Abstract
Epilepsy surgery is a well-established method of treatment for pharmacoresistant focal epilepsies, but it carries an inherent risk of damaging eloquent brain structures. This holds true in particular for visual system pathways, where the damage to, for example, the optic radiation may result in postoperative visual field defects. Such risk can be minimized by the identification and localization of visual pathways using diffusion magnetic resonance imaging (dMRI). The aim of this article is to provide an overview of the step-by-step process of reconstructing the visual pathways applying dMRI analysis. This includes data acquisition, preprocessing, identification of key structures of the visual system necessary for reconstruction, as well as diffusion modeling and the ultimate reconstruction of neural pathways. As a result, the reader will become familiar both with the ideas and challenges of imaging the visual system using dMRI and their relevance for planning the intervention.
Zusammenfassung
Epilepsiechirurgische Interventionen sind etablierte Ansätze zur Behandlung pharmakoresistenter fokaler Epilepsien, bergen jedoch das Risiko der Schädigung eloquenter Hirnstrukturen. In Bezug auf die Sehbahn kann dies, beispielsweise bei Läsionen im Bereich der Sehstrahlung, zu postoperativen Gesichtsfelddefekten führen. Die Identifikation und Lokalisierung der Sehbahnstrukturen mittels Diffusionsbildgebung (diffusionsgewichtete Magnetresonanztomographie, dMRT) ermöglicht es, diese Risiken einzuschätzen und zu reduzieren. Der vorliegende Artikel vermittelt eine Schritt-für-Schritt-Darstellung der Prozesse, die der dMRT-basierten Sehbahnrekonstruktionen zugrunde liegen. Dies umfasst die Datenakquise, Vorverarbeitung, Identifikation essenzieller Schlüsselstrukturen sowie die Diffusionsmodellierung und abschließend die Rekonstruktion der Sehbahn. So wird dem Leser ein Verständnis sowohl der Möglichkeiten und Herausforderungen der Bildgebung des Sehsystems mittels dMRT als auch ihrer Relevanz für die Interventionsplanung vermittelt.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
While epilepsy is a complex and not thoroughly understood phenomenon of brain malfunction, the key to its understanding is in some cases actually hidden in the structure of the brain. This applies, for instance, to acquired causes of epilepsy, such as serious brain trauma, strokes, tumors, and other lesions [5], as well as to observed interactions between structure and function [24]. The comprehensive and accurate description of brain anatomy and connectivity is even more important for surgical interventions, particularly those with a high risk of causing damage to the integrity of the visual system (e.g., anterior temporal lobe resections and amygdalohippocampectomy, see Fig. 1b). These examples demonstrate that research on and treatment of epilepsy greatly benefits from the integration of brain imaging techniques, such as diffusion magnetic resonance imaging [23]. Diffusion MRI is a noninvasive imaging technique capable of capturing microstructural tissue properties and of mapping the fiber architecture, e.g., trajectories of neural pathways, which has been demonstrated to significantly contribute to the field of epilepsy. Its capability of capturing the microstructural properties has made it possible to link the structural compromise of fibers tracts in temporal lobe epilepsy (TLE) with memory and language impairments [24] and, as demonstrated in some cases of focal cortical dysplasias, to reveal thinning of white matter fibers and reduced connectivity between subcortical gray matter and the dysplastic cortex [10]. The latter study employed also the second feature of dMRI, i.e., its capability of reconstruction of trajectories of neural pathways. This process, referred to as “tractography” or “fiber or streamline tracking” [26], uses estimates of local fiber orientations (calculated from dMRI data) to produce a tractogram, i.e., the reconstruction of the given pathways in the brain.
