Abstract
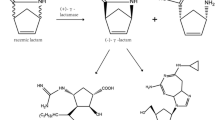
γ-Lactamases are versatile enzymes used for enzymatic kinetic resolution of racemic Vince lactam (2-azabicyclo[2.2.1]hept-5-en-3-one) in the industry. Optically pure enantiomers and their hydrolytic products are widely employed as key chemical intermediates for developing a wide range of carbocyclic nucleoside medicines, including US FDA-approved drugs peramivir and abacavir. Owing to the broad applications in the healthcare industry, the resolution process of Vince lactam has witnessed tremendous progress during the past decades. Some of the most important advances are the enzymatic strategies involving γ-lactamases. The strong industrial demand drives the progress in various strategies for discovering novel biocatalysts. In the past few years, several new scientific breakthroughs, including the genome-mining strategy and elucidation of several crystal structures, boosted the research on γ-lactamases. So far, several families of γ-lactamases for resolution of Vince lactam have been discovered, and their number is continuously increasing. The purpose of this mini-review is to describe the discovery strategy and classification of these intriguing enzymes and to cover our current knowledge on their potential biological functions. Moreover, structural properties are described in addition to their possible catalytic mechanisms. Additionally, recent advances in the newest approaches, such as immobilization to increase stability, and other engineering efforts are introduced.





Similar content being viewed by others
References
Boutureira O, Matheu MI, Diaz Y, Castillon S (2013) Advances in the enantioselective synthesis of carbocyclic nucleosides. Chem Soc Rev 42:5056–5072. https://doi.org/10.1039/c3cs00003f
Jeong LS, Lee JA (2004) Recent advances in the synthesis of the carbocyclic nucleosides as potential antiviral agents. Antiviral Chem Chemother 15:235–250
Marquez VE, Lim M-I (1986) Carbocyclic nucleosides. Med Res Rev 6:1–40. https://doi.org/10.1002/med.2610060102
Schneller SW (2002) Carbocyclic nucleosides (carbanucleosides) as new therapeutic leads. Curr Top Med Chem 2:1087–1092
Rodriguez JB, Comin MJ (2003) New progresses in the enantioselective synthesis and biological properties of carbocyclic nucleosides. Mini Rev Med Chem 3:95–114
Singh R, Vince R (2012) 2-Azabicyclo[2.2.1]hept-5-en-3-one: chemical profile of a versatile synthetic building block and its impact on the development of therapeutics. Chem Rev 112:4642–4686. https://doi.org/10.1021/cr2004822
Kusaka T, Yamamoto H, Shibata M, Muroi M, Kishi T (1968) Streptomyces citricolor nov. sp. and a new antibiotic, aristeromycin. J Antibiot 21:255–263
Yaginuma S, Muto N, Tsujino M, Sudate Y, Hayashi M, Otani M (1981) Studies on neplanocin A, new antitumor antibiotic. I. Producing organism, isolation and characterization. J Antibiot 34:359–366
Hervey PS, Perry CM (2000) Abacavir. Drugs 60:447–479. https://doi.org/10.2165/00003495-200060020-00015
Gupta RK, Hill A, Sawyer AW, Cozzi-Lepri A, von Wyl V, Yerly S, Lima VD, Gunthard HF, Gilks C, Pillay D (2009) Virological monitoring and resistance to first-line highly active antiretroviral therapy in adults infected with HIV-1 treated under WHO guidelines: a systematic review and meta-analysis. Lancet Infect Dis 9:409–417. https://doi.org/10.1016/S1473-3099(09)70136-7
