lineages present in many different bird groups. Interestingly, the lineages present in the examined gulls were located in both clusters. The most frequent lineage LARCAC02 was placed close to the passerine group, whereas the rarer CIAE02 lineage was located among lineages infecting birds of prey, owls and loons.
Evolutionary relationships of Leucocytozoon from the Herring and Caspian Gull and other bird species inferred using maximum likelihood with the GTR+G model. Numbers at nodes represent bootstrap values. Bootstrap values lower than 50 are not shown. All lineage names and host group species, except L_LARCAC02, are listed from MalAvi Database. The blue frames mark lineages detected in gulls examined in this study (colour figure online)
Discussion
Examined gulls from an inland breeding colony showed very high prevalence of Leucocytozoon parasites (95 %). Most of the gulls were infected with the lineage LARCAC02, which was not previously encountered in the literature, and many individual hosts showed multiple infections with another lineage of Leucocytozoon. The phylogenetic tree clustered the Leucocytozoon lineages into two well-separated groups that appear to infect different species of hosts, in general passerine and non-passerine birds. The lineages detected in gulls (CIAE02, LARCAC02) are placed in both of these clusters, indicating significant genetic differences between them. The CIAE02 lineage has been frequently recorded in birds of prey, such as the Red Kite and the Marsh Harrier, but also in the Corn Crakes which suggests that the lineage has a broad host range and could be cosmopolitan (Pérez-Rodríguez et al. 2013).
Parasites in the genus Leucocytozoon are distributed worldwide and occur in a broad range of host species; they are relatively rare in seabirds and have previously never been detected in the genus Larus (Valkiūnas 2005; Atkinson et al. 2008; Jenkins and Owens 2011; Quillfeldt et al. 2011). Among seabirds, high prevalence of Leucocytozoon was previously recorded in penguins (73.7 %; Argilla et al. 2013). One of hypotheses proposed to explain differences in blood parasite prevalence is the vector-density hypothesis (Bennett et al. 1992; Tella et al. 1996; Piersma 1997; Sol et al. 2000; Jovani et al. 2001; Martínez-Abraín and Urios 2002; Fokidis et al. 2008). Bird species associated with inland environments are expected to be more exposed to the malaria-like parasites than similar bird species inhabiting marine environments as the former provide more suitable breeding areas for vectors (Piersma 1997; Sol et al. 2000; Mendes et al. 2005). The low diversity or absence of blood parasites previously reported in seabirds is associated with the specific character of marine and coastal habitats that typically are relatively free of parasite vectors (Piersma 1997; Mendes et al. 2005). This is also consistent with a biogeographical perspective as the Leucocytozoon vectors, Simuliidae, are more abundant and diverse at higher latitudes (Valkiūnas 2005; Atkinson et al. 2008). We can expect that birds which inhabit more parasite-friendly environments, such as inland waterbodies, are more exposed to vectors and therefore to infections (but see Krams et al. 2012). At our inland colony in the Vistula River valley, we observed massive outbreaks of black flies during the study years, which is typical for this region (Bukaciński and Bukacińska 2000). In the years between 2002 and 2009, the abundance of black flies varied but was relatively high (K. Szpila and T. Kakareko, unpublished data). Three species have been reported: Simulium maculatum (55 % within sampled individuals-swarmed insects), S. erythrocephalum (42 %) and S. pusillum (3 %) (T. Kakareko, unpublished data). The observed swarms of black flies made host species highly exposed to vectors and could explain the high Leucocytozoon prevalence in gulls. High prevalence of blood parasites in gulls has been recorded once, in Yellow-legged Gull (Larus michahellis) where up to 100 % of individuals on one of the studied islands were infected by Haemoproteus lari (Martínez-Abraín et al. 2002). The examined gulls were breeding on four islands in the Mediterranean Sea located at different distance from the mainland. The study showed that the prevalence was inversely correlated to the distance to the coast, presumably depending on vector availability. However, vector abundance is not the only factor that influences bird parasite prevalence. An important factor that may influence parasite prevalence in seabirds is also the immunological capabilities of the host (Esparza et al. 2004; Krams et al. 2012). Krams et al. (2012) detected low prevalence of blood parasites in the Black-headed Gull (Chroicocephalus ridibundus) and absence of blood parasites in the Common Gull (Larus canus), both in inland and coastal colonies, despite the presence of appropriate vector environments. The lack or low prevalence of haemosporidians was suggested to be due to enhanced immunity in the examined gull species.
