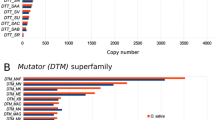
Abstract
Transposable elements (TEs) are distributed throughout the genome and play an important role in genome variation of the rice blast fungus, Magnaporthe oryzae. TE-associated molecular markers have been developed and used extensively for diversity analysis in natural populations. Here, we investigated the genomic distribution of a selected group of TEs that are dispersed as singletons, and each is of a size feasible for PCR validation, designated as SSTEs, in the genome of the reference laboratory strain, 70-15. The 75 SSTEs identified were distributed evenly on seven chromosomes of the M. oryzae genome. Approximately 40 % of SSTEs were located either in the coding or promoter regions of the predicted genes. The presence or absence of each SSTE at the respective locus was assessed, resolving significant presence/absence polymorphism among 11 rice blast strains collected from different locations worldwide. The presence/absence (P/A) polymorphism of SSTEs in different strains suggests that they may be useful for developing map-based land PATE markers for genetic analysis in M. oryzae.


Similar content being viewed by others
References
Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795
Chadha S, Gopalakrishna T (2005) Retrotransposon-microsatellite amplified polymorphism (REMAP) markers for genetic diversity assessment of the rice blast pathogen (Magnaporthe grisea). Genome 48:943–945
Chadha S, Gopalakrishna T (2007) Comparative assessment of REMAP and ISSR marker assays for genetic polymorphism studies in Magnaporthe grisea. Curr Sci 93:688–692
Chuma I, Isobe C, Hotta Y, Ibaragi K, Futamata N, Kusaba M, Yoshida K, Terauchi R, Fujita Y, Nakayashiki H et al (2011) Multiple translocation of the Avr-Pita effector gene among chromosomes of the rice blast fungus Magnaporthe oryzae and related species. PLoS Pathog 7:e1002147
Correll JC, Harp TL, Guerber JC, Zeigler RS, Liu B, Cartwright RD, Lee FN (2000) Characterization of Pyricularia grisea in the United States using independent genetic and molecular markers. Phytopathology 90:1396–1404
Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, Orbach MJ, Thon M, Kulkarni R, Xu JR, Pan H et al (2005) The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434:980–986
Farman ML, Taura S, Leong SA (1996a) The Magnaporthe grisea DNA fingerprinting probe MGR586 contains the 3′ end of an inverted repeat transposon. Mol Gen Genet 251:675–681
Farman ML, Tosa Y, Nitta N, Leong SA (1996b) MAGGY, a retrotransposon in the genome of the rice blast fungus Magnaporthe girsea. Mol Gen Genet 251:665–674
Flavell AJ, Knox MR, Pearce SR, Ellis THN (1998) Retrotransposon-based insertion polymorphisms (RBIP) for high throughput marker analysis. Plant J 16:643–650
George MLC, Nelson RJ, Zeigler RS, Leung H (1998) Rapid population analysis of Magnaporthe grisea by using rep-PCR and endogenous repetitive DNA sequences. Phytopathology 88:223–229
Hamer JE, Farrall L, Orbach MJ, Valent B, Chumley FG (1989) Host species-specific conservation of a family of repeated DNA sequences in the genome of a fungal plant pathogen. Proc Natl Acad Sci USA 86:9981–9985
Hayashi N, Ando I, Imbe T (1998) Identification of a new resistance gene to a Chinese blast fungus isolate in the Japanese rice cultivar Aichi Asahi. Phytopathology 88:822–827
Jantasuriyarat C, Gowda M, Haller K, Hatfield J, Lu G, Stahlberg E, Zhou B, Li H, Kim H, Yu Y et al (2005) Large-scale identification of expressed sequence tags involved in rice and rice blast fungus interaction. Plant Physiol 138:105–115