Bessieres M, Chevrier F, Roy V, Agrofoglio LA (2015) Recent progress for the synthesis of selected carbocyclic nucleosides. Future Med Chem 7:1809–1828. https://doi.org/10.4155/fmc.15.105
Zhu XF (2000) The latest progress in the synthesis of carbocyclic nucleosides. Nucleosides Nucleotides Nucleic Acids 19:651–690. https://doi.org/10.1080/15257770008035015
Agrofoglio LA, Gillaizeau I, Saito Y (2003) Palladium-assisted routes to nucleosides. Chem Rev 103:1875–1916. https://doi.org/10.1021/cr010374q
Boncel S, Gondela A, Walczak K (2010) Michael-type addition as a convenient method for regioselective N-alkylation of ambident uracils. Synthesis 2010:1573–1589
Jagt J, Van Leusen A (1974) Diels-Alder cycloadditions of sulfonyl cyanides with cyclopentadiene. Synthesis of 2-azabicyclo [2.2. 1] hepta-2, 5-dienes. J Org Chem 39:564–566
Kamlet AS, Préville C, Farley KA, Piotrowski DW (2013) Regioselective hydroarylations and parallel kinetic resolution of Vince lactam. Angew Chem Int Ed 52:10607–10610. https://doi.org/10.1002/anie.201304818
Walczak P, Pannek J, Boratynski F, Janik-Polanowicz A, Olejniczak T (2014) Synthesis and fungistatic activity of bicyclic lactones and lactams against Botrytis cinerea, Penicillium citrinum, and Aspergillus glaucus. J Argic Food Chem 62:8571–8578. https://doi.org/10.1021/jf502148h
Jia F, Hong J, Sun P-H, Chen J-X, Chen W-M (2013) Facile synthesis of the neuraminidase inhibitor peramivir. Synth Comm 43:2641–2647
Mulakayala N, Reddy ChU, Iqbal J, Pal M (2010) Synthesis of dipeptidyl peptidase-4 inhibitors: a brief overview. Tetrahedron 66:4919–4938. https://doi.org/10.1016/j.tet.2010.04.088
Wisniewski T, Bayne E, Flanagan J, Shao Q, Wnek R, Matheravidathu S, Fischer P, Forrest MJ, Peterson L, Song X, Yang L, Demartino JA, Struthers M (2010) Assessment of chemokine receptor function on monocytes in whole blood: in vitro and ex vivo evaluations of a CCR2 antagonist. J Immunol Methods 352:101–110. https://doi.org/10.1016/j.jim.2009.10.010
Sun H, Zhang H, Ang EL, Zhao H (2018) Biocatalysis for the synthesis of pharmaceuticals and pharmaceutical intermediates. Bioorg Med Chem 26:1275–1284. https://doi.org/10.1016/j.bmc.2017.06.043
Caner H, Groner E, Levy L, Agranat I (2004) Trends in the development of chiral drugs. Drug Discov Today 9:105–110. https://doi.org/10.1016/S1359-6446(03)02904-0
Taylor SJ, Brown RC, Keene PA, Taylor IN (1999) Novel screening methods–the key to cloning commercially successful biocatalysts. Bioorg Med Chem 7:2163–2168
Patel RN (2000) Microbial/enzymatic synthesis of chiral drug intermediates. Adv Appl Microbiol 47:33–78
Littlechild JA (2011) Thermophilic archaeal enzymes and applications in biocatalysis. Biochem Soc Trans 39:155–158. https://doi.org/10.1042/BST0390155
Littlechild JA (2015) Enzymes from extreme environments and their industrial applications. Front Bioeng Biotechnol 3:161. https://doi.org/10.3389/fbioe.2015.00161
Wang J, Zheng G, Wu S (2010) Advances in lactamases from microbes—a review. Wei Sheng Wu Xue Bao 50:988–994
Schmid A, Dordick JS, Hauer B, Kiener A, Wubbolts M, Witholt B (2001) Industrial biocatalysis today and tomorrow. Nature 409:258–268. https://doi.org/10.1038/35051736
Littlechild JA (2015) Archaeal enzymes and applications in industrial biocatalysts. Archaea 2015:147671. https://doi.org/10.1155/2015/147671