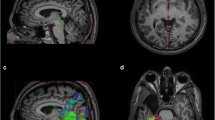
Relevance of visual system imaging in clinical routines. a Presurgical planning for LiTT on T1-weighted image in pseudo-sagittal view. Stereotactic trajectory (red line) and the aimed for lesion zone depicted by a brachytherapy planning tool (arbitrarily chosen: red contour resembles 50 Gy Isodose as desired limit of the irreversible damage zone, green contour resembles 100 Gy Isodose as core heat zone which needs to be monitored for peak temperatures). Neither trajectory nor lesion zone interfere with the visual tract (as defined by deterministic tractography; white area above trajectory), thus minimizing the risk of visual field loss. b Periventricular heterotopia (pink contour) adjacent to superimposed optic radiation (as defined by deterministic tractography; white streak adjacent to the pink contour) on T1-weighted image with gadolinium contrast. (Image courtesy of Dept. for Stereotactic Neurosurgery/Otto von Guericke University Magdeburg)
The inherent risk of surgical interventions for visual function is well known in standard procedures, such as anterior temporal lobectomy and amygdalohippocampectomy. Although mostly nondisabling, a significant numbers of visual field defects were reported (respectively 78% and 73%; [14]), as for temporal lobe resections, where postoperative upper quadrant visual field deficits were reported for 29 of 38 patients [36]. Between the aforementioned resective approaches, there is no significant difference concerning the risk for visual field deficit [14]. The indicated need for more tailored surgical approaches can be addressed by incorporating tractography in the routine [32, 45], which is known to predict postoperative visual field deficits based on pre- and postoperative tractography [8, 20]. Apart from resective interventions, dMRI can also be integrated for the prevention of visual function deficits in minimally invasive approaches, such as laser interstitial thermal therapy (LiTT; also known as “MRI-guided laser ablation” or “stereotactic laser-thermoablation”; [7]), e.g., for amygdalohippocampectomies in mesial temporal lobe epilepsies ([11, 31]; see Fig. 1a) or parieto-occipital lesionectomies typically performed for periventricular heterotopias ([44]; see Fig. 1b). At least for mesial temporal lobe epilepsies, more definitive information on risk can be awaited by larger prospective studies in the near future. However, initial results from consecutive case series suggest that the surgical risk for a visual field deficit after LiTT is less than that with the aforementioned resective procedures, particularly if an initial technical learning curve is acknowledged [17].
One risk-posing structure for all temporal surgical approaches is the individual anatomy of Meyer’s loop. Nilsson et al. examined the intra- and interindividual variability of Meyer’s loop in seven healthy volunteers and two patients undergoing anterior temporal lobe resection by measuring the distance between the anterior edge of the loop and the temporal lobe: the distance varied considerably between 34 and 51 mm (mean 44 mm; [27]) indicating the need for an individual approach during the presurgical planning (see section “Primary visual cortex”).
Tractography allows for the visualization and assessment of the connectivity between brain regions [10] and thus can also provide vital information for surgical intervention planning and risk assessment in epilepsy. In the following, the different concepts of imaging the visual system using dMRI are introduced.
Methods
In order to generate the tractogram of the visual system (Fig. 2), it is necessary to acquire data, select the analysis tools, preprocess the diffusion-weighted (DW) data, segment the structures of the visual system defining starting and ending points of reconstructed streamlines, fit the model to DW images, and perform the tractography. While each of these steps will be discussed separately, it should be noted that they should not be perceived as separate entities, but rather as interconnected and dependent.
Tractography of the visual pathways. a A 3D graphic in sagittal view displaying orientation of right optic tract and optic radiation streamlines. b T1-weighted images including optic tract (left) and optic tract streamlines (right) in false color (color code describes streamline orientation: green anterior–posterior; red left–right; blue inferior–superior). Streamlines are not cropped to the shown slices. (Image courtesy of Otto von Guericke University Magdeburg)
Overview of required MRI data
The optimal MRI dataset acquired for the purpose of tracking of the visual system should comprise (a) T1-weighted (Fig. 3a), and optionally a proton density (PD) map, to allow for the comprehensive delineation of brain tissue and structures, and (b) DW images necessary for the tractography (Fig. 3b). In the dMRI analysis, the T1-weighted images and PD maps are used in order to identify and segment regions of interest (ROIs) and white matter mask, which enhances tractography. The DW images provide information about microstructural properties (Fig. 3c) and what is critical for tractography, spatial organization of neural fibers (additionally, tractography may also provide its own contrast as shown in Figure 3d). The basic underlying concept is the measurement of dephasing of excited protons along the given direction simultaneously in all of the brain voxels [37]. The contrast in a DW image depends on the properties of diffusion processes occurring in the tissue and is controlled by two parameters set for MR scanning: b‑value (determines the sensitivity to diffusion; b0 is the lowest, where b‑value equal to, e.g., 3000 s/mm2 is considered high) and b‑vector (describes the direction along which the diffusion is measured). The selection of DW acquisition parameters is a complex issue, as one needs to consider MRI scanner capabilities, the focus of study (imaging of the whole brain or a single structure), data quality requirements (signal-to-noise ratio, spatial resolution, number of b‑values, and gradient directions) and external factors, e.g., scanning time (detailed in: [22, 40]). An efficient alternative solution is the adaptation of already validated and well-established protocols such as the Human Connectome Project (HCP) acquisition protocols [42]. Publicly available datasets should also be considered an option to tune the analysis procedures (see Table 1).
