Abstract
We examined the localization of probe-accessible chitin in Magnaporthe oryzae, which causes rice blast disease, during the early infection process and the functions of two genes encoding a chitin-binding domain (ChBD). Invasive hyphae in the first-invaded rice cell showed little staining with fluorescently labeled wheat germ agglutinin, a probe to detect chitin. However, in the second-invaded cell, hyphae showed strong fluorescence, and treatment with chitinase diminished the signal. Fourteen ChBD genes encoding family 18 carbohydrate-binding module (CBM18) were isolated from a Japanese strain of M. oryzae, Ina86-137. Reverse transcription-polymerase chain reaction analysis demonstrated that ChBD-1, ChBD-6, ChBD-8, ChBD-13, and ChBD-15 are expressed in the rice sheath. Gene-targeted disruptants of ChBD-1 and ChBD-15 had no significant differences in invasive growth, pathogenicity, or tolerance to chitinase compared to the wild type. These results suggest that M. oryzae has a mechanism to evade being a substrate for the host chitinase in the first-invaded cell, but neither ChBD-1 nor ChBD-15 contributes to this mechanism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chitin is a linear homopolymer of β-1,4-linked N-acetyl-d-glucosamine residues and is a major component of fungal cell walls. Although higher plants do not possess chitin, they produce various chitinases when infected by pathogens. Certain chitinases have antifungal activity, and transgenic plants with enhanced chitinase activity have increased resistance to fungal pathogens (e.g., Broglie et al. 1991; Nishizawa et al. 1999).
In many plants, chitin oligosaccharides have elicitor activity that induces various self-defense responses, including altered gene expression and the generation of reactive oxygen species and antimicrobial substances (Shibuya and Minami 2001). Indeed, pretreatment of rice plants with N-acetylchitoheptaose results in increased resistance to rice blast caused by M. oryzae (Tanabe et al. 2006). Receptors of the chitin elicitor have been identified in rice and Arabidopsis, and a lack of these receptors leads to a significant reduction in responses to the elicitor (Kaku et al. 2006; Miya et al. 2007). Moreover, perception of the chitin elicitor contributes to fungal disease resistance (Kishimoto et al. 2010; Miya et al. 2007; Wan et al. 2008), and genetically engineered enhancement of chitin elicitor signaling improves resistance to rice blast (Kishimoto et al. 2010). These studies strongly suggest that cell-wall chitin in the invading fungal pathogen is hydrolyzed by the host chitinase; the chitin oligosaccharides thus generated are recognized by the host cell via the receptor in the plasma membrane, which induces the defense responses that inhibit infection.
Thus, to establish successful infection, pathogenic fungi may need to evade attack by chitinase, suppress the induction of defense responses by the chitin elicitor, or both. Elucidating such a pathogenic mechanism should contribute to the control of plant diseases, but little is known about the molecular basis of the mechanism. Avr4, an effector of Cladosporium fulvum, the causal agent of tomato leaf mold disease, contains an invertebrate-type chitin-binding domain (van den Burg et al. 2003) and binds specifically in planta to chitin in fungal cell walls. Avr4 has been suggested to be capable of protecting fungal hyphae against tomato chitinases via this binding (van den Burg et al. 2006). Rice–M. oryzae interactions presumably involve a similar mechanism because genome sequence analyses have indicated that M. oryzae possesses highly abundant putative chitin-binding proteins, unlike the nonphytopathogenic fungi Aspergillus nidulans and Neurospora crassa (Dean et al. 2005). Labeling with wheat germ agglutinin (WGA), which binds chitin with high affinity, showed that in M. oryzae, cell wall chitin is reduced in the penetration peg formed in vitro compared to that in appressoria (Howard et al. 1991). Recently, Fujikawa et al. (2009) reported that chitin of M. oryzae infection hyphae in first-invaded rice cells is barely stained with a WGA probe. However, they did not examine the localization of chitin in cell walls at later stages, e.g., expansion into neighboring rice cells. In this study, we report the histochemical detection of chitin in hyphae of M. oryzae invading rice leaf sheath cells. We also characterized gene-targeted disruptants of M. oryzae genes encoding proteins that potentially bind chitin (designated ChBD genes).