Jiang N, Wessler SR (2001) Insertion preference of maize and rice miniature inverted repeat transposable elements as revealed by the analysis of nested elements. Plant Cell 13:2553–2564
Kachroo P, Leong SA, Chattoo BB (1994) Pot2, an inverted repeat transposon from the rice blast fungus Magnaporthe grisea. Mol Gen Genet 245:339–348
Kachroo P, Leong SA, Chattoo BB (1995) Mg-SINE: a short interspersed nuclear element from the rice blast fungus, Magnaporthe grisea. Proc Natl Acad Sci USA 92:11125–11129
Kang S (2001) Organization and distribution pattern of MGLR-3, a novel retrotransposon in the rice blast fungus Magnaporthe grisea. Fungal Genet Biol 32:11–19
Kito H, Takahashi Y, Sato J, Fukiya S, Sone T, Tomita F (2003) Occan, a novel transposon in the Fot1 family, is ubiquitously found in several Magnaporthe grisea isolates. Curr Genet 42:322–331
Lau GW, Chao CT, Ellingboe AH (1993) Interaction of genes controlling avirulence/virulence of Magnaporthe grisea on rice cultivar Katy. Phytopathology 83:375–382
Leung H, Borromeo ES, Bernardo MA, Notteghem JL (1988) Genetic analysis of virulence in the rice blast fungus Magnaporthe grisea. Phytopathology 78:1227–1233
Levy M, Romao J, Marchetti MA, Hamer JE (1991) DNA fingerprinting with a dispersed repeated sequence resolves pathotype diversity in the rice blast fungus. Plant Cell 3:95–102
Li W, Wang B, Wu J, Lu G, Hu Y, Zhang X, Zhang Z, Zhao Q, Feng Q, Zhang H et al (2009) The Magnaporthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t. Mol Plant Microbe Interact 22:411–420
Li P, Bai B, Zhang HY, Zhou H, Zhou B (2012) Genomic organization and sequence dynamics of the AvrPiz-t locus in Magnaporthe oryzae. J Zhejiang Univ Sci B 13:452–464
Liu G, Geurts AM, Yae K, Srinivasan AR, Fahrenkrug SC, Largaespada DA, Takeda J, Horie K, Olson WK, Hackett PB (2005) Target-site preference for Sleeping Beauty transposons. J Mol Biol 346:161–173
Ma LJ, Ibrahim AS, Skory C, Grabherr MG, Burger G, Butler M, Elias M, Idnurm A, Lang BF, Sone T et al (2009) Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet 5:e1000549
Nakayashiki H, Matsuo H, Chuma I, Ikeda K, Betsuyaku S, Kusaba M, Tosa Y, Mayama S (2001) Pyret, a Ty3/Gypsy retrotransposon in Magnaporthe grisea contains an extra domain between the nucleocapsid and protease domains. Nucleic Acids Res 29:4106–4113
Pan X, Li Y, Stein L (2005) Site preferences of insertional mutagenesis agents in Arabidopsis. Plant Physiol 137:168–175
Park SY, Milgroom MG, Han SS, Kang S, Lee YH (2003) Diversity of pathotypes and DNA fingerprint haplotypes in populations of Magnaporthe grisea in Korea over two decades. Phytopathology 93:1378–1385
Perrière G, Gouy M (1996) WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78:364–369
Rehmeyer C, Li W, Kusaba M, Kim YS, Brown D, Staben C, Dean R, Farman M (2006) Organization of chromosome ends in the rice blast fungus Magnaporthe oryzae. Nucleic Acids Res 34:4685–4701
Sanchez E Jr, Asano K, Sone T (2011) Characterization of Inago1 and Inago2 retrotransposons in Magnaporthe oryzae. J Gen Plant Pathol 77:239–242
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Thon MR, Martin SL, Goff S, Wing RA, Dean RA (2004) BAC end sequences and a physical map reveal transposable element content and clustering pattern in the genome of Magnaporthe grisea. Fungal Genet Biol 41:657–666
Thon MR, Pan H, Diener S, Papalas J, Taro A, Mitchell TK, Dean RA (2006) The role of transposable element clusters in genome evolution and loss of synteny in the rice blast fungus Magnaporthe oryzae. Genome Biol 7:R16
