Abstract
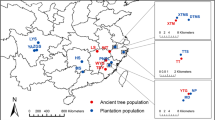
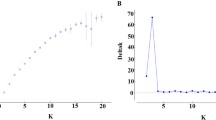
In order to improve the accessibility of genetic resources, a core collection needs to be constructed and appropriately evaluated. For Cryptomeria japonica, the most important forestry species in Japan, we constructed a core collection derived from 3,203 plus trees. First, genetic redundancy was removed based on SSR genotypes from the plus tree population. One diploid accession from each of the 539 groups that were identified using information on the original growing location was then selected considering the conservation conditions. We defined this population composed of 539 individuals as a core collection for C. japonica, and then evaluated the collection status of this core collection from multiple viewpoints. This core collection proved to contain on average one entry selected per 10 km radius (314 km2) from 10 × 10 km grid squares with an area of more than 40 % covered with C. japonica, and covered more than 85 % retention of all 12 environmental factors affecting plus trees, while maintaining almost the same genetic diversity as the complete set of plus trees. These methods of evaluation from multiple viewpoints, using a geographic information system and genetic markers, were efficient in determining whether the collection of resources was complete, and, if not, where additional efforts for balancing a deficit should be devoted.





Similar content being viewed by others
References
Balfourier F, Roussel V, Strelchenko P, Exbrayat-Vinson F, Sourdille P, Boutet G, Koenig J, Ravel C, Mitrofanova O, Beckert M, Charmet G (2007) A worldwide bread wheat core collection arrayed in a 384-well plate. Theor Appl Genet 114:1265–1275
Bhattacharjee R, Khairwal IS, Bramel PJ, Reddy KN (2007) Establishment of a pearl millet (Pennisetum glaucum (L.) R. Br.) core collection based on geographical distribution and quantitative traits. Euphytica 155:35–45
Brown AHD (1989) The case for core collections. In: Brown AHD, Frankel OH, Marshall DR, Williams JT (eds) The use of plant genetic resources. Cambridge University Press, Cambridge, pp 136–156
Brown AHD, Briggs JD (1991) Sampling strategies for genetic variation in ex situ collections of endangered plant species. In: Falk D, Holsinger KE (eds) Genetics and conservation of rare plants. Oxford University Press, New York
Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS (1991) ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res 19:4008
Dwivedi SL, Puppala N, Upadhyaya HD, Manivannan N, Singh S (2008) Developing a core collection of peanut specific to Valencia market type. Crop Sci 48:625–632
Ebana K, Kojima Y, Fukuoka S, Nagamine T, Kawase M (2008) Development of mini core collection of Japanese rice landrace. Breed Sci 58:281–291
El Bakkali A, Haouane H, Moukhli A, Costes E, Van Damme P, Khadari B (2013) Construction of core collections suitable for association mapping to optimize use of Mediterranean olive (Olea europaea L.) genetic resources. PLoS ONE 8:e61265
Environmental Agency (1999) Report of the vegetation survey for the 5th national survey on the natural environment. Asia Air Survey, Tokyo (in Japanese)
Escribano P, Viruelm A, Hormaza J (2008) Comparison of different methods to construct a core germplasm collection in woody perennial species with simple sequence repeat markers. A case study in cherimoya. Ann Appl Biol 153:25–32
Forest and Forestry Products Research Institute, Forest Tree Breeding Center (2013) Execution condition and statistics of forest tree breeding, in 2012 (in Japanese)
Forestry Agency (2007) Statistical Data 2007 (in Japanese)
Forestry Agency (2010) Annual report on forest and forestry in Japan Fiscal year 2010 (in Japanese)
Forestry Agency Guidance Department (1994) Notices related to forest tree breeding business (in Japanese)
Frankel OH (1984) Genetic perspectives of germplasm conservation. In: Arber W, Llimensee K, Peacock WJ, Starlinger P (eds) Genetic manipulation: impact on man and society. Cambridge University Press, Cambridge, pp 161–170
Gepts P (2006) Plant genetic resources conservation and utilization: the accomplishments and future of a societal insurance policy. Crop Sci 46:2278–2292