Materials and methods
Fungal isolates, rice plants, and culture conditions
Magnaporthe oryzae strains Ina86-137 (MAFF Gene Bank stock number MAFF101511) and TH68-126 (MAFF101517), Japanese pathogenic isolates from rice, were cultured on potato dextrose agar (PDA; Difco Laboratories, Detroit, MI, USA) at 25°C. To collect conidia, the fungi were cultured on oatmeal agar medium (5% oatmeal powder, 1.5% agar) for 7 days at 25°C in darkness. Production of conidia was induced by irradiation with a blacklight blue lamp for 4 days after the aerial mycelia were removed using a sterilized brush. Rice plants (Oryza sativa L. cv. Nipponbare BL no. 2, compatible with Ina86-137, and cv. Nipponbare BL no. 5, compatible with TH68-126) were grown hydroponically for 3–4 weeks in a nutrient solution as described previously (Tanabe et al. 2006), using a growth chamber with a 14-h light (28°C, 5,000 lx)/10-h dark (25°C) cycle.
Chitin staining
WGA-Alexa Fluor 488 conjugate (Molecular Probes, Eugene, OR, USA) was used according to Ramonell et al. (2005) to stain chitin in the infection structures of M. oryzae that formed on rice leaf sheaths. Trimmed leaf sheaths were washed in 1× phosphate-buffered saline (PBS; pH 7.4) containing 0.1% Triton X-100 for 20 min and stained with WGA-Alexa Fluor 488 at 10 μg/mL in 1× PBS (pH 7.4) containing 0.1% Triton X-100 for 4 h at 4°C. The sheaths were then washed in the PBS buffer for 3 h and in fresh buffer overnight. Propidium iodide (PI; 50 μg/mL) in 1× PBS (pH 7.4) was used as a counterstain. For enzymatic digestion of chitin, the inoculated rice leaf sheath was incubated with 0.5 U/mL of purified chitinase from Trichoderma viride (Sigma, St. Louis, MO, USA) in 1× PBS (pH 7.4) with 1% Tween 20 for 18 h at 25°C, then washed with 1× PBS (pH 7.4)–0.1% Triton X-100 three times before staining with WGA-Alexa Fluor 488.
Confocal microscopy
Confocal microscopy was performed using a TCS SP5 instrument (Leica, Wetzlar, Germany). Fluorescence was excited with an argon laser at 488 nm and detected at wavelengths of 500–520 nm [WGA-Alexa Fluor 488 and green fluorescent protein (GFP)] or 600–620 nm (PI). Images were processed and arranged using LAS AF software (Leica) and Adobe (San Jose, CA, USA) Photoshop CS4 software.
RT-PCR assay
Total RNA was extracted from mycelia growing in YG broth (0.5% yeast extract, 2% glucose) and from rice leaf sheaths at 40 h post-inoculation (hpi) using an RNeasy Mini Kit (Qiagen, Valencia, CA, USA). The mycelia and infected rice sheaths were placed in screw-capped microtubes containing 450 μL of the kit’s RLT buffer and five zirconia beads (2 mm diameter), then homogenized using a Micro Smash MS-100 system (Tomy Seiko, Tokyo, Japan) by shaking the tubes three times (30 s each, at 4,000 rpm). The extracted total RNA (500 ng) was used as the template for a reverse transcription reaction using a ReverTra Ace α Kit (Toyobo, Osaka, Japan) in a 20-μL reaction mixture. The mixture was diluted with 100 μL of sterile distilled water, and 3-μL samples were used for the PCR, which was performed with each of the primer sets listed in Table 1 at 400 nM and with 5% dimethyl sulfoxide, using a GoTaq kit (Promega, Madison, WI, USA). The initial denaturation step was 2 min at 95°C, followed by 30 (mycelia) or 35 (infected rice) cycles of amplification consisting of 30 s each at 95, 60, and 72°C.
Vectors and Agrobacterium-mediated transformation
Vectors used to knockout ChBD genes and for Agrobacterium-mediated transformation of M. oryzae were constructed using the GFP-tagging gene knockout (GGKO) method of Saitoh et al. (2008). Table 1 lists the primers used to construct the knockout vectors. Fungal DNA for PCR analysis was prepared using a simple method developed by Saitoh et al. (2006), and gene disruption was confirmed by PCR using the primers listed in Table 1.
Pathogenicity test
For the spray inoculation, 2-week-old rice seedlings grown in a greenhouse were sprayed with conidial suspensions (5 × 105 conidia/mL) of M. oryzae in 0.05% Triton X-100 using the method of Nishizawa et al. (1999). The inoculated seedlings were incubated in a humid chamber at 25°C for 20 h, then cultivated in a greenhouse. At least three independent mutant lines per target gene were analyzed, with the test carried out twice.