Zaks A (2001) Industrial biocatalysis. Curr Opin Chem Biol 5:130–136. https://doi.org/10.1016/S1367-5931(00)00181-2
Taylor SJC, Sutherland AG, Lee C, Wisdom R, Thomas S, Roberts SM, Evans C (1990) Chemoenzymatic synthesis of (−)-carbovir utilizing a whole cell catalysed resolution of 2-azabicyclo[2.2.1]hept-5-en-3-one. J Chem Soc Chem Comm. https://doi.org/10.1039/c39900001120
Taylor SJC, McCague R, Wisdom R, Lee C, Dickson K, Ruecroft G, O’Brien F, Littlechild J, Bevan J, Roberts SM, Evans CT (1993) Development of the biocatalytic resolution of 2-azabicyclo[2.2.1]hept-5-en-3-one as an entry to single-enantiomer carbocyclic nucleosides. Tetrahedron Asymmetry 4:1117–1128. https://doi.org/10.1016/S0957-4166(00)80218-9
Zhu S, Gong C, Song D, Gao S, Zheng G (2012) Discovery of a novel (+)-gamma-lactamase from Bradyrhizobium japonicum USDA 6 by rational genome mining. Appl Environ Microbiol 78:7492–7495. https://doi.org/10.1128/AEM.01398-12
Wang J, Zhu Y, Zhao G, Zhu J, Wu S (2015) Characterization of a recombinant (+)-gamma-lactamase from Microbacterium hydrocarbon oxydans which provides evidence that two enantiocomplementary gamma-lactamases are in the strain. Appl Microbiol Biotechnol 99:3069–3080. https://doi.org/10.1007/s00253-014-6114-8
Gao S, Huang R, Zhu S, Li H, Zheng G (2016) Identification and characterization of a novel (+)-gamma-lactamase from Microbacterium hydrocarbonoxydans. Appl Microbiol Biotechnol 100:9543–9553. https://doi.org/10.1007/s00253-016-7643-0
Wang J, Guo X, Zheng G, Wen C (2009) Purification and characterization of a novel (−) gamma-lactamase from Microbacterium hydrocarbonoxydans. Ann Microbiol 59:345. https://doi.org/10.1007/bf03178337
Yang M, Gao Q, Wu S, Wang J, Zheng G (2012) Characterization of a recombinant (−)gamma-lactamase from Microbacterium hydrocarbonoxydans. Biotechnol Lett 34:275–279. https://doi.org/10.1007/s10529-011-0758-6
Line K, Isupov MN, Littlechild JA (2004) The crystal structure of a (−) gamma-lactamase from an Aureobacterium species reveals a tetrahedral intermediate in the active site. J Mol Biol 338:519–532. https://doi.org/10.1016/j.jmb.2004.03.001
Nobeli I, Favia AD, Thornton JM (2009) Protein promiscuity and its implications for biotechnology. Nat Biotechnol 27:157–167. https://doi.org/10.1038/nbt1519
Scotto d’Abusco A, Ammendola S, Scandurra R, Politi L (2001) Molecular and biochemical characterization of the recombinant amidase from hyperthermophilic archaeon Sulfolobus solfataricus. Extremophiles 5:183–192
Toogood HS, Brown RC, Line K, Keene PA, Taylor SJC, McCague R, Littlechild JA (2004) The use of a thermostable signature amidase in the resolution of the bicyclic synthon (rac)-γ-lactam. Tetrahedron 60:711–716. https://doi.org/10.1016/j.tet.2003.11.064
Torres LL, Schliessmann A, Schmidt M, Silva-Martin N, Hermoso JA, Berenguer J, Bornscheuer UT, Hidalgo A (2012) Promiscuous enantioselective (−)-gamma-lactamase activity in the Pseudomonas fluorescens esterase I. Org Biomol Chem 10:3388–3392. https://doi.org/10.1039/c2ob06887g
Assaf Z, Eger E, Vitnik Z, Fabian WMF, Ribitsch D, Guebitz GM, Faber K, Hall M (2014) Identification and application of enantiocomplementary lactamases for vince lactam derivatives. ChemCatChem 6:2517–2521. https://doi.org/10.1002/cctc.201402077