González-Martínez SC, Wheeler NC, Ersoz E, Nelson CD, Neale DB (2007) Association genetics in Pinus taeda L. I. wood property traits. Genetics 175:399–409
Hayashi Y. (1960) Taxonomical and phytogeographical study of Japanese conifers Nohrin shuppan, Tokyo (in Japanese)
Hintum van TJL, Brown AHD, Spillane C, Hodgkin T (2000) Core collections of plant genetic resources. IPGRI Technical Bullletin No. 3. International Plant Genetic Resources Institute, Rome
Japan Forest Tree Breeding Association (2004) Project of forest tree breeding. Way of breeding for half a century, and use of improved tree. Japan Forest Tree Breeding Association, Tokyo (in Japanese)
Kaga A, Shimizu T, Watanabe S, Tsubokura Y, Katayose Y, Harada K, Vaughan DA, Tomooka N (2012) Evaluation of soybean germplasm conserved in NIAS genebank and development of mini core collections. Breed Sci 61:566–592
Kang CW, Kim SY, Lee SW, Mathur PN, Hodgkin T, Zhou MD, Lee RJ (2006) Selection of a core collection of Korean sesame germplasm by a stepwise clustering method. Breed Sci 56:85–91
Kira T (1977) A climatological interpretation of Japanese vegetation zones. In: Miyawaki A, Tuexen R (eds) Vegetation science and environmental protection. Maruzen, Tokyo, pp 21–30
Kojima Y, Ebana K, Fukuoka S, Nagamine T, Kawase M (2005) Development of an RFLP-based rice diversity research set of germplasm. Breed Sci 55:431–440
Mahalakshmi V, Ng Q, Atalobhor J, Ogunsola D, Lawson M, Ortiz R (2007) Development of a West African yam Dioscorea spp. core collection. Genet Resour Crop Evol 54:1817–1825
McKeand S, Kurinobu S (1998) Japanese tree improvement and forest genetics. J For 96:12–17
Meteorological Agency (1996) Climate normals for Japan. Tokyo (in Japanese)
Ministry of the Environment, Nature Conservation Bureau (2009) Collection and conservation manual of the seeds for endangered plants (in Japanese)
Mitchell A (2005) The ESRI guide to GIS analysis. Spatial measurements and statistics vol 2. ESRI, Redlands
Miyamoto N, Ono M, Watanabe A, Takahashi M (2008) Conversion of the address information into the numerical form in passport database: grading based on the reliability and character strings data into numerical data. For Forestry Prod Res Inst, For Tree Breed Cent, Annu Rep 2008:76–78 (in Japanese)
Moriguchi Y, Iwata H, Ujino-Ihara T, Yoshimura K, Taira H, Tsumura Y (2003) Development and characterization of microsatellite markers for Cryptomeria japonica D. Don. Theor Appl Genet 106:751–758
Mousadik EA, Petit RJ (1996) High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L) Skeels] endemic to Morocco. Theor Appl Genet 92:832–839
Nijensohn SE, Schaberg PG, Hawley GJ, DeHayes DH (2005) Genetic subpopulation structuring and its implications in a mature eastern white pine. Can J For Res 35:1041–1052
Ohba K, Yamate H, Rissen T, Kurinobu S, Toda T, Nishimura K (1978) Follow-up survey for plus trees of Cryptomeria japonica and Chamaechyparis obtusa. Kyushu J For Res 31:67–68 (in Japanese)
Oliveira MF, Nelson RL, Geraldi IO, Cruz CD, de Toledo JFF (2010) Establishing a soybean germplasm core collection. Field Crops Res 119:277–289
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pearson RG (2007) Species’ distribution modeling for conservation educators and practitioners. Synthesis. American Museum of Natural History. http://ncep.amnh.org
R Core Team (2013) R: a language and environment for statistical computing. R foundation for statistical computing, Vienna http://www.R-project.org
Rao VR, Hodgkin T (2002) Genetic diversity and conservation and utilization of plant genetic resources. Plant Cell Tiss Org 68:1–19
Reddy LJ, Upadhyaya HD, Gowda CLL, Singh S (2005) Development of core collection in pigeon pea [Cajanus cajan (L.) Millspaugh] using geographic and qualitative morphological descriptors. Genet Resour Crop Evol 52:1049–1056
Rioux PS (2013) http://cran.r-project.org/web/packages/PopGenKit/PopGenKit.pdf
Rodiño AP, Santalla M, DeRon AM, Singh SP (2003) A core collection of common bean from the Iberian peninsula. Euphytica 131:165–175
