Abstract
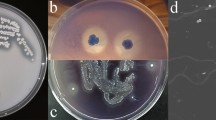
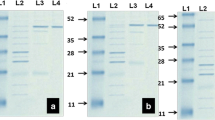
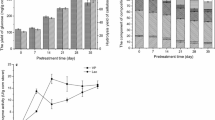
Lignin valorization can be obtained through cleavage of selected bonds by microbial enzymes, in which lignin is segregated from cellulose and hemicellulose and abundant phenolic compounds can be provided. In this study, Pseudomonas sp. Q18, previously isolated from rotten wood in China, was used to degrade alkali lignin and raw lignocellulosic material. Gel-permeation chromatography, field-emission scanning electron microscope, and GC–MS were combined to investigate the degradation process. The GC–MS results revealed that the quantities of aromatic compounds with phenol ring from lignin increased significantly after incubation with Pseudomonas sp. Q18, which indicated the degradation of lignin. According to the lignin-derived metabolite analysis, it was proposed that a DyP-type peroxidase (PmDyP) might exist in strain Q18. Thereafter, the gene of PmDyP was cloned and expressed, after which the recombinant PmDyP was purified and the enzymatic kinetics of PmDyP were assayed. According to results, PmDyP showed promising characteristics for lignocellulosic biodegradation in biorefinery.






Similar content being viewed by others
References
Abe M, Yamanaka S, Yamada H, Yamada T, Ohno H (2015) Almost complete dissolution of woody biomass with tetra-n-butylphosphonium hydroxide aqueous solution at 60 °C. Green Chem 17:4432–4438. https://doi.org/10.1039/c5gc00646e
Ahmad M, Roberts JN, Hardiman EM, Singh R, Eltis LD, Bugg TD (2011) Identification of DypB from Rhodococcus jostii RHA1 as a lignin peroxidase. Biochemistry 50:5096–5107. https://doi.org/10.1021/bi101892z
Ahmad M, Taylor CR, Pink D, Burton K, Eastwood D, Bending GD, Bugg TD (2010) Development of novel assays for lignin degradation: comparative analysis of bacterial and fungal lignin degraders. Mol BioSyst 6:815. https://doi.org/10.1039/b908966g
Azizi-Shotorkhoft A, Mohammadabadi T, Motamedi H, Chaji M, Fazaeli H (2016) Isolation and identification of termite gut symbiotic bacteria with lignocellulose-degrading potential, and their effects on the nutritive value for ruminants of some by-products. Anim Feed Sci Technol 221:234–242. https://doi.org/10.1016/j.anifeedsci.2016.04.016
Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54:519–546. https://doi.org/10.1146/annurev.arplant.54.031902.134938
Brissos V, Tavares D, Sousa AC, Robalo MP, Martins LO (2017) Engineering a bacterial Dyp-type peroxidase for enhanced oxidation of lignin-related phenolics at alkaline pH. ACS Catal 7:3454–3465. https://doi.org/10.1021/acscatal.6b03331
Brzonova I, Kozliak E, Kubátová A, Chebeir M, Qin W, Christopher L, Ji Y (2014) Kenaf biomass biodecomposition by basidiomycetes and actinobacteria in submerged fermentation for production of carbohydrates and phenolic compounds. Bioresour Technol 173:352–360. https://doi.org/10.1016/j.biortech.2014.09.057
Bu D, Zhou Y, Tang J, Jing F, Zhang W (2013) Expression and purification of a novel therapeutic single-chain variable fragment antibody against BNP from inclusion bodies of Escherichia coli. Protein Expression Purif 92:203–207. https://doi.org/10.1016/j.pep.2013.10.002
Bugg TD, Ahmad M, Hardiman EM, Rahmanpour R (2011) Pathways for degradation of lignin in bacteria and fungi. Nat Prod Rep 28:1883–1896. https://doi.org/10.1039/c1np00042j
Camarero S, Ibarra D, Martínez MJ, Martínez ÁT (2005) Lignin-derived compounds as efficient laccase mediators for decolorization of different types of recalcitrant dyes. Appl Environ Microbiol 71:1775–1784. https://doi.org/10.1128/aem.71.4.1775-1784.2005