Here, we report for the first time, to the best of our knowledge, blood parasites from Caspian Gulls (previously Larus cachinnans cachinnans and Larus cachinnans/argentatus michahellis were regarded as the same species) and gull hybrids. In our inland colony significant differences in prevalence between the Herring and Caspian Gull were not detected. Also we did not record any differences in parasite prevalence between the two gull species and their hybrids. While the coastal Herring Gull is not associated with environments typical for Leucocytozoon vectors (black flies), the Caspian Gull originally inhabited inland fresh and saltwater environments and river valleys in eastern Europe where black flies might be abundant. These different origins and therefore presumed different evolutionary exposure of these parasites may influence parasitemia in the two gull species; however, that would require analyses of blood smears or quantifying infection intensities with qPCR. There are no detailed data on the parasite prevalence from allopatric population (from the original breeding grounds) of these two species. To reveal a better picture of prevalence and parasitemia in these gull species it will be desirable to collect data from other colonies, explore the importance of environmental factors affecting parasite prevalence and also include vector monitoring and sampling.
In a review on blood parasites in seabirds, Quillfeldt et al. (2011) showed that the mean prevalence of blood parasites was 9.2 % and within 36 analysed Laridae species nearly 60 % were infected mainly with Haemoproteus, whereas Plasmodium was rare. We did not detect any Haemoproteus lineages, whereas we found one lineage of Plasmodium (SGS1), infecting three gulls in our sample. We applied a nested cytochrome b PCR to detect the parasites. As the method is sensitive it can amplify DNA from sporozoites of haemosporidians and DNA from aborted development in extra-erythrocytic tissues (Ricklefs et al. 2005; Valkiūnas et al. 2009) and therefore be misleading when identifying competent hosts. However, parasite gametocytes were observed in some of the blood smears, thus supporting the conclusion that the identified lineages can complete development in the analysed gull species. Co-infections by multiple parasite species are well documented across many animal systems (Rigaud et al. 2010; Knowles et al. 2013) and we found that a substantial proportion of the gulls were infected with two or more parasites. To what extent parasites in co-infections compete within the host is poorly investigated but it is possible that infections by one parasite may protect the host from other infections. Studies reporting multiple infections by Leucocytozoon lineages are scarce, and most of them have reported Leucocytozoon infections accompanied by other malaria parasite lineages (Argilla et al. 2013; Zhao et al. 2014; Oakgrove et al. 2014; van Rooyen et al. 2013). In a study of blood parasites in a community of birds in Alaska, it was suggested that the high Leucocytozoon prevalence may have restricted Haemoproteus occurrence in some of the host species through negative association between Haemoproteus and Leucocytozoon infections (Oakgrove et al. 2014). The lack of Haemoproteus parasites in our study population might thus partly be explained by the high prevalence of Leucocytozoon, a possibility that will require more thorough investigations.