For the evaluation of degree of hyphal growth, the midrib area of detached rice sheaths was inoculated with a conidial suspension (106 conidia/mL, 20 hpi, or 105 conidia/mL, 48 hpi) as described previously (Tanabe et al. 2006). After a 20 or 48 h incubation at 25°C in darkness, the inoculated sheath was hand-trimmed for microscopic observation. The degree of invasion for all appressoria on each 1.5-cm-long leaf sheath at 20 hpi and for ca. 150 intact appressoria at 48 hpi was assessed as “multicell invasion,” “single-cell invasion,” or “no invasion,” depending on whether appressoria had penetrated more than one rice cell, one cell, or no cells, respectively (Tanabe et al. 2006). Three to five leaf sheaths per line were observed. For assaying the sensitivity of germ tubes to chitinase, 20 μL of a conidial suspension (5.0 × 104 conidia/mL) was incubated on plastic coverslips for 16 h at 25°C in darkness. Appressorium-forming conidia were incubated with 1× PBS buffer with 0.5% Tween 20 containing 0.5 U/mL chitinase from T. viride for 8 h. Statistical analyses (Tukey’s test and Dunnett’s test) were performed using JMP8 software (SAS Institute, Cary, NC, USA).
Results
Localization of probe-accessible chitin in infection structures
To determine whether the surface components of the rice blast fungus change after invasion of rice cells, rice leaf sheaths were inoculated with a conidial suspension of a virulent isolate and stained with WGA-Alexa Fluor 488 probe, a chitin-binding lectin conjugated with a green fluorescent dye. The stained sheath sections were then observed using confocal laser microscopy. Surfaces of the germ tube and appressorium fluoresced strongly. During early infectious growth up to 30 hpi (Fig. 1a), the primary and invasive hyphae tended to be stained. However, the intensity of the fluorescence of the invasive hyphae inside the first-invaded epidermal cell diminished after 30 hpi. In contrast, once the invasive hyphae entered the second-invaded cell, they fluoresced strongly (Fig. 1b), and the fluorescent signals lasted for a longer time (Fig. 1c). Focal plane scanning confirmed that the hyphae were inside the epidermal cells. The test was performed using two combinations of compatible interactions (rice cv. Nipponbare BL no. 2 and blast strain Ina86-137, or cv. Nipponbare BL no. 5 and TH68-126), and similar staining patterns were observed for both combinations.
Wheat germ agglutinin (WGA) probe binding to the rice blast fungus growing in epidermal cells of rice leaf sheath. a–c Inoculated, detached rice sheaths were treated with WGA-Alexa Fluor 488 (green channel) and propidium iodide (PI) (red channel, staining nuclei and plant cell wall) at 30 hpi (a), 36 hpi (b), and 42 hpi (c). d, e Effect of pretreatment with chitinase on WGA binding. Samples were stained with WGA-Alexa Fluor 488 and PI after treatment without (d) and with chitinase (e) at 36 hpi. The fluorescence-field panels show superimposed confocal images in which stacked Z-series images corresponding to the appressoria and epidermis obtained in the green channel and an image in the red channel were merged. The bright-field panels are differential interference contrast images. In the rightmost panels in d and e, the fluorescence image and the bright field image are superimposed. Arrows indicate first-invaded epidermal cells. Arrowheads denote hyphae that had just invaded second cells. Asterisks show WGA-stained septa. Ap appressorium. Bar 10 μm
We next confirmed whether WGA-Alexa Fluor 488 probe stained chitin on M. oryzae. For treatment with chitinase, an 18-h incubation in buffer containing 1% Tween 20 was required prior to WGA staining. Figure 1d shows that hyphae in the second-invaded cells were successfully stained in this buffer without chitinase. In comparison, pretreatment of the inoculated rice sheaths with chitinase diminished the WGA staining of the invasive hyphae in the second-invaded cells (Fig. 1e), strongly suggesting that we had observed WGA probe-accessible chitin in the infection structures of M. oryzae.