T1-weighted and dMRI-derived sample images. a T1-weighted images. b Single post-processed diffusion-weighted volume acquired with b‑value = 1600 s/mm2. c Map of fractional anisotropy (FA) describing anisotropy of local diffusion. d Track-density imaging (TDI), tractography-derived contrast obtained by mapping density of streamlines. (Image courtesy of Otto von Guericke University Magdeburg)
Processing
Apart from the data acquisition parameters, it is important to consider the choice of tools for data preprocessing and tractography (see Table 2). Diffusion-weighted data preprocessing is an important stage, as it determines the data quality and may affect the final outcome of the study. While the applicability and/or necessity of constituent preprocessing steps depends on parameters of the data and specifics of the study, the preprocessing pipelines applied in the HCP project [16] are a commonly recognized standard. Table 3 provides a list of recommended preprocessing steps, created by extending the HCP preprocessing pipeline by more recent utilities, which result in a corrected DW dataset aligned to the T1-weighted image (Fig. 4). A recently emerging alternative to data preprocessing and analysis on local machines are online platforms, such as BrainLife [1]. BrainLife allows one to upload one’s own data, or to use existing publicly available datasets (which are either stored directly on BrainLife or can be downloaded from other repositories) and perform neuroimaging analysis in the cloud using online services.
Exemplary distortions affecting the quality of the diffusion-weighted (DW) b0 images. a Example of motion-induced distortions. First (left) and last (middle) b0 image in the DW series. Calculation of voxel-wise difference between volumes reveals displacement introduced by motion. b Example of Echo-Planar Imaging (EPI) distortions. Discontinuity of the medium causes the inhomogeneity of the magnetic field, which warps the image. These distortions are particularly pronounced in the region of the optic chiasm (white arrows) and near the ear canals, and depending on the phase-encoding direction (PED) are manifested either by squeezing (left) or stretching (middle). Images acquired with opposite PEDs can be combined in order to unwarp the distortion (right). c Alignment of the T1-weighted (T1w; middle) and DW image (left) ensures the correspondence between voxels (right). (Image courtesy of Otto von Guericke University Magdeburg)
Segmentation of visual system structures
While the previous sections can be generalized for any study involving dMRI data, the selection of regions of interest (ROIs) is determined individually for each study. In the case of tractography of the visual system, it is required to delineate at least the optic chiasm, lateral geniculate nucleus (LGN), and the primary visual cortex (V1), as depicted in Fig. 5. The idea here is to localize the structures being connected by visual pathways and to perform the tractography in order to reconstruct these connections. It should be noted that while tractography directly between the optic chiasm and V1 is theoretically possible, in practice it is extremely challenging due to its length and complexity. Therefore, it is recommended to incorporate the LGN in the tractography.
Segmentation of visual system structures. a A sagittal view of the 3D graphic displaying the optic chiasm (orange), left and right lateral geniculate nucleus (LGN; purple) and primary visual cortex (dark gray) and their location in the T1-weighted image. b Tilted axial slices displaying comparison of the optic chiasm masks obtained via FreeSurfer software (top) and manual segmentation (bottom). c Axial slices comparing LGN masks from FreeSurfer v6.0 (purple) or FreeSurfer v7.1 (cyan; overlap in blue). d Correspondence of the functional magnetic resonance imaging (fMRI)-derived retinotopic maps, their estimates derived from anatomy using Benson’s atlas and the volumetric representation of the latter. Left and right columns depict eccentricity and polar angle maps, respectively. Top row displays surface representation of retinotopic maps of the right hemisphere derived from fMRI data using population Receptive Field (pRF) models. Middle row displays analogical representation derived directly from T1-weighted image using Benson’s atlas. The bottom row displays Benson’s prediction in a volumetric representation. It should be noted that the representation is limited to the central visual field (14° radius). (Image courtesy of Otto von Guericke University Magdeburg)
In terms of segmentation there are two main approaches—manual, where the structures are identified and marked by hand by a trained user, or automated, which is performed by designated software. For the latter, the FreeSurfer segmentation software (Table 2) is widely used and functions as a semi-standard of automated segmentation. It should also be noted that recent rapid developments in deep learning (DL) methods, although not yet widely established, resulted in new emerging tools that are of great promise.
Optic nerves
If required, the currently recommended strategy would be a manual delineation based on T1-weighted images. This may change with emerging DL-based methods, which demonstrate good performance [25], but are not yet widely available.
Optic chiasm
A FreeSurfer software, regarded as a standard tool in automated brain segmentation, is capable of segmenting the optic chiasm, yet its results can be suboptimal (Fig. 5b, top). As such it is recommended to segment the optic chiasm manually in order to achieve optimal accuracy (Fig. 5b, bottom)—an approach widely used in optic chiasm imaging [30]. Similar to optic nerves, current DL developments allow for the segmentation of the optic chiasm, but they are still not widely tested and available—especially in the case of optic chiasm malformations.