Zhu S, Ren L, Yu S, Gong C, Song D, Zheng G (2014) Enzymatic preparation of optically pure (+)-2-azabicyclo[2.2.1]hept-5-en-3-one by (-)-gamma-lactamase from Bradyrhizobium japonicum USDA 6. Bioorg Med Chem Lett 24:4899–4902. https://doi.org/10.1016/j.bmcl.2014.08.057
Gao S, Zhu S, Huang R, Lu Y, Zheng G (2015) Efficient synthesis of the intermediate of abacavir and carbovir using a novel (+)-gamma-lactamase as a catalyst. Bioorg Med Chem Lett 25:3878–3881. https://doi.org/10.1016/j.bmcl.2015.07.054
Wang J, Zhu J, Wu S (2015) Immobilization on macroporous resin makes E. coli RutB a robust catalyst for production of (−) Vince lactam. Appl Microbiol Biotechnol 99:4691–4700. https://doi.org/10.1007/s00253-014-6247-9
Xue TY, Xu GC, Han RZ, Ni Y (2015) Soluble expression of (+)-gamma-lactamase in Bacillus subtilis for the enantioselective preparation of abacavir precursor. Appl Biochem Biotechnol 176:1687–1699. https://doi.org/10.1007/s12010-015-1670-7
Ren L, Zhu S, Shi Y, Gao S, Zheng G (2015) Enantioselective resolution of gamma-lactam by a novel thermostable type II (+)-gamma-lactamase from the hyperthermophilic archaeon Aeropyrum pernix. Appl Biochem Biotechnol 176:170–184. https://doi.org/10.1007/s12010-015-1565-7
Zhu S, Huang R, Gao S, Li X, Zheng G (2016) Discovery and characterization of a second extremely thermostable (+)-gamma-lactamase from Sulfolobus solfataricus P2. J Biosci Bioeng 121:484–490. https://doi.org/10.1016/j.jbiosc.2015.09.012
Yin J-G, Gong Y, Zhang X-Y, Zheng G-W, Xu J-H (2016) Green access to chiral vince lactam in a buffer-free aqueous system using a newly identified substrate-tolerant (−)-[gamma]-lactamase. Catal Sci Technol 6:6305–6310. https://doi.org/10.1039/C6CY00786D
Assaf Z, Faber K, Hall M (2016) Scope, limitations and classification of lactamases. J Biotechnol 235:11–23. https://doi.org/10.1016/j.jbiotec.2016.03.050
Brabban AD, Littlechild J, Wisdom R (1996) Stereospecific γ-lactamase activity in a Pseudomonas fluorescens species. J Ind Microbiol 16:8–14. https://doi.org/10.1007/bf01569915
Bernhardt P, Hult K, Kazlauskas RJ (2005) Molecular basis of perhydrolase activity in serine hydrolases. Angew Chem Int Ed 44:2742–2746. https://doi.org/10.1002/anie.200463006
Sun Y, Zhao H, Wang J, Zhu J, Wu S (2015) Identification and regulation of the catalytic promiscuity of (−)-gamma-lactamase from Microbacterium hydrocarbonoxydans. Appl Microbiol Biotechnol 99:7559–7568. https://doi.org/10.1007/s00253-015-6503-7
Yin DL, Bernhardt P, Morley KL, Jiang Y, Cheeseman JD, Purpero V, Schrag JD, Kazlauskas RJ (2010) Switching catalysis from hydrolysis to perhydrolysis in Pseudomonas fluorescens esterase. Biochemistry 49:1931–1942. https://doi.org/10.1021/bi9021268
Yan X, Wang J, Sun Y, Zhu J, Wu S (2016) Facilitating the evolution of esterase activity from a promiscuous enzyme (Mhg) with catalytic functions of amide hydrolysis and carboxylic acid perhydrolysis by engineering the Substrate entrance tunnel. Appl Environ Microbiol 82:6748–6756. https://doi.org/10.1128/AEM.01817-16
Ohtaki A, Murata K, Sato Y, Noguchi K, Miyatake H, Dohmae N, Yamada K, Yohda M, Odaka M (2010) Structure and characterization of amidase from Rhodococcus sp. N-771: insight into the molecular mechanism of substrate recognition. BBA Proteins Proteom 1804:184–192. https://doi.org/10.1016/j.bbapap.2009.10.001