Chandra R, Raj A, Purohit H, Kapley A (2007) Characterisation and optimisation of three potential aerobic bacterial strains for kraft lignin degradation from pulp paper waste. Chemosphere 67:839–846. https://doi.org/10.1016/j.chemosphere.2006.10.011
Chang YC, Choi D, Takamizawa K, Kikuchi S (2014) Isolation of Bacillus sp. strains capable of decomposing alkali lignin and their application in combination with lactic acid bacteria for enhancing cellulase performance. Bioresour Technol 152:429–436. https://doi.org/10.1016/j.biortech.2013.11.032
Chen HZ, Liu ZH, Qu Y, Wu WT (2015) Steam explosion and its combinatorial pretreatment refining technology of plant biomass to bio-based products. Biotechnol J 10:866. https://doi.org/10.1002/biot.201400705
Ćilerdžić J, Stajić M, Vukojević J (2016) Degradation of wheat straw and oak sawdust by Ganoderma applanatum. Int Biodeterior Biodegrad 114:39–44. https://doi.org/10.1016/j.ibiod.2016.05.024
Cserzö M, Wallin E, Simon I, Von HG, Elofsson A (1997) Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng 10:673–676. https://doi.org/10.1093/protein/10.6.673
Duan J, Huo X, Du W, Liang J, Wang D, Yang S (2016) Biodegradation of kraft lignin by a newly isolated anaerobic bacterial strain, Acetoanaerobium sp. WJDL-Y2. Lett Appl Microbiol 62:55–62. https://doi.org/10.1111/lam.12508
Dunford HB (1999) Heme peroxidases. Encycl Biophys. https://doi.org/10.1007/978-3-642-16712-6_52
Gardy JL, Laird MR, Chen F, Rey S, Walsh CJ, Ester M, Brinkman FS (2005) PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21:617–623. https://doi.org/10.1093/bioinformatics/bti057
Gilca IA, Ghitescu RE, Puitel AC, Popa VI (2014) Preparation of lignin nanoparticles by chemical modification. Iran Polym J 23:355–363. https://doi.org/10.1007/s13726-014-0232-0
Haq I, Kumar S, Kumari V, Singh SK, Raj A (2016) Evaluation of bioremediation potentiality of ligninolytic Serratia liquefaciens for detoxification of pulp and paper mill effluent. J Hazard Mater 305:190–199. https://doi.org/10.1016/j.jhazmat.2015.11.046
Huang XF, Santhanam N, Badri DV, Hunter WJ, Manter DK, Decker SR, Vivanco JM, Reardon KF (2013) Isolation and characterization of lignin-degrading bacteria from rainforest soils. Biotechnol Bioeng 110:1616–1626. https://doi.org/10.1002/bit.24833
Jackson CA, Couger MB, Prabhakaran M, Ramachandriya KD, Canaan P, Fathepure BZ (2017) Isolation and characterization of Rhizobium sp. strain YS-1r that degrades lignin in plant biomass. J Appl Microbiol 122:940–952. https://doi.org/10.1111/jam.13401
Karim M, Daryaei MG, Torkaman J, Oladi R, Ghanbary MAT, Bari E (2016) In vivo investigation of chemical alteration in oak wood decayed by Pleurotus ostreatus. Int Biodeterior Biodegrad 108:127–132. https://doi.org/10.1016/j.ibiod.2015.12.012
Kirk TK, Obst JR (1988) Lignin determination. Methods Enzymol 161:87–101. https://doi.org/10.1016/0076-6879(88)61014-7
Ko JJ, Shimizu Y, Ikeda K, Kim SK, Park CH, Matsui S (2009) Biodegradation of high molecular weight lignin under sulfate reducing conditions: lignin degradability and degradation by-products. Bioresour Technol 100:1622–1627. https://doi.org/10.1016/j.biortech.2008.09.029
Kumar M, Singh J, Singh MK, Singhal A, Thakur IS (2015) Investigating the degradation process of kraft lignin by β-proteobacterium, Pandoraea sp.ISTKB. Environ Sci Pollut Res 22:15690–15702. https://doi.org/10.1007/s11356-015-4771-5
Kumar P, Barrett DM, Delwiche MJ, Stroeve P (2009) Methods for pretreatment of lignocellulosic biomass for efficient hydrolysis and biofuel production. Ind Eng Chem Res 48:3713–3729. https://doi.org/10.1021/ie801542g