Structure of genes encoding chitin-binding domains
Carbohydrate-binding modules (CBMs) are classified into 54 families based on sequence similarity and protein folds (http://www.cazy.org/; Cantarel et al. 2009). Several of these modules, such as CBM5, 12, 14, and 18, possess chitin-binding activity. A search of the M. oryzae genome sequence database (ver. 4; Magnaporthe Sequencing Project. R. Dean, Fungal Genomics Laboratory, North Carolina State University, http://www.fungalgenomics.ncsu.edu; and MIT/Harvard Broad Institute, http://www.broad.mit.edu) revealed 15 genes encoding CBM18 motifs (designated ChBD-1–ChBD-15; Table 2), including seven lectin-type ChBD genes (ChBD-1–ChBD-6, ChBD-15), one class III chitinase gene (ChBD-7), three class V chitinase genes (ChBD-8–ChBD-10), and two chitin deacetylase genes (ChBD-11 and ChBD-12). Because all 15 proteins possess a signal sequence, they appear to be secreted. The ChBD found in the Avr4 protein of C. fulvum belongs to the CBM14 family (van den Burg et al. 2003), but we found no genes encoding CBM14, CBM5, or CBM12 in the M. oryzae database.
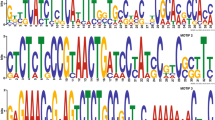
We compared the genomic sequences corresponding to the 15 ChBD genes in a Japanese isolate, Ina86-137, with the sequence of experimental strain 70-15 (Dean et al. 2005). ChBD-5 from Ina86-137 was not amplified, even though the gene was successfully isolated from another M. oryzae strain, Guy11. The nucleotide sequences of the other 14 ChBD genes obtained from Ina86-137 have been registered in DDBJ under accession numbers AB513115–AB513128. Figure 2a shows their putative protein structures. As summarized in Table 2, ChBD-1 of Ina86-137 carries a 174-bp insert, resulting in the introduction of another CBM18 motif, and ChBD-4 and ChBD-11 have a 261-bp insert and a 117-bp deletion, respectively. The sequences of open reading frames, introns, and adjacent regions up to 500–1,900 bp of the other genes, which were confirmed to produce knockout mutants, showed more than 99% identity. All amino acid sequences of CBM18, except those of ChBD-1 and ChBD-11, were conserved between strains 70-15 and Ina86-137. Figure 2b shows an alignment of the CBM18 amino acid sequences found in ChBD-1–ChBD-15, except for ChBD-5, along with the CBM18 consensus sequence and the sequence for CBM18 of Urtica dioica isolectin, for which the crystal structure and amino acid residues participating in chitin binding have been determined (Harata and Muraki 2000). CBM18 contains several disulfide bonds, and the corresponding cysteine residues, as well as adjoining glycine residues, are well conserved in the ChBD-1–ChBD-15 proteins. The central region of CBM18, which contains three aromatic amino acid residues and one serine residue that participate in chitin binding (Harata and Muraki 2000), is especially highly conserved.
Position and sequence of CBM18 in ChBD genes. a Schematic of structures of proteins encoded by the ChBD-1–ChBD-15 genes in M. oryzae Ina86-137. Conserved domains detected by Pfam analysis (http://pfam.sanger.ac.uk/search) are color-coded. Each CBM18 is numbered beginning at the N-terminus. b Comparison of CBM18 amino acid sequences found in ChBD-1–ChBD-15. The amino acid sequences were aligned with the CBM18 consensus sequence and the CBM18 of Urtica dioica isolectin. The triad of aromatic residues and the serine residue are indicated by yellow and red boxes, respectively. Conserved cysteines and glycines are indicated by green and blue boxes, respectively
Expression analysis of ChBD genes using RT-PCR
To analyze the expression of the ChBD genes at the infection stage, we conducted RT-PCR using primer sets designed to specifically amplify each of the 14 ChBD genes. Five ChBD genes, ChBD-1, ChBD-6, ChBD-8, ChBD-13, and ChBD-15, were expressed in the rice sheath at 40 hpi, but only ChBD-8 was expressed in vegetatively growing mycelia (Fig. 3). No expression of other ChBD genes was detected at the stages tested. These genes may be expressed under specific conditions or may be pseudogenes.
Knockout of ChBD genes
We selected ChBD-1 and ChBD-15 from the three lectin-type ChBD genes expressed in the inoculated rice sheath and generated gene-targeted knockout mutants of each using the GGKO method of Saitoh et al. (2008). Successful gene-targeted disruption was confirmed by PCR (Fig. 4). No significant differences in radial growth on PDA medium up to 10 days or sporulation on oatmeal agar were detected between the two knockout mutants and wild-type controls (n = 3; Tukey’s test and Dunnett’s test). Appressorium formation was examined on cover glasses made of plastic or glass, on cellophane, and on plant epidermis samples, and no morphological aberrations or reductions in frequency of appressoria formation were observed.