References
Argilla LS, Howe L, Gartrell BD, Alley MR (2013) High prevalence of Leucocytozoon spp. in the endangered yellow-eyed penguin (Megadyptes antipodes) in the sub-Antarctic regions of New Zealand. Parasitology 140:672–682
Atkinson CT, Thomas N, Hunter DB (2008) Parasitic disease in wild birds. Wiley, Ames
Bennett GF, Montgomere R, Seutin G (1992) Scarcity of haematozoa in birds breeding on the Arctic tundra of North America. Condor 94:289–292
Bensch S, Åkesson S (2003) Temporal and spatial variation of hematozoans in Scandinavian willow warblers. J Parasitol 89:388–391
Bensch S, Stjernman M, Hasselquist D, Östman Ö, Hansson B, Westerdahl H, Pinheiro RT (2000) Host specificity in avian blood parasites: a study of Plasmodium and Haemoproteus mitochondrial DNA amplified from birds. Proc R Soc B 267:1583–1589
Bensch S, Hellgren O, Pérez-Tris J (2009) MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol Ecol Res 9:1353–1358
Berdyev KS (1979) K faune kroveparazitov dikikh ptits Yuzhnogo Turkmenistana. Blood parasites of wild birds in South Turkmenistan. Razvitie Parazitologicheskoi Naukiv Turkmenistane, Ashkhabad, pp 156–162 (In Russian)
Bukaciński D, Bukacińska M (2000) The impact of mass outbreaks of black flies (Simuliidae) on the parental behaviour and breeding output of colonial common gulls (Larus canus). Ann Zool Fenn 37:43–49
Bunbury N, Barton E, Jones CG, Greenwood AG, Tyler KM, Bell DJ (2007) Avian blood parasites in an endangered columbid: Leucocytozoon marchouxi in the Mauritian Pink Pigeon Columba mayeri. Parasitology 134:797–804
Coatney GR (1938) Some blood parasites from birds of the Lake Okaboji region. Am Midl Nat 20:336–340
Dezfoulian O, Zibaei M, Nayebzadeh H, Haghgoo M, Emami-Razavi A, Kiani K (2013) Leucocytozoonosis in domestic birds in southwestern Iran: an ultrastructural study. Iran J Parasitol 8:171–176
Earle RA, Underhill LG (1993) Absence of haematozoa in some Charadriiformes breeding in the Taimyr Peninsula, Russia. Ardea 81:21–24
Engström H, Dufva R, Olsson G (2000) Absence of haematozoa and ectoparasites in a highly sexually ornamented species, the crested auklet. Waterbirds 23:486–488
Esparza B, Martínez-Abraín A, Merino S, Oro D (2004) Immunocompetence and the prevalence of haematozoan parasites in two long-lived seabirds. Ornis Fenn 81:40–46
Figuerola J (1999) Effects of salinity on rates of infestation of waterbirds by haematozoa. Ecography 22:681–685
Fokidis BH, Greiner EC, Deviche P (2008) Interspecific variation in avian blood parasites and haematology associated with urbanization in a desert habitat. J Avian Biol 39:300–310
Gay L, Neubauer G, Zagalska-Neubauer M, Bebain C, Pons JM, David P, Crochet PA (2007) Molecular and morphological patterns of introgression between two large white-headed gull species in a zone of recent secondary contact. Mol Ecol 16:3215–3227
González-Solís J, Abella JC (1997) Negative records of haematozoan parasites on Cory’s shearwater Calonectris diomedea. Ornis Fenn 74:153–155
Hall TA (1999) BioEdit: a user-friendly biological sequences alignment editor and analysis program for Windows 95/98/NT. Nucleic Acid Symp Ser 41:95–98
Hellgren O, Waldenström J, Bensch S (2004) A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium and Haemoproteus from avian blood. J Parasitol 90:797–802
Ishak HD, Dumbacher JP, Anderson NL, Keane JJ, Valkiūnas G, Haiq SM, Tell LA, Sehqal RN (2008) Blood parasites in owls with conservation implications for the Spotted Owl (Strix occidentalis). PLoS ONE 3:e2304. doi:10.1371/journal.pone.0002304
Ishtiaq F, Gering E, Rappole JH, Rahmani AR, Jhala YV, Dove CJ, Milensky C, Olson SL, Peirce MA, Fleischer RC (2007) Prevalence and diversity of avian hematozoan parasites in Asia: a regional survey. J Wildl Dis 43:382–398
Jenkins T, Owens IP (2011) Biogeography of avian blood parasites (Leucocytozoon spp.) in two residents hosts across Europe: phylogeographic structuring or the abundance-occupancy relationship? Mol Ecol 20:3910–3920
Jones HI, Shellam GR (1999) The occurrence of blood-inhabiting protozoa in captive and free-living penguins. Polar Biol 21:5–10