Tosa Y, Uddin W, Viji G, Kang S, Mayama S (2007) Comparative genetic analysis of Magnaporthe oryzae isolates causing gray leaf spot of perennial ryegrass turf in the United States and Japan. Plant Dis 91:517–524
Xue M, Yang J, Li Z, Hu S, Yao N, Dean RA, Zhao W, Shen M, Zhang H, Li C et al (2012) Comparative analysis of the genomes of two field isolates of the rice blast fungus Magnaporthe oryzae. PLoS Genet 8:e1002869
Zeigler RS, Cuoc LX, Scott RP, Bernardo MA, Chen DH, Valent B, Nelson RJ (1995) The relationship between lineage and virulence in Pyricularia grisea in the Philippines. Phytopathology 85:443–451
Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214
Acknowledgments
We are grateful to Drs. Jong-Seong Jeon at the Kyung Hee University, Korea, Guo-Liang Wang at the Ohio State University, USA, Yulin Jia at USDA-ARS Dale Bumpers National Rice Research Center, USA, and Chenyun Li at Yunnan Agriculture University, China, for providing the strains listed in Tables 1 and S4. We thank Mr. Neal Rasmi for a grammatical review of this manuscript. This project was supported by grants from Natural Science Foundation in China (30971878), Open Project of State Key Laboratory Breeding Base for Zhejiang Sustainable Pest and Disease Control (2010DS700124-KF1108) and partial funding from the Faculty of Science and the Graduate School, Kasetsart University.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10327_2013_428_MOESM5_ESM.ppt
Fig. S1. Chromosomal distribution of SSTEs. The supercontigs indicated in grey bars were aligned on each chromosome in the same order as the one in the integrated map of M. grisea (http://www.broadinstitute.org/annotation/genome/magnaporthe_grisea/maps/ViewMap.html?sp=0http://www.broadinstitute.org/annotation/genome/magnaporthe_grisea/maps/ViewMap.html?sp=0). The SSTEs corresponding to the PATE loci are indicated by lines with different abbreviated names. Abbreviated name for each SSTE family: Mg-SINE = MS; Pot2-A = P2A; Pot2-B = P2B; Pot3 = P3; MINE = MN; solo-LTR = SR; Occan = ON; MGL = ML. The location of SSTE in each supercontig is listed in Table 2. The expected size of PCR product at each PATE locus is listed in Table 4. The map was drawn to scale. (PPT 672 kb)
10327_2013_428_MOESM6_ESM.ppt
Fig. S2. Cladistic analyses of Mg-SINE (A) and Pot2 (B) elements used in this study. The nucleotide sequences of TEs were used for multiple sequence alignments in the program Clustal_X 1.81 (Thompson et al. 1997). The derived phylogenetic tree was viewed with the program Njplot (Perrière and Gouy 1996). (PPT 116 kb)
10327_2013_428_MOESM7_ESM.ppt
Fig. S3. Multiple sequence alignments of Pot2 elements. Two loci, 6-194-M10 and 3-191-M1, were used as representatives for Pot2-A and Pot2-B elements, respectively. Locus 6-169-M3 is a chimera of Pot2-A and -B. (A). Nucleotide sequences of these three Pot2 elements were used for the multiple sequence alignment. The terminal inverted repeat (TIR) sequences are indicated by the upper lines at both termini. (B). Amino-acid sequences of these three Pot2 elements used for the multiple sequence alignment. The central DDE domain is indicated by line above sequences. (PPT 265 kb)
Rights and permissions
About this article
Cite this article
Zhang, Hy., He, Dy., Kasetsomboon, T. et al. Analysis of selected singleton transposable elements (SSTEs) and their application for the development of land PATE markers in Magnaporthe oryzae . J Gen Plant Pathol 79, 96–104 (2013). https://doi.org/10.1007/s10327-013-0428-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10327-013-0428-8