Kuuskeri J, Häkkinen M, Laine P, Smolander OP, Tamene F, Miettinen S, Nousiainen P, Kemell M, Auvinen P, Lundell T (2016) Time-scale dynamics of proteome and transcriptome of the white-rot fungus Phlebia radiata: growth on spruce wood and decay effect on lignocellulose. Biotechnol Biofuels 9:192. https://doi.org/10.1186/s13068-016-0608-9
Li J, Liu C, Li B, Yuan H, Yang J, Zheng B (2012) Identification and molecular characterization of a novel DyP-type peroxidase from Pseudomonas aeruginosa PKE117. Appl Biochem Biotechnol 166:774–785. https://doi.org/10.1007/s12010-011-9466-x
Mann DGJ, Labbé N, Sykes RW, Gracom K, Kline L, Swamidoss IM, Burris JN, Davis M, Stewart CN Jr, Haigler C (2009) Rapid assessment of lignin content and structure in switchgrass (Panicum virgatum L.) grown under different environmental conditions. Bioenergy Res 2:246–256. https://doi.org/10.1007/s12155-009-9054-x
Masai E, Katayama Y, Fukuda M (2007) Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci Biotechnol Biochem 71:1–15. https://doi.org/10.1271/bbb.60437
Mathews SL, Pawlak JJ, Grunden AM (2014) Isolation of Paenibacillus glucanolyticus from pulp mill sources with potential to deconstruct pulping waste. Bioresour Technol 164:100–105. https://doi.org/10.1016/j.biortech.2014.04.093
Mnich E, Vanholme R, Oyarce P, Liu S, Lu F, Goeminne G, Jørgensen B, Motawie MS, Boerjan W, Ralph J (2016) Degradation of lignin β-aryl ether units in Arabidopsis thaliana expressing LigD, LigF and LigG from Sphingomonas paucimobilis SYK-6. Plant Biotechnol J 15:581–593. https://doi.org/10.1111/pbi.12655
Nielsen H, Engelbrecht J, Brunak S, Von HG (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10:1–6. https://doi.org/10.1093/protein/10.1.1
Ogola H, Kamiike T, Hashimoto N, Ashida H, Ishikawa T, Shibata H, Sawa Y (2009) Molecular characterization of a novel peroxidase from the cyanobacterium Anabaena sp. strain PCC 7120. Appl Environ Microbiol 75:7509. https://doi.org/10.1128/aem.01121-09
Pamidipati S, Ahmed A (2017) Degradation of lignin in agricultural residues by locally isolated fungus Neurospora discreta. Appl Biochem Biotechnol 181:1561–1572. https://doi.org/10.1007/s12010-016-2302-6
Pandey MP, Kim CS (2011) Lignin depolymerization and conversion: a review of thermochemical methods. Chem Eng Technol 34:29–41. https://doi.org/10.1002/ceat.201000270
Pollegioni L, Tonin F, Rosini E (2015) Lignin-degrading enzymes. FEBS J 282:1190–1213. https://doi.org/10.1111/febs.13224
Ragauskas AJ, Beckham GT, Biddy MJ, Chandra R, Chen F, Davis MF, Davison BH, Dixon RA, Gilna P, Keller M (2014) Lignin valorization: improving lignin processing in the biorefinery. Science 344:1246843. https://doi.org/10.1126/science.1246843
Rahmanpour R, Bugg TD (2015) Characterisation of Dyp-type peroxidases from Pseudomonas fluorescens Pf-5: oxidation of Mn(II) and polymeric lignin by Dyp1B. Arch Biochem Biophys 574:93–98. https://doi.org/10.1016/j.abb.2014.12.022
Rahmanpour R, Rea D, Jamshidi S, Fulop V, Bugg TD (2016) Structure of Thermobifida fusca DyP-type peroxidase and activity towards Kraft lignin and lignin model compounds. Arch Biochem Biophys 594:54–60. https://doi.org/10.2210/pdb5fw4/pdb
Raj A, Chandra R, Reddy MMK, Purohit HJ, Kapley A (2007) Biodegradation of kraft lignin by a newly isolated bacterial strain, Aneurinibacillus aneurinilyticus from the sludge of a pulp paper mill. World J Microbiol Biotechnol 23:793–799. https://doi.org/10.1007/s11274-006-9299-x
Raj A, Kumar S, Haq I, Singh SK (2014) Bioremediation and toxicity reduction in pulp and paper mill effluent by newly isolated ligninolytic Paenibacillus sp. Ecol Eng 71:355–362. https://doi.org/10.1016/j.ecoleng.2014.07.002