Lateral geniculate nucleus
The segmentation of the LGN is particularly challenging, as this structure does not stand out in T1-weighted images. While it is possible to use automated segmentation tools (e.g., FreeSurfer, SPM, FSL, GIF, MALP-EM, Mipav) to this end, this approach has limitations, e.g., LGN segmentation can even vary between versions (as shown for FreeSurfer in Fig. 5c). Furthermore, the lack of validation undermines the study’s reliability. Therefore, it is recommended to obtain PD maps during the data acquisition, as their contrast allows for an LGN identification. In the absence of this information, an alternative strategy is the tractography-based identification of the LGN [28]. We propose that this strategy would be even further improved by simultaneous tractography from both the optic chiasm and V1 with the target in the thalamus. The intersection of the streamline endpoints might serve to identify the LGN, which can be combined with the previously introduced FreeSurfer segmentation.
Primary visual cortex
For the identification of the primary visual cortex (V1), three approaches are available: (1) the use of automated segmentation tools, (2) individualized retinotopic mapping, and (3) the estimation of retinotopic maps from anatomical priors. (1) Automated segmentation tools in principle allow for an estimation of V1; however, individual differences from the general template introduce errors in the results of the V1 ROI definition. Further, this approach does not provide an estimate of the retinotopy of V1. (2) The alternative is to obtain retinotopic maps of the visual cortex specifically for the respective individuals with dedicated functional magnetic resonance imaging (fMRI) measurements [32]. Due to the retinotopic organization of the visual cortex, visual areas can be identified via fMRI-based retinotopic mapping (reviewed in [18, 19, 43]). T2*-weighted BOLD gradient-EPI sequences are acquired in fMRI scans during visual stimulation, typically with a contrast-inverting or moving checkerboard pattern section that travels through the visual field in a systematic manner to create a specific spatiotemporal response pattern on the visual cortex. Conventional phase-encoded retinotopic mapping [33] or the neuro-computationally more demanding population receptive field mapping [13] can be applied to obtain cortical maps of the eccentricity and polar angle representations of the visual field. These are typically visualized on the computationally inflated or flattened surface of the visual cortex as derived from high-resolution T1 images. Visual area boundaries can be delineated from these maps [33] as depicted in Fig. 5d (top row) for the primary visual cortex (V1). (3) Based on the evidence of qualitatively consistent organization of primary (V1) and extra-striate (V2 and V3) visual area topography [12], Benson et al. [4] demonstrated the ability of an anatomical template to predict the retinotopic organization of the visual cortex with high accuracy using only a participant’s brain anatomy, i.e., the patterns of the gyral and sulcal curvatures. In the absence of retinotopic mapping data, a viable alternative is therefore the use of Benson’s atlas, which utilizes known anatomical priors in order to estimate V1 location, as well as polar angle and eccentricity map (see Fig. 5d, middle row). This information can be used to specify ROIs for the purpose of tractography.
White matter
In order to enhance the accuracy of the tractography it is recommended to use the known anatomical priors, such as the limitation of the reconstructed pathways to white matter only. Such white matter masks can be extracted using, e.g., FSL or FreeSurfer software. This approach can be further extended by taking into account other types of tissues, such as in five-tissue-type segmentation (implemented in MRtrix), which is a critical component in their proposed anatomically constrained tractography (ACT; [34]).
DW data modeling
Subsequent to DW data preprocessing, it is necessary to fit the diffusion model to the data (Fig. 6b). The available modeling approaches depend on the quality and properties of DW data, such as the number of gradient directions, number of shells etc. As the in-depth discussion of available models is beyond the scope of this paper, the following section will cover a sample of two well-established models.