Mayaux JF, Cerbelaud E, Soubrier F, Yeh P, Blanche F, Petre D (1991) Purification, cloning, and primary structure of a new enantiomer-selective amidase from a Rhodococcus strain: structural evidence for a conserved genetic coupling with nitrile hydratase. J Bacteriol 173:6694–6704
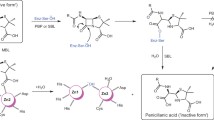
Li H, Zhu S, Zheng G (2018) Promiscuous (+)-gamma-lactamase activity of an amidase from nitrile hydratase pathway for efficient synthesis of carbocyclic nucleosides intermediate. Bioorg Med Chem Lett 28:1071–1076. https://doi.org/10.1016/j.bmcl.2018.02.019
Parsons JF, Calabrese K, Eisenstein E, Ladner JE (2003) Structure and mechanism of Pseudomonas aeruginosa PhzD, an isochorismatase from the phenazine biosynthetic pathway. Biochemistry 42:5684–5693. https://doi.org/10.1021/bi027385d
Fukuta Y, Koizumi S, Komeda H, Asano Y (2010) A new aryl acylamidase from Rhodococcus sp. strain Oct1 acting on ω-lactams: its characterization and gene expression in Escherichia coli. Enzyme Microb Technol 46:237–245. https://doi.org/10.1016/j.enzmictec.2009.09.010
Erna X, Jiaxin W, Hong Z, Ge C, Hong Y, Jimin Z, Lei W, Zhi W (2013) Resolution of N-hydroxymethyl vince lactam catalyzed by lipase in organic solvent. J Chem Technol Biotechnol 88:904–909. https://doi.org/10.1002/jctb.3919
Forró E, Fülöp F (2008) Enzymatic method for the synthesis of blockbuster drug intermediates—synthesis of five-membered cyclic γ-amino acid and γ-lactam enantiomers. Eur J Org Chem 2008:5263–5268. https://doi.org/10.1002/ejoc.200800723
Li F, Zhao D, Chen G, Zhang H, Yue H, Wang L, Wang Z (2013) Enantioselective transesterification of N-hydroxymethyl vince lactam catalyzed by lipase under ultrasound irradiation. Biocatal Biotransform 31:299–304. https://doi.org/10.3109/10242422.2013.857314
Nakano H, Iwasa K, Okuyama Y, Hongo H (1996) Lipase-catalyzed resolution of 2-azabicyclo[2.2.1]hept-5-en-3-ones. Tetrahedron Asymmetry 7:2381–2386. https://doi.org/10.1016/0957-4166(96)00293-5
Zhu L, Zhu F, Qin S, Wu B, He B (2016) Highly efficient resolution of N-hydroxymethyl vince lactam by solvent stable lipase YCJ01. J Mol Catal B Enzym 133:S150–S156. https://doi.org/10.1016/j.molcatb.2016.12.009
Connelly S, Line K, Isupov MN, Littlechild JA (2005) Synthesis and characterisation of a ligand that forms a stable tetrahedral intermediate in the active site of the Aureobacterium species (−) gamma-lactamase. Org Biomol Chem 3:3260–3262. https://doi.org/10.1039/b511078e
Gao S, Zhou Y, Zhang W, Wang W, Yu Y, Mu Y, Wang H, Gong X, Zheng G, Feng Y (2017) Structural insights into the gamma-lactamase activity and substrate enantioselectivity of an isochorismatase-like hydrolase from Microbacterium hydrocarbonoxydans. Sci Rep 7:44542. https://doi.org/10.1038/srep44542
Gonsalvez IS, Isupov MN, Littlechild JA (2001) Crystallization and preliminary X-ray analysis of a gamma-lactamase. Acta Crystallogr D 57:284–286
Mateo C, Palomo JM, Fernandez-Lorente G, Guisan JM, Fernandez-Lafuente R (2007) Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzyme Microb Technol 40:1451–1463. https://doi.org/10.1016/j.enzmictec.2007.01.018
Hickey AM, Ngamsom B, Wiles C, Greenway GM, Watts P, Littlechild JA (2009) A microreactor for the study of biotransformations by a cross-linked gamma-lactamase enzyme. Biotechnol J 4:510–516. https://doi.org/10.1002/biot.200800302