Confirmation of knockout mutants by genomic PCR. a Schematic representation of two PCR sets to confirm gene disruption. PCR1 was performed with primers corresponding to areas outside the upstream flanking regions (UFRs) and downstream flanking regions (DFRs; solid arrows). Knockout mutants were expected to generate a single band in PCR1 that differed in size from that of the wild-type Ina86-137 and ectopic transformants. PCR2 consisted of multiplex PCR using primers specific for the target gene (open arrows) and the hygromycin phosphatase gene (HPT; gray arrows). The HPT fragment should be about 500 bp. In PCR2, the wild type was expected to amplify the target gene, whereas the knockout mutants were expected to amplify HPT and a disrupted target gene fragment from the ectopic transformants, thus generating products from both genes. b Results of both PCR sets. In the names of the isolates, “K” indicates a knockout mutant, and “E” indicates an ectopic transformant. In PCR2, the binary vector DNA for the target gene disruption was also used (V). M DNA size marker
Expression analysis of selected ChBD genes by GFP fluorescence
Because the gene-disrupted lines possessed the GFP gene under the control of their own promoter, we were able to observe stage- and site-specific expression of each gene using GFP fluorescence. Neither ChBD-1 nor ChBD-15 was expressed before appressoria formed. During the appressorium formation and intracellular invasive growth, both ChBD-1 and ChBD-15 were expressed in appressoria (Fig. 5), suggesting that the two genes function before penetration. ChBD-1, but not ChBD-15, was also expressed in the primary hyphae (16 hpi). Neither of the genes was expressed in expanded invasive hyphae (36 hpi), suggesting that they function during an early stage of infection.
Fluorescence microscopy of knockout mutants growing in rice leaf sheaths. All panels are superimposed images of the stacked Z-series confocal GFP fluorescent images corresponding to the appressoria and epidermis and a differential interference contrast image. Arrowheads show primary hyphae. Bar 10 μm
Pathogenicity and tolerance to chitinase of ΔChBD-1 and ΔChBD-15
To analyze the pathogenicity of the ChBD-1 and ChBD-15 knockout mutants (ΔChBD-1 and ΔChBD-15) after spray inoculation, we compared lesion areas at 8 days post-inoculation (dpi). Typical susceptible lesions developed in the knockout lines (Fig. 6a), and no significant differences were observed in the degree of lesion formation among them.
Evaluation of pathogenicity of Magnaporthe oryzae and rice cell wall sensitivity to chitinase of ChBD knockout mutants. a Lesions on rice leaf blade at 8 dpi. Two-week-old rice seedlings (16–20 plants per mutant line) were sprayed with a conidial suspension of each mutant. Inoculation tests were performed twice, and representative lesions are shown. Bar 1 mm. b Degree of hyphal penetration into rice leaf sheath epidermal cells at 20 hpi. c Degree of hyphal growth in rice leaf sheath epidermal cells at 48 hpi. d Percentage of nonlysed germ tubes after 8 h incubation in reaction buffer without and with chitinase. Mutant line names correspond to those in Fig. 4. Bars indicate standard deviation. Statistical analyses (Tukey’s test and Dunnett’s test) demonstrated no significant difference between wild-type controls and mutants
We next evaluated infection ability more precisely with a leaf sheath assay. Rice leaf sheaths were inoculated with a conidial suspension of each knockout mutant (two lines per disruptant), and the extent of elongation of the infection hyphae was assessed at 20 and 48 hpi. No significant differences were observed between mutants and the wild type (Fig. 6b, c). No morphological alterations during germination and appressoria formation on the rice sheath were detected.
To examine the effect of gene disruption of ChBD-1 or ChBD-15 on tolerance to chitinase, appressorium-forming conidia of ΔChBD-1 and ΔChBD-15 were treated with chitinase and the percentage of lysis of germ tubes was assessed. As shown in Fig. 6d, ca. 50% of the germ tubes were lysed, which was not significantly different from the percentage for the wild-type strain.
Discussion
Chitin and its fragments, N-acetylchitooligosaccharides, are representative microbe-associated molecular patterns of fungi that trigger various defense responses in plants at subnanomolar levels (Shibuya and Minami 2001). Thus, pathogenic fungi are very likely to have mechanisms to avoid generating the chitin elicitor during the infection process. Because M. oryzae possesses more genes encoding putative chitin-binding proteins than do nonphytopathogenic fungi (Dean et al. 2005), we speculated that M. oryzae has a mechanism to prevent being a substrate of rice chitinases and that chitin-binding proteins may contribute to this mechanism.