Raj A, Reddy MK, Chandra R (2007) Identification of low molecular weight aromatic compounds by gas chromatography–mass spectrometry (GC-MS) from kraft lignin degradation by three Bacillus sp. Int Biodeterior Biodegrad 59:292–296. https://doi.org/10.1016/j.ibiod.2006.09.006
Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz PF, Marita JM, Hatfield RD, Ralph SA, Christensen JH (2004) Lignins: natural polymers from oxidative coupling of 4-hydroxyphenyl-propanoids. Phytochem Rev 3:29–60. https://doi.org/10.1023/b:phyt.0000047809.65444.a4
Rashid GM, Taylor CR, Liu Y, Zhang X, Rea D, Fülöp V, Bugg TD (2015) Identification of manganese superoxide dismutase from Sphingobacterium sp. T2 as a novel bacterial enzyme for lignin oxidation. ACS Chem Biol 10:2286–2294. https://doi.org/10.1021/acschembio.5b00298
Reiter J, Strittmatter H, Wiemann LO, Schieder D, Sieber V (2013) Enzymatic cleavage of lignin β-O-4 aryl ether bonds via net internal hydrogen transfer. Green Chem 15:1373–1381. https://doi.org/10.1039/c3gc40295a
Salvachúa D, Karp EM, Nimlos CT, Vardon DR, Beckham GT (2015) Towards lignin consolidated bioprocessing: simultaneous lignin depolymerization and product generation by bacteria. Green Chem 17:4951–4967. https://doi.org/10.1039/c5gc01165e
Santos A, Mendes S, Brissos V, Martins LO (2013) New dye-decolorizing peroxidases from Bacillus subtilis and Pseudomonas putida MET94: towards biotechnological applications. Appl Microbiol Biotechnol 98:2053–2065. https://doi.org/10.1007/s00253-013-5041-4
Santos RB, Capanema EA, Balakshin MY, Chang H, Jameel H (2012) Lignin structural variation in hardwood species. J Agric Food Chem 60:4923. https://doi.org/10.1021/jf301276a
Schmidt O, Bahmani M, Koch G, Potsch T, Brandt K (2016) Study of the fungal decay of oil palm wood using TEM and UV techniques. Int Biodeterior Biodegrad 111:37–44. https://doi.org/10.1016/j.ibiod.2016.04.014
Shi Y, Yan X, Li Q, Wang X, Xie S, Chai L, Yuan J (2017) Directed bioconversion of Kraft lignin to polyhydroxyalkanoate by Cupriavidus basilensis B-8 without any pretreatment. Process Biochem 52:238–242. https://doi.org/10.1016/j.procbio.2016.10.004
Singh R, Grigg JC, Qin W, Kadla JF, Murphy ME, Eltis LD (2013) Improved manganese-oxidizing activity of DypB, a peroxidase from a lignolytic bacterium. ACS Chem Biol 8:700–706. https://doi.org/10.1021/cb300608x
Sugano Y (2009) DyP-type peroxidases comprise a novel heme peroxidase family. Cell Mol Life Sci 66:1387. https://doi.org/10.1007/s00018-008-8651-8
Sugano Y, Muramatsu R, Ichiyanagi A, Sato T, Shoda M (2007) DyP, a unique dye-decolorizing peroxidase, represents a novel heme peroxidase family: ASP171 replaces the distal histidine of classical peroxidases. J Biol Chem 282:36652–36658
Sugawara K, Nishihashi Y, Narioka T, Yoshida T, Morita M, Sugano Y (2017) Characterization of a novel DyP-type peroxidase from Streptomyces avermitilis. J Biosci Bioeng 123:425–430. https://doi.org/10.1016/j.jbiosc.2016.12.001
Suman SK, Dhawaria M, Tripathi D, Raturi V, Adhikari DK, Kanaujia PK (2016) Investigation of lignin biodegradation by Trabulsiella sp. isolated from termite gut. Int Biodeterior Biodegrad 112:12–17. https://doi.org/10.1016/j.ibiod.2016.04.036
Sun Y, Qiu X, Liu Y (2013) Chemical reactivity of alkali lignin modified with laccase. Biomass Bioenergy 55:198–204. https://doi.org/10.1016/j.biombioe.2013.02.006
Taylor CR, Hardiman E, Ahmad M, Sainsbury P, Norris P, Bugg TD (2012) Isolation of bacterial strains able to metabolize lignin from screening of environmental samples. J Appl Microbiol 113:521–530. https://doi.org/10.1111/j.1365-2672.2012.05352.x