Wang J, Zhang X, Min C, Wu S, Zheng G (2011) Single-step purification and immobilization of γ-lactamase and on-column transformation of 2-azabicyclo [2.2.1] hept-5-en-3-one. Process Biochem 46:81–87. https://doi.org/10.1016/j.procbio.2010.07.018
Wang J, Zhu J, Min C, Wu S (2014) CBD binding domain fused gamma-lactamase from Sulfolobus solfataricus is an efficient catalyst for (−) gamma-lactam production. BMC Biotechnol 14:40. https://doi.org/10.1186/1472-6750-14-40
Wang J, Zhao G, Zhang Z, Liang Q, Min C, Wu S (2014) Autodisplay of an archaeal gamma-lactamase on the cell surface of Escherichia coli using Xcc_Est as an anchoring scaffold and its application for preparation of the enantiopure antiviral drug intermediate (−) vince lactam. Appl Microbiol Biotechnol 98:6991–7001. https://doi.org/10.1007/s00253-014-5704-9
Li W, Wen H, Shi Q, Zheng G (2016) Study on immobilization of (+) γ-lactamase using a new type of epoxy graphene oxide carrier. Process Biochem 51:270–276. https://doi.org/10.1016/j.procbio.2015.11.030
Li H, Zheng G, Zhu S (2018) Construction of an organelle-like nanodevice via supramolecular self-assembly for robust biocatalysts. Microb Cell Fact 17:26. https://doi.org/10.1186/s12934-018-0873-3
Bornscheuer UT, Huisman GW, Kazlauskas RJ, Lutz S, Moore JC, Robins K (2012) Engineering the third wave of biocatalysis. Nature 485:185–194. https://doi.org/10.1038/nature11117
Davids T, Schmidt M, Bottcher D, Bornscheuer UT (2013) Strategies for the discovery and engineering of enzymes for biocatalysis. Curr Opin Chem Biol 17:215–220. https://doi.org/10.1016/j.cbpa.2013.02.022
Gao S, Zhu S, Huang R, Li H, Wang H, Zheng G (2018) Engineering the enantioselectivity and thermostability of a (+)-gamma-lactamase from Microbacterium hydrocarbonoxydans for kinetic resolution of vince lactam (2-Azabicyclo[2.2.1]hept-5-en-3-one). Appl Environ Microbiol. https://doi.org/10.1128/aem.01780-17
Sheldon RA, van Pelt S (2013) Enzyme immobilisation in biocatalysis: why, what and how. Chem Soc Rev 42:6223–6235. https://doi.org/10.1039/C3CS60075K
Kettle AJ, Carere J, Batley J, Benfield AH, Manners JM, Kazan K, Gardiner DM (2015) A gamma-lactamase from cereal infecting Fusarium spp. catalyses the first step in the degradation of the benzoxazolinone class of phytoalexins. Fungal Genet Biol 83:1–9. https://doi.org/10.1016/j.fgb.2015.08.005
Wang J, Cheng D, Wang J, Jin L (2016) Isolation of Pseudomonas granadensis B6 with (+)-γ-lactamase and whole cell resolution of racemic γ-lactam. J Mol Catal B Enzym 133:S114–S118. https://doi.org/10.1016/j.molcatb.2016.12.004
Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC, Grant No. 21706005), China Postdoctoral Science Foundation (Grant No. 2017M610747), the Fundamental Research Funds for the Central Universities (No. XK1802-8), and National Great Science and Technology Projects (2018ZX09721001).
Author information
Authors and Affiliations
Contributions
SZ and GZ wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest associated with the contents of this article.
Rights and permissions
About this article
Cite this article
Zhu, S., Zheng, G. Dynamic kinetic resolution of Vince lactam catalyzed by γ-lactamases: a mini-review. J Ind Microbiol Biotechnol 45, 1017–1031 (2018). https://doi.org/10.1007/s10295-018-2093-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-018-2093-6