In the first-invaded rice cell, invasive hyphae had little fluorescence from fluorescently labeled WGA, consistent with the results reported by Fujikawa et al. (2009). Our present study demonstrated that when the hyphae expanded to neighboring cells, they fluoresced strongly. Because invasive hyphae, which develop from primary hyphae, are sealed within the plant membrane, designated an extrainvasive hyphal membrane (EIHM) (Kankanala et al. 2007), access of the WGA probe to the cell walls of the invasive hyphae could be restricted. However, the septae of invasive hyphae inside the first-invaded epidermal cell fluoresced (Fig. 1b), indicating that the WGA probe had accessed hyphae within the rice cell. Invasive hyphae growing in neighboring cells are also sheathed in EIHM (Kankanala et al. 2007); therefore, the accessibility of WGA probe to fungal cell walls should be similar in first- and second-invaded rice cells. Pretreatment with chitinase diminished WGA binding to the hyphae in the second-invaded cells (Fig. 1e), indicating that the observed fluorescence resulted from the binding of the WGA probe to chitin. Thus, our data strongly suggest that chitin in the invasive hyphae within the first-invaded epidermal cell is masked or modified, whereas chitin in the hyphae in neighboring cells is accessible to the WGA probe. These observations imply that the rice blast fungus has a mechanism to protect the cell wall against host chitinase and prevent the release of chitin elicitor in the early infection stages, as suggested for other host–pathogen interactions (El Gueddari et al. 2002; van den Burg et al. 2006). In contrast, hyphae in second- and later-invaded cells may not need to limit chitin exposure to be virulent. Invasive hyphae in second- and later-invaded rice cells have been reported to differ in behavior and often in appearance from those in first-invaded cells (Kankanala et al. 2007). Our results indicate that the cell wall properties of invasive hyphae in the first-invaded rice cell also differ from those in the second- and later-invaded rice cells, which implies that host–pathogen interactions in neighboring cells also differ from those in the first-invaded cell. Kishimoto et al. (2010) reported that transgenic rice plants with enhancement of chitin elicitor signaling suppress multi-cell invasion of M. oryzae, but do not influence the degree of one-cell invasion, which probably reflects the distribution of chitin in the M. oryzae cell wall.
We found 15 ChBD genes encoding CBM18 motifs in the genome sequence database, of which 14 were successfully cloned from a Japanese isolate. RT-PCR analysis of the 14 genes indicated that ChBD-1, ChBD-6, ChBD-8, ChBD-13, and ChBD-15 are expressed in the rice sheath. We produced gene-disrupted mutants of the two lectin-type ChBD genes using the GGKO method (Saitoh et al. 2008), which allows observation of stage- and site-specific expression of the target gene using GFP fluorescence. Unlike RT-PCR analysis, GFP fluorescence observations revealed that only ChBD-1 is expressed in rice cells. This inconsistency is likely because the infection process of each conidium proceeds asynchronously, and the RNA sample was obtained from tissue containing fungi at different stages of infection. Thus, live-cell imaging of the mutants generated by the GGKO method is advantageous in analyzing stage- and site-specific gene expression.
Expression data suggest that ChBD-1 and ChBD-15 function in the penetration stage. However, neither ΔChBD-1 nor ΔChBD-15 showed defects in appressoria formation, invasive growth, pathogenicity, or tolerance to chitinase. The staining pattern of fluorescently labeled WGA at 30, 36 and 42 hpi also did not differ from that of the wild-type strain (data not shown). Although these ChBD genes might have functional redundancy, our data strongly suggest that neither gene contributes to pathogenicity or protection from chitinase, which implies that the participation of chitin-binding proteins with CBM18 in virulence is low in M. oryzae.
A secretory protein, Ecp6, which harbors three lysin motifs (LysMs), was identified as a virulence factor of C. fulvum (Bolton et al. 2008), and quite recently, Ecp6 was found to sequester chitin oligosaccharides, preventing elicitation of host immunity (de Jonge et al. 2010). LysMs, recognized in prokaryotes and plants as a family of CBMs, are found in receptors for chitin elicitors CEBiP and CERK1 (Kaku et al. 2006; Miya et al. 2007). Putatively secreted LysM-containing proteins are widespread in the fungal kingdom, including M. oryzae (Bolton et al. 2008). Secretory proteins containing LysM domains rather than CBM18 may play important roles in virulence of M. oryzae.