Jovani R, Tella JL, Forero MG, Bertellotti M, Blanco G, Ceballos O, Donázar JA (2001) Apparent absence of blood parasites in the Patagonian seabird community: is it related to the marine environment? Waterbirds 24:430–433
Knowles SCL, Fenton A, Petchey OL, Jones TR, Barber R, Pedersen AB (2013) Stability of within-host–parasite communities in a wild mammal system. Proc R Soc B 280:20130598. doi:10.1098/rspb.2013.0598
Krams I, Suraka V, Rattiste K, Āboliņš-Ābols M, Krama T, Rantala MJ, Mierauskas P, Cīrule D, Saks L (2012) Comparative analysis reveals a possible immunity-related absence of blood parasites in Common Gulls (Larus canus) and Black-headed Gulls (Chroicocephalus ridibundus). J Ornithol 153:1245–1252
Lenda M, Zagalska-Neubauer M, Neubauer G, Skórka P (2010) Do invasive species undergo metapopulation dynamics? a case study of the invasive Caspian Gull Larus cachinnans in Poland. J Biogeogr 37:1824–1834
Lowery RS (1971) Blood parasites of vertebrates on Aldabra. Philos Trans R Soc Lond B 260:577–580
Martínez-Abraín A, Urios G (2002) Absence of blood parasites in nestlings of the Eleonora’s falcon (Falco eleonorae). J Raptor Res 36:139–141
Martínez-Abraín A, Merino S, Oro D, Esparza B (2002) Prevalence of blood parasites in two western-Mediterranean local populations of the Yellow-legged Gull Larus cachinnans michahellis. Ornis Fenn 79:34–40
Martínez-Abraín A, Esparza B, Oro D (2004) Lack of blood parasites in birds species: does absence of blood parasite vectors explain it all? Ardeola 51(1):225–232
Mendes L, Piersma T, Lecoq M, Spaans B, Ricklefs RE (2005) Disease-limited distribution? Contrast in the prevalence of avian malaria in shorebirds species using marine and freshwater habitats. Oikos 109:396–404
Merino S, Minguez E (1998) Absence of haematozoa in a breeding colony of the Storm Petrel Hydrobates pelagicus. Ibis 140:180–181
Neubauer G, Zagalska-Neubauer M, Gwiazda R, Faber M, Bukaciński D, Betleja J, Chylarecki P (2006) Breeding large gulls in Poland: distribution, numbers, trends and hybridisation. Vogelwelt 127:11–22
Neubauer G, Zagalska-Neubauer M, Pons JM, Crochet PA, Chylarecki P, Przystalski A, Gay L (2009) Assortative mating without complete reproductive isolation in a zone of recent secondary contact between Herring Gulls (Larus argentatus) and Caspian Gulls (L. cachinnans). Auk 126:409–419
Oakgrove KS, Harrigan RJ, Loieseau C, Guers S, Seppi B, Sehgal RNM (2014) Distribution, diversity and drivers of blood-borne co-infections in Alaska bird population. Int J Parasitol 44:717–727
Padilla LR, Whiteman NK, Merkel J, Huyvert KP, Parker PG (2006) Health assessment of seabirds on Isla Genovesa, Galápagos. Ornithol Monogr 60:86–97
Peirce MA (1981) Haematozoa of British birds. VI. Redescription of Haemoproteus larae Yakunin from the lesser black-backed gull Larus fuscus. J Nat Hist 15:459–462
Pérez- Rodríguez A, de la Puente J, Onrubia A, Pérez-Tris J (2013) Molecular characterization of haemosporidian parasites from kites of the genus Milvus (Aves: Accipitridae). International J Parasitol 43:381–387
Pérez-Tris J, Bensch S (2005) Diagnosing genetically diverse avian malaria infections using mixed-sequence analysis and TA-cloning. Parasitology 131:15–23
Piersma T (1997) Do global patterns of habitat use and migration strategies coevolve with relative investment in immunocompetence due to spatial variation in parasites pressure? Oikos 80:623–631
Quillfeldt P, Martínez J, Hennicke J, Ludynia K, Gladbach A, Masello JF, Riou S, Merino S (2010) Hemosporidian blood parasites in seabirds—a comparative genetic study of species from Antarctic to tropical habitats. Naturwissenschaften 97:809–817
Quillfeldt P, Arriero E, Martínez J, Masello JF, Merino S (2011) Prevalence of blood parasites in seabirds – a review. Front Zool 8:26. doi:10.1186/1742-9994-8-26
Ricklefs RE, Swanson BL, Fallon SM, Martínez-Abraín A, Scheuerlein A, Gray J, Latta SC (2005) Community relationships of avian malaria parasites in southern Missouri. Ecol Monogr 75:543–559
Rigaud T, Perrot-Minnot MJ, Brown MJF (2010) Parasite and host assemblages: embracing the reality will improve our knowledge of parasite transmission and virulence. Proc R Soc B 277:3693–3702