Wang B, Rezenom YH, Cho KC, Tran JL, Lee DG, Russell DH, Gill JJ, Young R, Chu KH (2014) Cultivation of lipid-producing bacteria with lignocellulosic biomass: effects of inhibitory compounds of lignocellulosic hydrolysates. Bioresour Technol 161:162–170. https://doi.org/10.1016/j.biortech.2014.02.133
Wang Y, Liu Q, Yan L, Gao Y, Wang Y, Wang W (2013) A novel lignin degradation bacterial consortium for efficient pulping. Bioresour Technol 139:113–119. https://doi.org/10.1016/j.biortech.2013.04.033
Yadav S, Chandra R (2015) Syntrophic co-culture of Bacillus subtilis and Klebsiella pneumonia for degradation of kraft lignin discharged from rayon grade pulp industry. J Environ Sci 33:229–238. https://doi.org/10.1016/j.jes.2015.01.018
Yang CX, Wang T, Gao LN, Yin HJ, Lu X (2017) Isolation, identification and characterization of lignin-degrading bacteria from Qinling, China. J Appl Microbiol 123:1447–1460. https://doi.org/10.1111/jam.13562
Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y (2015) The I-TASSER Suite: protein structure and function prediction. Nat Methods 12:7–8. https://doi.org/10.1038/nmeth.3213
Yang S, Yuan TQ, Li MF, Sun RC (2015) Hydrothermal degradation of lignin: products analysis for phenol formaldehyde adhesive synthesis. Int J Biol Macromol 72:54–62. https://doi.org/10.1016/j.ijbiomac.2014.07.048
Yang Y, Zhou J, Lu H, Yuan Y, Zhao L (2011) Isolation and characterization of a fungus Aspergillus sp. strain F-3 capable of degrading alkali lignin. Biodegradation 22:1017–1027. https://doi.org/10.1007/s10532-011-9460-6
Ye W, Liu J, Wang H, Wang J, Wang X (2012) Cloning, expression, purification, and characterization of a glutamate-specific endopeptidase from Bacillus licheniformis. Protein Expr Purif 82:138–143. https://doi.org/10.1016/j.pep.2011.12.001
Zakzeski J, Bruijnincx PC, Jongerius AL, Weckhuysen BM (2010) The catalytic valorization of lignin for the production of renewable chemicals. Chem Rev 110:3552–3599. https://doi.org/10.1021/cr900354u
Zheng Y, Chai L, Yang Z, Chen Y, Shi Y, Wang Y (2014) Environmentally safe treatment of black liquor with Comamonas sp. B-9 under high-alkaline conditions. J Basic Microbiol 54:152–161. https://doi.org/10.1002/jobm.201200340
Zhu D, Zhang P, Xie C, Zhang W, Sun J, Qian WJ, Yang B (2017) Biodegradation of alkaline lignin by Bacillus ligniniphilus L1. Biotechnol Biofuels 10:44. https://doi.org/10.1186/s13068-017-0735-y
Zubieta C, Krishna SS, Kapoor M, Kozbial P, McMullan D, Axelrod HL, Miller MD, Abdubek P, Ambing E, Astakhova T, Carlton D, Chiu HJ, Clayton T, Deller MC, Duan L, Elsliger MA, Feuerhelm J, Grzechnik SK, Hale J, Hampton E, Han GW, Jaroszewski L, Jin KK, Klock HE, Knuth MW, Kumar A, Marciano D, Morse AT, Nigoghossian E, Okach L, Oommachen S, Reyes R, Rife CL, Schimmel P, van den Bedem H, Weekes D, White A, Xu Q, Hodgson KO, Wooley J, Deacon AM, Godzik A, Lesley SA, Wilson IA (2007) Crystal structures of two novel dye-decolorizing peroxidases reveal a beta-barrel fold with a conserved heme-binding motif. Proteins 69:223–233. https://doi.org/10.1002/prot.21550
Acknowledgements
The authors thank the financial support of Special Fund for Agro-scientific Research in the Public Interest (Grant no. 201503135).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, C., Yue, F., Cui, Y. et al. Biodegradation of lignin by Pseudomonas sp. Q18 and the characterization of a novel bacterial DyP-type peroxidase. J Ind Microbiol Biotechnol 45, 913–927 (2018). https://doi.org/10.1007/s10295-018-2064-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-018-2064-y