Exemplary diffusion models, tractography algorithms, and tracking strategies. a The axial slice of the T1-weighted image with highlighted LGN (cyan) and V1 (red). b Two exemplary models fit to the data: constrained spherical deconvolution (CSD; left) and diffusion tensor (DT; right). c Sample streamlines connecting LGN and V1 generated using different tracking algorithms (from left to right): second-order Integration over Fiber Orientation Distrbutions (iFOD2) based on CSD model, Parallel Transport Tractography (PTT) based on CSD model, deterministic algorithm based on DT, and probabilistic algorithm based on DT. d Impact of incorporating regions of interest (ROIs) in the iFOD2-based tractography. From left to right: streamlines connecting LGN to V1, but not limited to white matter (WM); streamlines generated in LGN and limited to white matter but not requested to reach V1; streamlines connecting LGN and V1 and limited to white matter; streamlines connecting LGN and V1, limited to white matter and requested to pass through user-defined ROI (enforcing anatomical plausibility of streamlines). (Image courtesy of Otto von Guericke University Magdeburg)
Diffusion tensor
Basic, yet successful and well-established, is the DT [3] model (Fig. 6b, right), which’s popularity caused confusion of Diffusion Tensor Imaging (DTI; application of DT model) with the general dMRI term (covering all models). The DT model represents diffusion as a 3 × 3 matrix with six independent terms (i.e., from at least six volumes with unique gradient directions), which can be graphically represented as an ellipsoid. This model has limitations, as it models only one diffusion direction per voxel, and as such fails to represent more complex structures, such as the optic chiasm (due to the presence of crossing fibers), Meyer’s loop (due to its curvature), or posterior parts of optic radiation (due to fanning).
Constrained spherical deconvolution
Among other alternatives (such as Q‑ball imaging, diffusion spectrum imaging), we would like to discuss in detail CSD [39], which treats signal in each voxel as a convolution of the response from a single fiber population and distribution of the local fiber’s orientations. As such, CSD is capable of resolving multiple fiber bundles crossing a single voxel, which are described by orientation distribution functions (ODFs). The fitting of the CSD model requires more than six gradient directions (typically 30–60), which grants noise reduction and higher angular resolution at the cost of longer scanning time.
Tractography
In addition to the chosen diffusion model, the outcome tractogram depends on multiple parameters governing the tracking process (Fig. 6). These options are introduced and briefly discussed here.
Probabilistic vs. deterministic algorithms
Generally, there are two classes of tracking algorithms—deterministic and probabilistic. Deterministic algorithms assume that fibers in each voxel are oriented in only one direction as determined by the given model (see Fig. 6c)—as such they offer robust results, but fail to grasp complex architectures. Alternative probabilistic algorithms at each step of tracking sample the final direction from the distribution of all possible directions. This approach makes it possible to uncover connections missed by the deterministic algorithms (Fig. 6c) and has been proven to be superior to deterministic tracking. Consequently, modern tracking algorithms relying on advanced models, such as CSD, employ probabilistic tracking algorithms.
Algorithm types
Even within the class of probabilistic algorithms, there exists a wide range of possible choices, which impacts the outcome tractogram. The difference between two probabilistic algorithm—iFOD2 (implemented in MRtrix) and parallel transport tractography (implemented in Trekker)—is demonstrated in Fig. 6c.
Seeding region
The term “seed” refers to locations chosen as starting points for reconstructed streamlines. Global seeding describes allowance to track from any brain voxel (usually white matter voxel), while ROI seeding refers to limiting seeds to defined ROIs. Global seeding allows for the reconstruction of all possible pathways, which is important in studies on the connectivity within the whole brain, but at the same time is much more computationally demanding. It also does not guarantee that the pathways of interest will indeed be reconstructed. The ROI seeding limits the tractography only to the selected structures, which is faster, but may, at the same time, limit the application of streamline post-processing options (see next section).
Target ROIs
For the recommended probabilistic algorithms, the generated streamlines will not only be limited to the “true” pathways between seeds, but will also cover a wide range of positive, but anatomically implausible, connections (Fig. 6d). In order to limit the tractography outcome to only valid streamlines, it is advised to employ information about the destination of the streamlines, known as a “target.” A combination of information about start (seed) and end (target) ROIs greatly improves the accuracy of tractography (Fig. 6d).
Additional inclusion/exclusion ROIs
Still, the definition of seed and target ROIs does not necessarily unambiguously determine the reconstructed streamlines, especially in the case of complex neural connections, such as the optic radiation. Therefore, it is necessary to always visually inspect the tractography results. For uncertain results, it is recommended to introduce further inclusion or exclusion ROIs (based on anatomical knowledge) in order to impose other restrictions on generated streamlines and remove implausible connections (Fig. 6d). A good and widely used example of the incorporation of anatomical priors in tractography is using the white matter mask as a limitation for the tractography. This option is supported in all tracking software, as well as its extensions to tissue types other than WM (such as in ACT; [34]).
Other parameters and ensemble tractography
Apart from the aforementioned points, it is also necessary to choose the parameters governing tracking, such as the number of steps, maximal angle between consecutive steps, and possible thresholds preventing tractography from entering false regions etc. A detailed discussion of this topic requires a separate study, as the selection of fixed parameters introduces tracking bias. A solution to this problem is the repetition of tractography multiple times (using different algorithms, sets of parameters, and possibly even models) and combining all these into one outcome [38]. This will, however, naturally extend the complexity of the analysis and its duration.