Ruiz X, Oro D, González-Solís J (1995) Incidence of a Haemoproteus lari parasitemia in a threatened gull: Larus audouinii. Ornis Fenn 72(4):159–164
Scheuerlein A, Ricklefs RE (2004) Prevalence of blood parasites in European passeriform birds. Proc R Soc Lond B 271:1363–1370
Snow DW, Perrins CM (1998) The birds of the Western Palearctic, concise edn. Oxford University Press, Oxford
Sol D, Jovani R, Torres J (2000) Geographical variation in blood parasites in feral pigeons: the role of vectors. Ecography 23:307–314
Swanson BL, Lyons A, Bouzat JL (2014) Distribution, prevalence and host specificity of avian malaria parasites across the breeding range of the migratory lark sparrow. Genetica 142:235–249
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tella JL, Forero MG, Gajón A, Hiraldo F, Donázar JA (1996) Absence of blood-parasitization effects on lesser kestrel fitness. Auk 113(1):253–256
Tella JL, Blanco G, Forero MG, Gajon A, Donazar JA, Hiraldo F (1999) Habitat, world geographic range, and embryonic development of hosts explain the prevalence of avian hematozoa at small spatial and phylogenetic scales. Proc Natl Acad Sci USA 96(4):1785–1789
Valkiūnas G (2005) Avian malaria parasites and other haemosporidia. CRC, Boca Raton
Valkiūnas G, Iezhova TA, Loiseau C, Sehgal RNM (2009) Nested cytochrome b polymerase chain reaction diagnostics detect sporozoites of hemosporidian parasites in peripheral blood of naturally infected birds. J Parasitol 95:1512–1515
van Rooyen J, Lalubin F, Glaizot O, Christe P (2013) Avian haemosporidian persistence and co-infection in great tit at the individual level. Malar J 12:40. doi:10.1186/1475-2875-12-40
Waldenstöm J, Bensch S, Hasselquist D, Östman Ö (2004) A new nested polymerase chain reaction method very efficient in detecting Plasmodium and Haemoproteus infections from avian blood. J Parasitol 90:191–194
Wojczulanis-Jakubas K, Svoboda A, Kruszewicz A, Johnsen A (2010) No evidence of blood parasites in Little Auk (Alle alle) breeding on Svalbard. J Wildl Dis 46:574–578
Wood MJ, Cosgrove CL, Wilkin TA, Knowles SCL, Day KP, Sheldon BC (2007) Within-population variation in prevalence and lineage distribution of avian malaria in blue tits, Cyanistes caeruleus. Mol Ecol 16:3263–3273
Zagalska-Neubauer M, Neubauer G (2012) Reproductive performance and changes in relative species abundance in a mixed colony of Herring and Caspian Gulls, Larus argentatus and L. cachinnans. Acta Ornithol 47:185–194
Zhao W, Cai B, Qi Y, Liu S, Hong L, Lu M, Chen X, Qiu C, Peng W, Li J, Su X (2014) Multi-Strain infections and ‘relapse’ of Leucocytozoon sabrazesi gametocytes in domestic chickens in Southern China. PLoS ONE 9(4):e94877. doi:10.1371/journal.pone.0094877
Acknowledgments
We would like to thank the anonymous reviewers for their helpful suggestions for improving this manuscript. We thank also Dimitar Dimitrov for comments on earlier versions of this manuscript. We thank Grzegorz Neubauer and Magda Zadrąg for help with organization and assistance in the field, Krzysztof Szpila and Tomasz Kakareko for sharing the data on Simuliidae occurrence. We thank Kate Petty for improving the manuscript’s English. Additionally, we thank Jane Jönsson for assistance with laboratory studies. This work was funded by grants from the Swedish Research Council to S.B. and to the Centre for Animal Movement Research (CAnMove), Polish Research Grant of the Ministry of Science and Higher Education (N304 073 31/2805) to M.Z.-N. A travel grant was partially funded by the Museum and Institute of Zoology PAS. The study was performed following the Guidelines of the European Union Council and the current laws in Poland, according to the Local Ethical Committee (permission number: 01/2007).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. C. Klasing.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Zagalska-Neubauer, M., Bensch, S. High prevalence of Leucocytozoon parasites in fresh water breeding gulls. J Ornithol 157, 525–532 (2016). https://doi.org/10.1007/s10336-015-1291-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10336-015-1291-5