Recent studies have revealed that cell wall chitin is physically masked by other cell wall components. Rappleye et al. (2007) reported the presence of α-1,3-glucan, which masks the trigger of the host immune system, in the outermost layer of the cell wall of Histoplasma capsulatum, a pathogenic fungus of mammals. Macrophages recognize the infection by H. capsulatum in part through the interaction of β-glucan from the fungal cell wall and the receptor dectin-1, and thus α-1,3-glucan blocks recognition by covering β-glucan (Rappleye et al. 2007). Similarly in M. oryzae, Fujikawa et al. (2009) showed that α-1,3-glucan masks both chitin and β-1,3-glucan in the cell walls of M. oryzae at 24 hpi. They also demonstrated that the presence of α-1,3-glucan results in increased tolerance to chitinase digestion. Thus, masking chitin by α-1,3-glucan is a major strategy of M. oryzae to evade attack by chitinase.
Another possible explanation for the unaltered pathogenicity is that chitin may be modified to avoid hydrolysis by chitinase. One mechanism that protects the fungal cell wall from the host chitinase may be the conversion of chitin to chitosan by chitin deacetylase secreted from the pathogen. For example, El Gueddari et al. (2002) found that chitin was exposed on surfaces of infection structure of the rust fungi Puccinia graminis and Uromyces fabae that form on plant cuticles, whereas chitosan was exposed on surfaces of the infection hyphae. Deuteromycetes that produce chitin deacetylase are all plant pathogens and secrete chitin deacetylase during plant cell penetration (Hekmat et al. 2003). Determining whether the reduced affinity of WGA for the invasive hyphae of M. oryzae is also due to the conversion of chitin to chitosan will be revealing. Fujikawa et al. (2009) have also reported that a major polysaccharide labeled at the accessible surface of M. oryzae invasive hyphae is chitosan, in addition to α-1,3-glucan. Of the ChBD genes listed in Table 2, ChBD-11 and ChBD-12 encode a putative chitin deacetylase. ΔChBD-12 showed no defects in invasive growth, pathogenicity or staining pattern of the fluorescently labeled WGA (K. Saitoh, S. Mochizuki, and Y. Nishizawa, unpublished data). However, we were unable to obtain ΔChBD-11, because the mutation may be lethal. Examination of the stage and site of ChBD-11 expression should provide information on the contribution of chitosan in the protection of the M. oryzae cell wall during infection.
References
Bolton MD, van Esse HP, Vossen JH, de Jonge R, Stergiopoulos I, Stulemeijer IJE, van den Berg GCM, Borrás-Hidalgo O, Dekker HL, de Koster CG, de Wit PJGM, Joosten MHAJ, Thomma BPHJ (2008) The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol Microbiol 69:119–136
Broglie K, Chet I, Holliday M, Cressman R, Biddle P, Knowlton S, Mauvais CJ, Broglie R (1991) Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science 254:1194–1197
Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238
de Jonge R, van Esse HP, Kombrink A, Shinya T, Desaki Y, Bours R, van der Krol S, Shibuya N, Joosten MHAJ, Thomma BPHJ (2010) Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 329:953–955
Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, Orbach MJ, Thon M, Kulkarni R, Xu JR, Pan H, Read ND, Lee YH, Carbone I, Brown D, Oh YY, Donofrio N, Jeong JS, Soanes DM, Djonovic S, Kolomiets E, Rehmeyer C, Li W, Harding M, Kim S, Lebrun MH, Bohnert H, Coughlan S, Butler J, Calvo S, Ma LJ, Nicol R, Purcell S, Nusbaum C, Galagan JE, Birren BW (2005) The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434:980–986
El Gueddari NE, Rauchhaus U, Moerschbacher BM, Deising HB (2002) Developmentally regulated conversion of surface-exposed chitin to chitosan in cell walls of plant pathogenic fungi. New Phytol 156:103–112
Fujikawa T, Kuga Y, Yano S, Yoshimi A, Tachiki T, Abe K, Nishimura M (2009) Dynamics of cell wall components of Magnaporthe grisea during infectious structure development. Mol Microbiol 73:553–570
Harata K, Muraki M (2000) Crystal structures of Urtica dioica agglutinin and its complex with tri-N-acetylchitotriose. J Mol Biol 297:673–681
Hekmat O, Tokuyasu K, Withers SG (2003) Subsite structure of the endo-type chitin deacetylase from a deuteromycete, Colletotrichum lindemuthianum: an investigation using steady-state kinetic analysis and MS. Biochem J 374:369–380