Streamline post-processing— editing and filtering
Once the tractogram is generated, it can be subjected to further editing and adjustments. All approaches can generally be grouped into two classes: (a) User-informed tractograms can be merged, divided with respect to the number of streamlines or their properties (such as length), split using newly defined ROIs, transformed to different templates etc. The common denominator here is the user-made decision about the action. (b) Signal-informed tractograms are automated methods that refine the selection of streamlines using initially measured signal as a reference, a validation process referred to as filtering. As an example, linear fascicle evaluation (LiFE; [29]) calculates the predicted signal based on generated streamlines and compares it with the currently measured signal. Redundant streamlines with zero contribution to the currently measured signal are subsequently discarded. Other filtering methods include, e.g., Spherical-deconvolution Informed Filtering of Tractograms (SIFT; [35]) or Convex Optimization Modeling for Microstructure Informed Tractography (COMMIT; [9]).
Outlook
The previous sections presented the procedure involved in the tractography of the visual system and its relevance for presurgical planning. From a more general viewpoint, dMRI with its unparalleled capability of capturing the architecture and microstructural properties is a versatile tool also in research on epilepsy, or any kind of neuroscientific research involving brain anatomy. Neurosurgery is continuously seeking to be less invasive, yet attempts in this direction are often hindered by limitations in knowledge, e.g., of the function of higher-order visual cortices. This is expected to eventually change with the scientific progress regarding structure–function relationships in the visual system and thus foster its integration with clinical applications and presurgical planning. A fine example of such developments are research initiatives aiming to design visual prosthesis by microelectrode stimulation of the V1 [6]. Here, it is anticipated that dMRI will be of assistance in deciphering retinal projection fields in V1 a priori to the application of the high-resolution stimulation grids. As such, this would be a meaningful step in the development of visual prosthesis, even though it is still a long way from creating phosphenes capable of inducing meaningful visual impressions by neuronal stimulation.
Practical conclusion
-
Surgical planning and risk assessment in epilepsy benefit greatly from an individualized reconstruction of the visual pathways.
-
Integrated diffusion magnetic resonance imaging (dMRI)-based tractography allows for the individualized identification of the visual pathways, including optic tracts and optic radiation.
-
To acquire the information essential for successful tractography and to cope with imaging artifacts, dMRI requires careful consideration of data acquisition settings and preprocessing tools.
-
Tractography requires the segmentation of seed structures, i.e., optic chiasm, lateral geniculated nucleus, primary visual cortex, and white matter masks.
-
The choice of the correct dMRI data modeling framework is critical for the successful tractography-based reconstruction of the visual pathways.
References
Avesani P, McPherson B, Hayashi S et al (2019) The open diffusion data derivatives, brain data upcycling via integrated publishing of derivatives and reproducible open cloud services. Sci Data 6:69. https://doi.org/10.1038/s41597-019-0073-y
Aydogan DB, Shi Y (2019) A novel fiber-tracking algorithm using parallel transport frames
Basser PJ, Mattiello J, LeBihan D (1994) MR diffusion tensor spectroscopy and imaging. Biophys J 66:259–267
Benson NC, Butt OH, Brainard DH, Aguirre GK (2014) Correction of distortion in flattened representations of the cortical surface allows prediction of V1-V3 functional organization from anatomy. PLoS Comput Biol 10:e1003538. https://doi.org/10.1371/journal.pcbi.1003538
Berkovic SF, Mulley JC, Scheffer IE, Petrou S (2006) Human epilepsies: interaction of genetic and acquired factors. Trends Neurosci 29:391–397. https://doi.org/10.1016/j.tins.2006.05.009
Bosking WH, Beauchamp MS, Yoshor D (2017) Electrical stimulation of visual cortex: relevance for the development of visual cortical prosthetics. Annu Rev Vis Sci 3:141–166. https://doi.org/10.1146/annurev-vision-111815-114525