Santesteban LG, Miranda C, Royo JB (2009) Assessment of the genetic and phenotypic diversity maintained in apple core collections constructed by using either agro-morphologic or molecular marker data. Span J Agric Res 7:572–584
Shehzad T, Okuizumi H, Kawase M, Okuno K (2009) Development of SSR-based sorghum (Sorghum bicolor (L.) Moench) diversity research set of germplasm and its evaluation by morphological traits. Genet Resour Crop Evol 56:809–827
Shiraishi S, Watanabe A (1995) Identification of chloroplast genome between Pinus densiflora Sieb. et Zucc and P. thunbergii Parl. based on the polymorphism in rbcL gene. J Jpn For Soc 77:429–436 (in Japanese with English summary)
Soto-Cerda BJ, Diederichsen A, Ragupathy R, Cloutier S (2013) Genetic characterization of a core collection of flax (Linum usitatissimum L.) suitable for association mapping studies and evidence of divergent selection between fiber and linseed types. BMC Plant Biol 13:78
Suzuki T, Suzuki K (1971) Japan sea index and Setouchi index. Jpn J Ecol 20:252–255 (in Japanese)
Tani N, Takahashi T, Ujino-Ihara T, Iwata H, Yoshimura K, Tsumura Y (2004) Development and characteristics of microsatellite markers for sugi (Cryptomeria japonica D. Don) derived from microsatellite-enriched libraries. Ann For Sci 61:569–575
Thumma BR, Nolan MF, Evans R, Moran GF (2005) Polymorphisms in cinnamoyl CoA reductase (CCR) are associated with variation in microfibril angle in Eucalyptus spp. Genetics 171:1257–1265
Tomaru N (1994) Studies on genetic variation in plantations and plus-trees of Cryptomeria japonica using isozyme markers. Memoirs of the Institute of Agriculture and Forestry, University of Tsukuba, Tsukuba 6:1–66
Tsumura Y (2006) Description for revegetation guideline study—relation of geographical genetic variation and morphological variation for plants. In: Kobayashi T, Kuramoto N (eds) Handbook of revegetation for biodiversity conservation. New approach of planning and technique for ecologically sustainable landscape. Chijinshokan, Tokyo, pp 59–73
Uchiyama K, Miyamoto N, Takahashi M, Watanabe A, Tsumura Y (2014) Population genetic structure and the effect of historical human activity on the genetic variability of Cryptomeria japonica core collection, in Japan. Tree Genet Genomes (accepted)
Upadhyaya HD (2003) Development of a groundnut core collection using taxonomical, geographical and morphological descriptors. Genet Resour Crop Evol 50:139–148
Volk GM, Richards CM, Reilley AA, Henk AD, Forsline PL, Aldwinkle HS (2005) Ex situ conservation of vegetatively propagated species: development of a seed-based core collection for Malus sieversii. J Am Soc Hortic Sci 130:203–210
Wright S (1965) The interpretation of population structure by F-Statistics with special regard to systems of mating. Evolution 19:395–420
Xiurong Z, Yingzhong Z, Yong C, Xiangyun F, Qingyuan G, Mingde Z, Hodgkin T (2000) Establishment of sesame germplasm core collection in China. Genet Resour Crop Evol 47:273–279
Yan W, Rutger JN, Bryant RJ, Bockelman HE, Fjellstrom RG, Chen MH, Tai TH, McClung AM (2007) Development and evaluation of a core subset of the USDA rice germplasm collection. Crop Sci 47:869–878
Zenkoku Ringyo Kairyo Fukyu Kyokai (1969) All about Cryptomeria japonica, Tokyo (in Japanese)
Zenkoku Ringyo Kairyo Fukyu Kyokai (1983) All about Cryptomeria japonica, new edn. Tokyo (in Japanese)
Acknowledgments
We thank the Tohoku, Kansai, and Kyusyu Regional Breeding Offices for providing genetic resources. We are also grateful to K. Tanaka, H. Hatakeyama, T. Kaminaga, and M. Shibata for their helpful assistance in DNA analysis. We would like to thank the anonymous reviewers for their valuable comments and suggestions to improve the quality of the paper. Financial support was provided by the Forest Tree Breeding Center.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
About this article
Cite this article
Miyamoto, N., Ono, M. & Watanabe, A. Construction of a core collection and evaluation of genetic resources for Cryptomeria japonica (Japanese cedar). J For Res 20, 186–196 (2015). https://doi.org/10.1007/s10310-014-0460-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10310-014-0460-3