Abstract
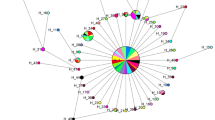
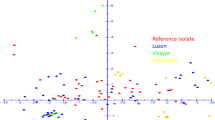
Population structure of Eleusine isolates of Pyricularia oryzae (Magnaporthe oryzae) was examined using DNA markers. On the basis of rDNA sequences, Eleusine isolates were divided into two groups. One group clustered with Triticum isolates, while the other clustered with Eragrostis isolates. This grouping was supported by DNA fingerprinting with three repetitive elements: MGR586, MGR583, and grasshopper. These results suggest that the population of Eleusine isolates is composed of at least two groups that evolved independently from the original population of P. oryzae. Most of the isolates that were collected just after an outbreak of finger millet blast in the 1970s had almost identical fingerprint profiles although they were collected in distant prefectures. This result supports the idea that the outbreak was caused by seed transmission of a particular strain of Eleusine isolates.




Similar content being viewed by others
References
Couch BC, Kohn LM (2002) A multilocus gene genealogy concordant with host preference indicates segregation of a new species, Magnaporthe oryzae, from M. grisea. Mycologia 94:683–693
Dobinson KF, Harris RE, Hamer JE (1993) Grasshopper, a long terminal repeat (LTR) retroelement in the phytopathogenic fungus Magnaporthe grisea. Mol Plant Microbe Interact 6:114–126
Eto Y, Ikeda K, Chuma I, Kataoka T, Kuroda S, Kikuchi N, Don LD, Kusaba M, Nakayashiki H, Tosa Y, Mayama S (2001) Comparative analyses of the distribution of various transposable elements in Pyricularia and their activity during and after the sexual cycle. Mol Gen Genet 264:565–577
Farman ML, Taura S, Leong SA (1996) The Magnaporthe grisea DNA fingerprinting probe MGR586 contains the 3′ end of an inverted repeat transposon. Mol Gen Genet 251:675–681
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Felsenstein J (2005) PHYLIP: phylogenetic inference package. Version 3.65. Department of Genome Sciences, University of Washington, Seattle
Hamer JE, Farrall L, Orbach MJ, Valent B, Chumley FG (1989) Host species-specific conservation of a family of repeated DNA sequences in the genome of a fungal plant pathogen. Proc Natl Acad Sci USA 86:9981–9985
Hayashi N, Kato H (1988) Viability and aggressiveness of Pyricularia cultures preserved by silica gel-drying grain method (in Japanese with English summary). Proc Kanto-Tosan Plant Prot Soc 35:12–13
Hirata K, Kusaba M, Chuma I, Osue J, Nakayashiki H, Mayama S, Tosa Y (2007) Speciation in Pyricularia inferred from multilocus phylogenetic analysis. Mycol Res 111:799–808
Holliday P (1989) A dictionary of plant pathology. Cambridge University Press, Cambridge
Kachroo P, Ahuja M, Leong SA, Chattoo BB (1997) Organization and molecular analysis of repeated DNA sequences in the rice blast fungus Magnaporthe grisea. Curr Genet 31:361–369
Kato H (1978) Biological and genetic aspects in the perfect state of rice blast fungus, Pyricularia oryzae Cav. and its allies. Gamma Field Symp 17:1–19
Kato H, Yamaguchi T, Nishihara N (1976) The perfect state of Pyricularia oryzae Cav. in culture. Ann Phytopathol Soc Jpn 42:507–510
Kato H, Yamaguchi T, Nishihara N (1977) Seed transmission, pathogenicity and control of ragi blast fungus and susceptibility of ragi to Pyricularia spp. from grasses, cereals and mioga (in Japanese with English summary). Ann Phytopathol Soc Jpn 43:392–401
Kato H, Yamamoto M, Yamaguchi-Ozaki T, Kadouchi H, Iwamoto Y, Nakayashiki H, Tosa Y, Mayama S, Mori N (2000) Pathogenicity, mating ability and DNA restriction fragment length polymorphisms of Pyricularia populations isolated from Gramineae, Bambusideae and Zingiberaceae plants. J Gen Plant Pathol 66:30–47
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Nakayashiki H, Kiyotomi K, Tosa Y, Mayama S (1999) Transposition of the retrotransposon MAGGY in heterologous species of filamentous fungi. Genetics 153:693–703
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Swofford DL (2002) PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4.0. Sinauer, Sunderland
Taga M, Nakagawa H, Tsuda M, Ueyama A (1978) Ascospore analysis of kasugamycin resistance in the perfect stage of Pyricularia oryzae. Phytopathology 68:815–817
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Ueyama A, Tsuda M (1975) Formation of the perfect state in culture of Pyricularia sp. from some graminaceous plants (preliminary report). Trans Mycol Soc Jpn 16:420–422
Urashima AS, Hashimoto Y, Don LD, Kusaba M, Tosa Y, Nakayashiki H, Mayama S (1999) Molecular analysis of the wheat blast population in Brazil with a homolog of retrotransposon MGR583. Ann Phytopathol Soc Jpn 65:429–436
Valent B (1990) Rice blast as a model system for plant pathology. Phytopathology 80:33–36
Valent B, Chumley FG (1991) Molecular genetic analysis of the rice blast fungus, Magnaporthe grisea. Annu Rev Phytopathol 29:443–467
Valent B, Crawford MS, Weaver CG, Chumley FG (1986) Genetic studies of fertility and pathogenicity in Magnaporthe grisea (Pyricularia oryzae). Iowa State J Res 60:569–594
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322
Yaegashi H (1978) Inheritance of pathogenicity in crosses of Pyricularia isolates from weeping lovegrass and finger millet. Ann Phytopathol Soc Jpn 44:626–632
Yaegashi H, Nishihara N (1976) Production of the perfect stage in Pyricularia from cereals and grasses. Ann Phytopathol Soc Jpn 42:511–515
Acknowledgments
We thank Dr. N. Hayashi, National Institute of Agrobiological Sciences, Japan, for providing several isolates, especially the isolate from weeping lovegrass. Special thanks are due to Dr. H. Kato, a former professor at Kobe University, for providing most of the test isolates, the information on these isolates, and valuable suggestions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tanaka, M., Nakayashiki, H. & Tosa, Y. Population structure of Eleusine isolates of Pyricularia oryzae and its evolutionary implications. J Gen Plant Pathol 75, 173–180 (2009). https://doi.org/10.1007/s10327-009-0158-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10327-009-0158-0