Büntjen L, Voges J, Heinze HJ et al (2017) Stereotaktische Laserablation – Technische Konzepte und klinische Anwendungen. Z Epileptol 30:152–161. https://doi.org/10.1007/s10309-016-0099-5
Chen X, Weigel D, Ganslandt O et al (2009) Prediction of visual field deficits by diffusion tensor imaging in temporal lobe epilepsy surgery. Neuroimage 45:286–297. https://doi.org/10.1016/j.neuroimage.2008.11.038
Daducci A, Dal Palù A, Lemkaddem A, Thiran J‑P (2015) COMMIT: convex optimization modeling for microstructure informed tractography. IEEE Trans Med Imaging 34:246–257. https://doi.org/10.1109/TMI.2014.2352414
Diehl B, Tkach J, Piao Z et al (2010) Diffusion tensor imaging in patients with focal epilepsy due to cortical dysplasia in the temporo-occipital region: electro-clinico-pathological correlations. Epilepsy Res 90:178–187. https://doi.org/10.1016/j.eplepsyres.2010.03.006
Donos C, Rollo P, Tombridge K et al (2020) Visual field deficits following laser ablation of the hippocampus. Neurology 94:e1303–e1313. https://doi.org/10.1212/WNL.0000000000008940
Dougherty RF, Koch VM, Brewer AA et al (2003) Visual field representations and locations of visual areas V1/2/3 in human visual cortex. J Vis 3:586–598. https://doi.org/10.1167/3.10.1
Dumoulin SO, Wandell BA (2008) Population receptive field estimates in human visual cortex. Neuroimage 39:647–660. https://doi.org/10.1016/j.neuroimage.2007.09.034
Egan RA, Shults WT, So N et al (2000) Visual field deficits in conventional anterior temporal lobectomy versus amygdalohippocampectomy. Neurology 55:1818–1822. https://doi.org/10.1212/wnl.55.12.1818
Fischl B, Salat DH, Busa E et al (2002) Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33:341–355. https://doi.org/10.1016/s0896-6273(02)00569-x
Glasser MF, Sotiropoulos SN, Wilson JA et al (2013) The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 80:105–124. https://doi.org/10.1016/j.neuroimage.2013.04.127
Gross RE, Stern MA, Willie JT et al (2018) Stereotactic laser amygdalohippocampotomy for mesial temporal lobe epilepsy. Ann Neurol 83:575–587. https://doi.org/10.1002/ana.25180
Hoffmann MB, Dumoulin SO (2015) Congenital visual pathway abnormalities: a window onto cortical stability and plasticity. Trends Neurosci 38:55–65. https://doi.org/10.1016/j.tins.2014.09.005
Hoffmann MB, Kaule F, Grzeschik R et al (2011) Retinotopic mapping of the human visual cortex with functional magnetic resonance imaging—basic principles, current developments and ophthalmological perspectives. Klin Monatsbl Augenheilkd 228:613–620. https://doi.org/10.1055/s-0029-1245625
James JS, Radhakrishnan A, Thomas B et al (2015) Diffusion tensor imaging tractography of Meyer’s loop in planning resective surgery for drug-resistant temporal lobe epilepsy. Epilepsy Res 110:95–104. https://doi.org/10.1016/j.eplepsyres.2014.11.020
Jenkinson M, Beckmann CF, Behrens TEJ et al (2012) FSL. Neuroimage 62:782–790. https://doi.org/10.1016/j.neuroimage.2011.09.015
Jones DK, Knösche TR, Turner R (2013) White matter integrity, fiber count, and other fallacies: the do’s and don’ts of diffusion MRI. Neuroimage 73:239–254. https://doi.org/10.1016/j.neuroimage.2012.06.081
Le Bihan D, Breton E, Lallemand D et al (1986) MR imaging of intravoxel incoherent motions: application to diffusion and perfusion in neurologic disorders. Radiology 161:401–407. https://doi.org/10.1148/radiology.161.2.3763909
McDonald CR, Ahmadi ME, Hagler DJ et al (2008) Diffusion tensor imaging correlates of memory and language impairments in temporal lobe epilepsy. Neurology 71:1869–1876. https://doi.org/10.1212/01.wnl.0000327824.05348.3b
Mlynarski P, Delingette H, Alghamdi H et al (2020) Anatomically consistent CNN-based segmentation of organs-at-risk in cranial radiotherapy. J Med Imaging 7:14502. https://doi.org/10.1117/1.JMI.7.1.014502
Mori S, van Zijl PCM (2002) Fiber tracking: principles and strategies—a technical review. NMR Biomed 15:468–480. https://doi.org/10.1002/nbm.781
Nilsson D, Starck G, Ljungberg M et al (2007) Intersubject variability in the anterior extent of the optic radiation assessed by tractography. Epilepsy Res 77:11–16. https://doi.org/10.1016/j.eplepsyres.2007.07.012
Ogawa S, Takemura H, Horiguchi H et al (2014) White matter consequences of retinal receptor and ganglion cell damage. Invest Ophthalmol Vis Sci 55:6976–6986. https://doi.org/10.1167/iovs.14-14737