Howard RJ, Bourett TM, Ferrari MA (1991) Infection by Magnaporthe: an in vitro analysis. In: Mendgen K, Lesemann DE (eds) Electron microscopy of plant pathogens. Springer, Berlin, pp 251–264
Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyama C, Dohmae N, Takio K, Minami E, Shibuya N (2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci USA 103:11086–11091
Kankanala P, Czymmek K, Valent B (2007) Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19:706–724
Kishimoto K, Kouzai Y, Kaku H, Shibuya N, Minami E, Nishizawa Y (2010) Perception of the chitin oligosaccharides contributes to disease resistance to blast fungus Magnaporthe oryzae in rice. Plant J 64:343–354
Li L, Ding SL, Sharon A, Orbach M, Xu JR (2007) Mirl is highly upregulated and localized to nuclei during infectious hyphal growth in the rice blast fungus. Mol Plant Microbe Interact 20:448–458
Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA 104:19613–19618
Nishizawa Y, Nishio Z, Nakazono K, Soma M, Nakajima E, Ugaki M, Hibi T (1999) Enhanced resistance to blast (Magnaporthe grisea) in transgenic rice by constitutive expression of rice chitinase. Theor Appl Genet 99:383–390
Ramonell K, Berrocal-Lobo M, Koh S, Wan J, Edwards H, Stacey G, Somerville S (2005) Loss-of-function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum. Plant Physiol 138:1027–1036
Rappleye CA, Eissenberg LG, Goldman WE (2007) Histoplasma capsulatum α-(1,3)-glucan blocks innate immune recognition by the β-glucan receptor. Proc Natl Acad Sci USA 104:1366–1370
Saitoh K, Togashi K, Arie T, Teraoka T (2006) A simple method for a mini-preparation of fungal DNA. J Gen Plant Pathol 72:348–350
Saitoh K, Nishimura M, Kubo Y, Hayashi N, Minami E, Nishizawa Y (2008) Construction of a binary vector for knockout and expression analysis of rice blast fungus genes. Biosci Biotechnol Biochem 72:1380–1383
Shibuya N, Minami E (2001) Oligosaccharide signalling for defense responses in plant. Physiol Mol Plant Pathol 59:223–233
Tanabe S, Okada M, Jikumaru Y, Yamane H, Kaku H, Shibuya N, Minami E (2006) Induction of resistance against rice blast fungus in rice plants treated with a potent elicitor, N-acetylchitooligosaccharide. Biosci Biotechnol Biochem 70:1599–1605
van den Burg HA, Westerink N, Francoijs KJ, Roth R, Woestenenk E, Boeren S, de Wit PJGM, Joosten MHAJ, Vervoort J (2003) Natural disulfide bond-disrupted mutants of AVR4 of the tomato pathogen Cladosporium fulvum are sensitive to proteolysis, circumvent Cf-4-mediated resistance, but retain their chitin binding ability. J Biol Chem 278:27340–27346
van den Burg HA, Harrison SJ, Joosten MHAJ, Vervoort J, de Wit PJGM (2006) Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol Plant Microbe Interact 19:1420–1430
Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G (2008) A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20:471–481
Acknowledgments
We are grateful to Drs. Nagao Hayashi and Shigeru Tanabe at the National Institute of Agrobiological Sciences, Tsukuba, Japan; Dr. Hayashi provided strains of M. oryzae (MAFF101511 and MAFF10517) and Dr. Tanabe provided technical advice regarding inoculation. We are also grateful to Dr. Hiroyuki Satoh at the National Institute of Crop Science, Tsukuba, Japan, for providing rice seeds of Nipponbare BL no. 2 and BL no. 5. We also thank Kyoko Iwasaki, Emi Nakajima, and Hiroko Kurano for daily assistance. This research was supported by the program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN). The experiments performed comply with the current laws of Japan.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
S. Mochizuki and K. Saitoh contributed equally to this work.
The nucleotide sequence data reported are available in the DDBJ/EMBL/GenBank databases under the accession numbers AB513115–AB513128.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Mochizuki, S., Saitoh, Ki., Minami, E. et al. Localization of probe-accessible chitin and characterization of genes encoding chitin-binding domains during rice–Magnaporthe oryzae interactions. J Gen Plant Pathol 77, 163–173 (2011). https://doi.org/10.1007/s10327-011-0310-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10327-011-0310-5