Pestilli F, Yeatman JD, Rokem A et al (2014) Evaluation and statistical inference for living connectomes. Nat Methods 11:1058–1063. https://doi.org/10.1038/nmeth.3098
Puzniak RJ, Ahmadi K, Kaufmann J et al (2019) Quantifying nerve decussation abnormalities in the optic chiasm. Neuroimage Clin 24:102055. https://doi.org/10.1016/j.nicl.2019.102055
Schmitt FC, Büntjen L, Schütze H et al (2020) Stereotaktische Laserthermoablation bei mesialer Temporallappenepilepsie mit Hippocampussklerose rechts – Patientenentscheidung, Durchführung und Visualisierung von Gedächtnisfunktion. Z Epileptol 33:42–49. https://doi.org/10.1007/s10309-020-00313-z
Schmitt FC, Kaufmann J, Hoffmann MB et al (2014) Case report: practicability of functionally based tractography of the optic radiation during presurgical epilepsy work up. Neurosci Lett 568:56–61. https://doi.org/10.1016/j.neulet.2014.03.049
Sereno MI, Dale AM, Reppas JB et al (1995) Borders of multiple visual areas in humans revealed by functional magnetic resonance imaging. Science 268:889–893. https://doi.org/10.1126/science.7754376
Smith RE, Tournier J‑D, Calamante F, Connelly A (2012) Anatomically-constrained tractography: improved diffusion MRI streamlines tractography through effective use of anatomical information. Neuroimage 62:1924–1938. https://doi.org/10.1016/j.neuroimage.2012.06.005
Smith RE, Tournier J‑D, Calamante F, Connelly A (2013) SIFT: spherical-deconvolution informed filtering of tractograms. Neuroimage 67:298–312. https://doi.org/10.1016/j.neuroimage.2012.11.049
Steensberg AT, Olsen AS, Litman M et al (2018) Visual field defects after temporal lobe resection for epilepsy. Seizure 54:1–6. https://doi.org/10.1016/j.seizure.2017.11.011
Stejskal EO, Tanner JE (1965) Spin diffusion measurements: spin echoes in the presence of a time-dependent field gradient. J Chem Phys 42:288–292. https://doi.org/10.1063/1.1695690
Takemura H, Caiafa CF, Wandell BA, Pestilli F (2016) Ensemble tractography. PLoS Comput Biol 12:e1004692. https://doi.org/10.1371/journal.pcbi.1004692
Tournier J‑D, Calamante F, Connelly A (2007) Robust determination of the fibre orientation distribution in diffusion MRI: non-negativity constrained super-resolved spherical deconvolution. Neuroimage 35:1459–1472. https://doi.org/10.1016/j.neuroimage.2007.02.016
Tournier J‑D, Calamante F, Connelly A (2013) Determination of the appropriate b value and number of gradient directions for high-angular-resolution diffusion-weighted imaging. NMR Biomed 26:1775–1786. https://doi.org/10.1002/nbm.3017
Tournier J‑D, Smith R, Raffelt D et al (2019) MRtrix3: a fast, flexible and open software framework for medical image processing and visualisation. Neuroimage 202:116137. https://doi.org/10.1016/j.neuroimage.2019.116137
Van Essen DC, Smith SM, Barch DM et al (2013) The WU-Minn Human Connectome Project: an overview. Neuroimage 80:62–79. https://doi.org/10.1016/j.neuroimage.2013.05.041
Wandell BA, Winawer J (2015) Computational neuroimaging and population receptive fields. Trends Cogn Sci 19:349–357. https://doi.org/10.1016/j.tics.2015.03.009
Whiting AC, Bingaman JR, Catapano JS et al (2020) Laser interstitial thermal therapy for epileptogenic periventricular nodular heterotopia. World Neurosurg 138:e892–e897. https://doi.org/10.1016/j.wneu.2020.03.133
Winston GP, Mancini L, Stretton J et al (2011) Diffusion tensor imaging tractography of the optic radiation for epilepsy surgical planning: a comparison of two methods. Epilepsy Res 97:124–132. https://doi.org/10.1016/j.eplepsyres.2011.07.019
Funding
This work was supported by European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement (No. 641805) and by the German Research Foundation (DFG, HO 2002 10-3) to M.B.H.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
R.J. Puzniak, G.T. Prabhakaran, L. Büntjen and M.B. Hoffmann declare that they have no competing interests. F.C. Schmitt received reimbursement for expenses and travel from Medtronic Inc.
All procedures performed in studies involving human participants or on human tissue were in accordance with the ethical standards of the institutional and/or national research committee and with the 1975 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Puzniak, R.J., Prabhakaran, G.T., Buentjen, L. et al. Tracking the visual system—from the optic chiasm to primary visual cortex. Z. Epileptol. 34, 57–66 (2021). https://doi.org/10.1007/s10309-020-00384-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10309-020-00384-y